13.3
Impact Factor
Theranostics 2016; 6(11):1877-1886. doi:10.7150/thno.15284 This issue Cite
Research Paper
Aptamer-Mediated Transparent-Biocompatible Nanostructured Surfaces for Hepatocellular Circulating Tumor Cells Enrichment
1. Department of Oncology, Zhongnan Hospital of Wuhan University; Hubei Key Laboratory of Tumor Biological Behaviors & Hubei Cancer Clinical Study Center, Wuhan 430071, P. R. China;
2. Department of Oncology, Peking University Shenzhen Hospital, No. 1120, Lianhua Road, Futain District, Shenzhen 518036, Guangdong Province, P. R. China;
3. Department of Breast Cancer, Cancer Center, Guangdong General Hospital, Guangzhou 510050, Guangdong Province, P.R. China.
*These authors contribute to this study equally.
Received 2016-2-15; Accepted 2016-7-7; Published 2016-8-7
Abstract
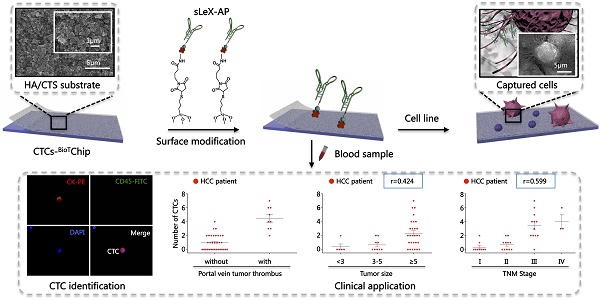
Circulating tumor cells (CTCs) have been considered as the origin of cancer metastasis. Thus, detection of CTCs in peripheral blood is of great value in different types of solid tumors. However, owing to extremely low abundance of CTCs, detection of them has been technically challenging. To establish a simple and efficient method for CTCs detection in patients with hepatocellular carcinoma (HCC), we applied biocompatible and transparent HA/CTS (Hydroxyapatite/chitosan) nanofilm to achieve enhanced topographic interactions with nanoscale cellular surface components, and we used sLex-AP (aptamer for carbohydrate sialyl Lewis X) to coat onto HA/CTS nanofilm for efficient capture of HCC CTCs, these two functional components combined to form our CTC-BioTChip platform. Using this platform, we realized HCC CTCs' capture and identification, the average recovery rate was 61.6% or more at each spiking level. Importantly, our platform identified CTCs (2±2 per 2 mL) in 25 of 42 (59.5%) HCC patients. Moreover, both the positivity rate and the number of detected CTCs were significantly correlated with tumor size, portal vein tumor thrombus, and the TNM (tumor-node-metastasis) stage. In summary, our CTC-BioTChip platform provides a new method allowing for simple but efficient detection of CTCs in HCC patients, and it holds potential of clinically usefulness in monitoring HCC prognosis and guiding individualized treatment in the future.
Keywords: Circulating tumor cells, Cell capture, Aptamer, Hepatocellular carcinoma, Nanomaterial, Hydroxyapatite/chitosan.
Introduction
Solid tumor metastasis is the most lethal clinical relevance and causes over 90% of cancer-related death in patients [1, 2]. As a complex process, cancer metastasis remains the most poorly understood aspect of cancer progression, while one important step of metastasis is tumor cells disseminate into blood stream and circulate [3], there is a considerable body of evidence indicating that tumor cells shed from primary tumor mass contribute to this malignant progression [4]. These 'break-away' cancer cells are known as circulating tumor cells (CTCs) which circulate in the blood stream, adhere to local vascular endothelium and invade different tissues of the body as the cellular origin of metastasis [5-7]; thus, to get more insights into metastasis-associated progression, CTCs detection in peripheral blood is of great value in different types of solid tumors. By now, CTCs can be regarded as the “liquid biopsy” of the tumor to provide convenient access to tumor cells, and earlier access to potentially fatal metastasis [8]. In the past decade, abundant of CTCs studies have demonstrated that CTCs could serve as prognostic indicators for poor clinical outcomes [9-11] and treatment responses [12-15]; thus, detection of CTCs has the potential to realize early diagnosis, personalized therapy, and therapeutic efficacy monitoring. However, owing to the extremely low abundance of CTCs [16] (one tumor cell in millions of blood cells), detection and characterization of CTCs has been technically challenging.
Over the past decade, a diversity of CTCs detection technologies has been developed based on different working mechanisms. Especially, nanostructured materials [17] have been widely used as a new set of tools for CTC capture and characterization such as silicon nanopillars/nanowires [18-21], nanoparticles [22-25], nanofibers [26, 27], and nanoroughened surfaces [28]. Recently, our group have established biocompatible transparent nanostructured substrates made of hydroxyapatite/chitosan (HA/CTS) (CTC-BioTChip) to capture CTCs, and also demonstrated its potential clinical use in efficiently capturing CTCs [25, 29], we explored the concept of “Biocompatible” of CTC-BioTChip, after capture and culture, CTCs on the CTC-BioTChip still showed normal cell morphology, excellent viability and potential proliferation ability, also we studied the “Transparent” feather of CTC-BioTChip, and found that this remarkable property could realize accurate positioning for immobilized CTCs and facilitate downstream analysis such as immunocytochemistry, fluorescence in situ hybridization; furthermore, enhanced local topographic interactions between cancer cells surface (e.g., microvilli) and the antibody (anti-EpCAM) coated lead to improved CTCs capture efficiency, CTC-BioTChip was capable of enriching, identifying and enumerating CTCs in whole-blood samples collected from colorectal, gastric, lung cancer patients. However, further applications of this platform for enrichment of hepatocellular carcinoma (HCC) CTCs were constrained by the fact that most HCC cells exhibited low expression of EpCAM [30, 31]. Although EpCAM-positive HCC CTCs were identified as cancer stem-like cells in HCC patients [32, 33], anti-EpCAM as bait for capturing is not appropriate to get a more comprehensive spectrum of HCC CTCs [34]. Thus, it is essential to develop a new method for HCC CTCs detections with sufficient sensitivity and lower cost.
To improve our CTC-BioTChip for HCC CTCs detection, the functional molecules immobilized should be emphatically reconsidered. These functional molecules, such as antibodies and DNA aptamers [35, 36], are derived from aberrantly expressed biomarkers that distinguish cancer cells from normal ones. Notably, DNA aptamers were single-stranded oligonucleotides had unique secondary or tertiary structures, and showed highly specific affinity to recognize surface molecules on targeted cells. Aptamers had been used as functional molecules in several kinds of solid tumors and realized efficient and sensitive capture of CTCs [19, 23, 37-41]. Furthermore, compared with antibodies, DNA aptamers could be easily and reproducibly synthesized, they had low immunogenicity and high stability. Besides, cell-surface carbohydrate sialyl Lewis X (sLex) upregulated on several cancer cell surfaces, acts as a critical factor for cancer metastasis and correlates with poor survival of cancer patients [42]; it involves in the adhesion of cancer cells to vascular endothelial cells through sLex/E-selectin interaction which is a key starting point of cancer metastasis in liver and lung cancer [43]. Also, sLex antibodies (α-sLex) used to control CTCs adhesion to endothelial cells exhibits its potential for possible CTCs capture [44]. These findings inspired us to use sLex aptamer (sLex-AP) as the functional molecule to work with CTC-BioTChip. The whole platform was designed to mimic the biological procedure of interaction between cancer cells and vascular endothelial cells to achieve efficient capture of HCC CTCs.
In this study, we focus on two aspects: 1) sLeX-AP coated CTC-BioTChip can achieve HCC CTCs detection; 2) sLeX-AP coated CTC-BioTChip may have potential for clinical use. Herein, we report CTC-BioTChip combined with cell affinitive sLex-AP for HCC CTCs detection. This method shows efficient capture of HCC CTCs, we conclude that this improved method is capable of isolating HCC CTCs for subsequent basic and clinical researches.
Materials and Methods
Preparation of CTC-BioTChip and sLex-AP
CTC-BioTChip was prepared as previous description [25]. Simply, the glass slid was firstly plasma treated and washed by piranha solution at 100°C for 1 hour, rinsed in deionized (DI) water, then treated with oxygen plasma (PDC- 32G, Harrick Plasma, Ithaca, NY, USA) for 2 minutes, and dried with nitrogen gas for further use. Chitosan aqueous solution of 2 weight percentage (wt%) was prepared by dissolving chitosan powder in distilled water containing 2% acetic acid, then 5 wt% nano-hydroxyapatite (HA) powder was added slowly, mixed solution was blended with a magnetic stirrer for 12 hours and then uniformly coated on the clean glass using a photoresist spinner. Then, the coated solution was heated on a bake table for 1 hour. After baking, the CTC-BioTChip was soaked in 10 wt% sodium hydroxide (NaOH) solution for 10 hours. Finally, the substrate was washed with DI water and dried for use.
The sLeX-AP (biotin-5′- ATG ACC ATG ACC CTC CAC ACG TTT TTG TGT GCA TGT GAC GCT TGT ATG ATT CAG ACT GTG GCA GGG AAA C -3′) were harvested from Professor Maohui Feng's group in Zhongnan Hospital of Wuhan University. The SELEX experiment for sLeX-AP screen was carried out as reported by Professor Feng's lab (detailed in Supporting information).
Surface modification with sLeX-AP [25]
First, CTC-BioTChip was modified with 4% (v/v) 3-mercaptopropyl trimethoxysilane (Sigma-Aldrich Co.) in ethanol at room temperature for 1 hour. Then, the substrate was treated with the coupling agent N-y-maleimidobutyryloxy succinimide ester (GMBS) 0.25 mM (Sigma-Aldrich Co.) for 1 hour. Afterwards, the substrate was treated with 20 μg mL-1 of streptavidin (SA) (Invitrogen, Carlsbad, CA) at room temperature overnight, and subsequently washed with 1× phosphate buffered saline (PBS) to remove excess SA. Then, CTC-BioTChip was modified with sLeX-AP for specific cancer cell capture.
Cells and blood sample
The hepatocellular cancer cell line (HepG2), breast cancer cell line (MCF-7), colorectal cancer cell line (HCT116), gastric carcinoma cell line (MGC803), cervical cancer cell line (HeLa), were harvested from Hubei Key Laboratory of Tumor Biological Behaviors (Hubei, People's Republic of China). Cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) (HyClone, China) and supplemented with 10% fetal bovine serum (HyClone, China) and 1% penicillin/streptomycin solution at 37°C under a humidified 5% CO2 atmosphere. Whole blood samples from healthy donors and patients were obtained from Department of Clinical Laboratory, Zhongnan Hospital of Wuhan University according to a protocol by the Institutional Review Board (IRB). All blood specimens were collected into anticoagulant tubes (EDTA K2, 2 ml) (Wuhan Zhiyuan Medical Technology Co, Ltd, Wuhan, China).
Cell capture and identification
Prior to cell capture, CTC-BioTChip was placed into a size-matched chamber, and then 10 μL sLex-AP (in Dulbecco's Phosphate Buffered saline, DPBS) (Gibco, China) or anti-EpCAM was added, after 2 hours' incubation, CTC-BioTChip was gently washed with PBS, 1 mL of cell suspensions (105 cells mL-1) was loaded, then placing in a cell incubator under the conditions of 37°C and 5% CO2. After the substrate was gently washed with PBS, the captured cells on the substrate were fixed with 4% paraformaldehyde (PFA) (Sigma-Aldrich) in PBS for 10 minutes, blocking solution (2% donkey serum in PBS/0.02% triton X-100) was then added on to the chip for intracellular staining, after incubated for 30 min at room temperature, captured cells were stained with 4′,6-diamidino-2- phenylindole dihydrochloride (DAPI; 0.2 μg mL-1 in DI water) for 10 minutes. Finally, we imaged and detected targeted cells using a fluorescence microscope (IX81; Olympus, Tokyo, Japan). CTCs captured on substrates were photographed using IPP software (Media Cybernetics Inc., Silver Spring, MD, USA).
Cell capture from artificial blood and patient blood samples
Cell capture efficiency of the optimal capture condition was validated using sLex-AP coated CTC-BioTChip to capture target cells (HepG2) from the two kinds of artificial blood sample: 1) 1 mL DMEM containing HepG2 cells, and 2) 1 mL human blood (healthy donor) containing HepG2 cells. After rinsing, followed by staining of anti-CK, anti-CD45, and DAPI, specifically captured HepG2 cells were identified and counted on the substrates.2mL patient peripheral blood sample was introduced onto our chip according to the procedure described above.
Statistical analysis
Categorical data displayed in a contingency table were analyzed using Fisher's exact test. Continuous data were analyzed by using Mann-Whitney test and Kruskal-Wallis test. Spearman rank correlation analysis was used for nonparametric correlation analysis. All statistical analyses were carried out with the IBM SPSS statistical software package (IBM Inc.). P < 0.05 was considered statistically significant.
Results
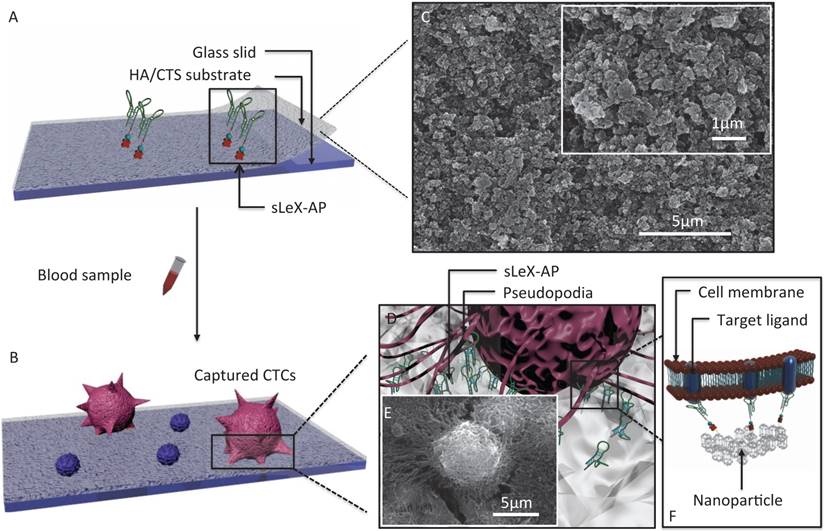
The principle for cancer cell capture by using CTC-BioTChip (detailed dimension in Supporting information) was schematically described (Figure 1). CTC-BioTChip was designed to be platform was composed of two functional components: 1) cell-preferred nanostructured substrate (HA/CTS), 2) capture agent sLeX-AP (Figure 1A-B). We used HA/CTS as the substrate material due to its excellent cell-preferred nanoscale topography (Figure 1C), and great biocompatibility, which can improve CTC-capture efficiency; the captured cancer cells on HA/CTS substrate appeared as many extended pseudopodia (Figure 1 D-E). This feather supplied CTC-BioTChip with solid fundamental for CTCs detection. In our work, synthesized sLeX- AP we use as capture agent was a kind of single stranded DNA with unique structure to recognize surface sLex on HepG2 cells (Figure 1F), and exhibited excellent affinity to HepG2 cells. HA/CTS substrate through fabrication can be coated with capture agent (i.e. anti-EpCAM) to further improve CTC-capture efficiency. In this study, we replaced the antibodies with sLeX-AP for HCC CTC capture. These two functional components supported the working mechanism of our cell-capture approach.
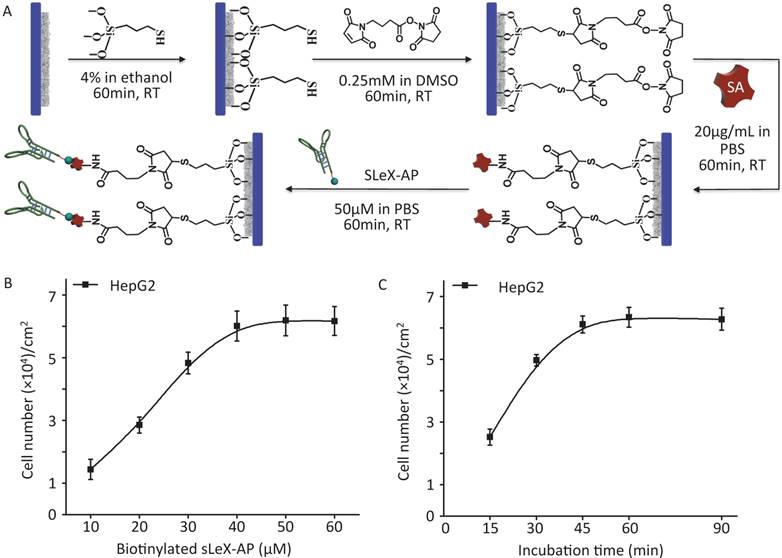
As illustrated procedure of fabrication (Figure 2A) (details in Materials and Methods), CTC-BioTChip platform was prepared for cancer cell capture. Roughness of the nanofilm substrates was controlled by changing the HA: CTS ratio and this ratio (5:2 by weight) had already been optimized [25]. Before capture efficiency studies, optimization studies were conducted in order to determine a reasonable sLeX-AP concentration that allowed for effective capture of HepG2 cells in DMEM. Artificial samples were prepared by spiking 105 HepG2 cells into 1 mL of DMEM, and were utilized to CTC-BioTChip platform, sLeX-AP concentration set at 10, 20, 30, 40, 50, 60 μM in DPBS. As observed in Figure 2B, the number of captured HepG2 cells elevated along with the increasing concentration of sLeX-AP, and reached the plateau at sLeX-AP concentration of 50 μM. This observation indicated that 50 μM will be enough for surface modification to achieve satisfied capture efficiency. During our optimization studies, we noticed that another important factor may influence capture efficiency of CTC-BioTChip platform: incubation time. The viable cells need appreciable amount of time to form stable contact with the substrate. They need time for both the mobility of the cell membrane and extracellular matrix exposure, also for the firm combination between the capture agent and their specific surface components. Thus, we then validated the cell capture efficiency of CTC-BioTChip using optimized sLeX-AP concentration in different incubation time (Figure 2C). At first, the number of captured cells increased with longer incubation time, then the number of captured cancer cells plateaued at maximum value at 60 min and no more cells were captured longer than 60 min, which was consistent with our previous work [25, 29].
(A-B) Schematic workflow of the CTC-BioTChip. The biotinylated-sLeX-AP coated on HA/CTS substrate for HCC CTCs capture. (C) SEM image of HA/CTS substrate. (D) Conceptual illustration of HCC CTC interacting with sLeX-AP coated CTC-BioTChip. (E) SEM image of HepG2 cell captured on sLeX-AP coated CTC-BioTChip. (F) Conceptual illustration of the molecular mechanism of capturing HCC CTC by sLeX-AP coated CTC-BioTChip. Abbreviations: CTCs, circulating tumor cells; SEM, scanning electron microscope; HA/CTS, hydroxyapatite/chitosan.
(A) Schematic workflow of CTC-BioTChip fabrication. HepG2 capture yields of sLeX-AP coated CTC-BioTChip (B) at different sLeX-AP concentration (with concentration of 10, 20, 30, 40, 50, 60 μM) and (C) at different incubation time (15, 30, 45, 60, 90min). The error bar represents standard deviation from three repeats.
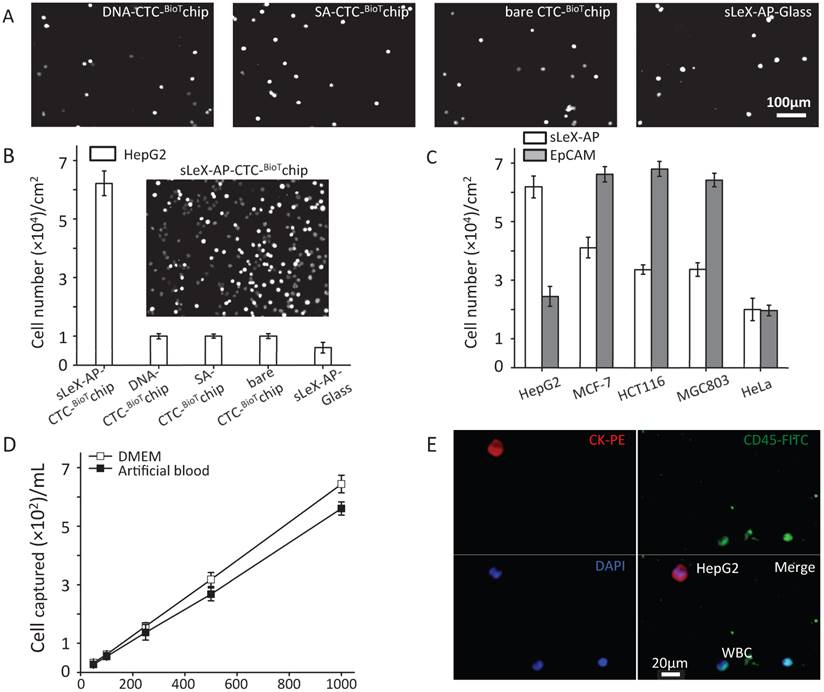
Under the optimized condition, series of studies were conducted to assess the capture performance of CTC-BioTChip platform. Firstly, five control studies to capture prepared HepG2 cell suspension (105 cells mL-1 in DMEM): 1) SA- CTC-BioTChip, 2) bare CTC-BioTChip, 3) random-DNA coated CTC-BioTChip, 4) sLeX-AP coated glass slid, 5) anti-EpCAM coated CTC-BioTChip were carried out in parallel with sLeX-AP coated CTC-BioTChip. After modification, the sLeX-AP coated CTC-BioTChip can provide a much higher capture efficiency compared to bare CTC-BioTChip and random DNA (detailed in supporting information) -coated CTC-BioTChip as evidenced by DAPI- staining fluorescence images (Figure 3A-B), and also the similar phenomenon was observed when comparing sLeX-AP coated CTC-BioTChip with other two control groups. Moreover we also observed that bare CTC-BioTChip captured more HepG2 cells than sLeX-AP coated glass captured. All of these results illustrated that both sLeX-AP as capture agents and the HA/CTS substrate played an essential role in achieving the superb HepG2 capture performance. Next, to illustrate the cell capture specificity and optimized cell capture performance of CTC-BioTChip for HepG2, we used three cancer cell lines MCF-7 (breast cancer cell line), HCT116 (colorectal cancer cell line) and MGC803 (gastric carcinoma cell line) for specific cell capture as cell control with high expression of EpCAM, HeLa (cervical cancer cell line) as non-specific cell control, to compare the performance between sLex-AP and anti-EpCAM coated CTC-BioTChip. Anti-EpCAM coated CTC-BioTChip reaches higher capture yields than that of sLex-AP coated CTC-BioTChip when capturing MCF-7, HCT116 and MGC803. However, for HepG2 capturing, sLex-AP coated CTC-BioTChip reached significantly higher capture yield than anti-EpCAM coated CTC-BioTChip. These results (Figure 3C) support the idea that CTC-BioTChip coated with sLex-AP was capable of enhancing HepG2 capture, but unable to achieve improved performance of capturing MCF-7, HCT116, and MGC803 cells.
Then, we validated the capture performance of sLeX-AP coated CTC-BioTChip. Series of artificial blood samples were prepared by spiking HepG2 cells into DMEM and healthy donor's blood at densities of approximately 50, 100, 250, 500, 1000 cells mL-1. A few to hundreds mL-1 HepG2 cells were captured from DMEM and healthy donor's blood with a yield of 67.3% and 61.6%, respectively (Figure 3D). And, we also compared the capture performance of anti-EpCAM coated CTC-BioTChip with slex-AP coated CTC-BioTChip using artificial blood (detailed in supporting information). PE-labeled anti-CK, FITC-labeled anti-CD45 and DAPI were introduced onto the CTC-BioTChip for artificial CTCs' identification [24] (Figure 3E). These results indicate that sLeX-AP coated CTC-BioTChip exhibited sufficient capture efficiency to manage CTCs capture and identification from clinical samples which normally had a few to hundreds CTCs mL-1.
In order to validate the clinical utility of our platform, we applied optimized sLex-AP coated CTC-BioTChip for rare CTCs capture from HCC patients' peripheral blood samples. 2.0 mL peripheral blood sample (preserved in EDTA-K2 tubes) from 42 HCC patients was introduced onto CTC-BioTChip for CTCs enrichment. The clinical characteristics of 42 HCC patients were summarized in Table 1, and were classified according to the criteria of the sixth edition of International Union Against Cancer (UICC) tumor-node-metastasis (TNM) staging system [45]. Captured cells were rinsed with PBS, fixed and permeabilized, and then they were stained by PE-labeled anti-CK, FITC-labeled anti-CD45 and DAPI. Specifically captured CTCs (CK+/ CD45-/ DAPI+, 10 µm<cell sizes<30 µm) were identified from WBCs (CK- /CD45+ /DAPI+, sizes<15 µm) using fluorescence microscopy (Figure 4A).
(A-B) Five control studies and their corresponding fluorescent micrographs (DAPI nuclear staining only): 1) sLex-AP coated CTC-BioTChip, 2) random DNA coated CTC-BioTChip, 3) SA coated CTC-BioTChip, 4) bare CTC-BioTChip, 5)sLeX-AP coated glass were examined. (C) Cell capture efficiences on sLeX-AP and anti-EpCAM coated CTC-BioTChip were validated using HepG2 (EpCAM-), MCF7 (EpCAM+), HCT116 (EpCAM+), MGC803 (EpCAM+), HeLa (EpCAM-). (D)The captured cell number of CTCs was validated from two kinds of artificial blood samples. (E)The fluorescent micrographs of cancer cells captured from the artificial blood samples. Three-color immunocytochemistry method based on FITC-labeled anti-CD45, PE-labeled anti-CK, and DAPI nuclear staining was applied to identify and enumerate CTCs from non-specially trapped WBCs. Scale bars are 20μm. All capture efficiency experiments were under optimal condition. The error bar represents standard deviation from three repeats. Abbreviations: CTCs, circulating tumor cells; FITC, fluorescein isothiocyanate; PE, phycoerythrin; DAPI, 4 ,6-diamidino-2-phenylindole dihydrochloride; WBCs, white blood cells.
(A) CTCs isolated from a HCC patient. Three-color immunocytochemistry method based on FITC-labeled anti-CD45, PE-labeled anti-CK, and DAPI nuclear staining was applied to identify and enumerate CTCs from non-specifically trapped WBCs. (B) CTC enumeration results obtained from 42 cancer patients. Scatter plot for CTCs number of HCC patients(C) with and without portal vein tumor thrombus, (D) different tumor size and (E) different TNM stage, each red dots stand for one HCC patient. The error bar represents standard error of the mean. Abbreviations: CTCs, circulating tumor cells; FITC, fluorescein isothiocyanate; PE, phycoerythrin; DAPI, 4 ,6-diamidino-2-phenylindole dihydrochloride; WBCs, white blood cells.
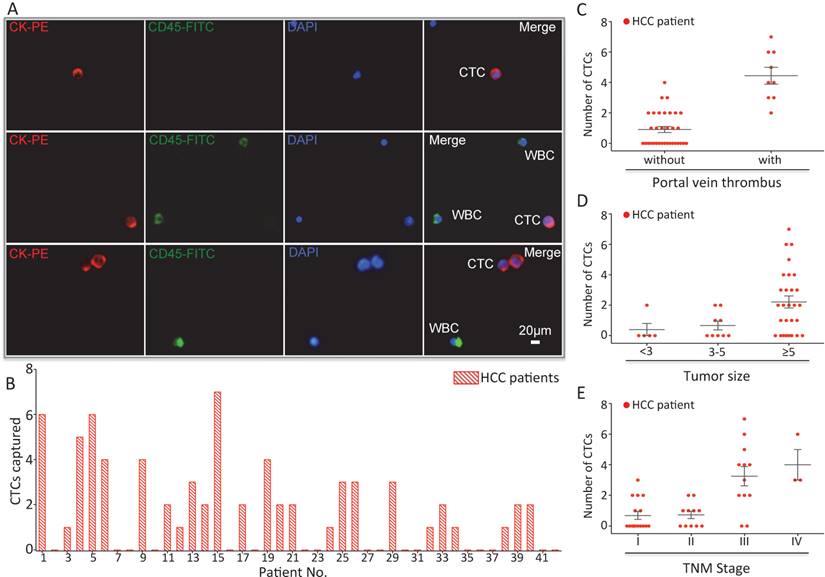
After using CTC-BioTChip for CTCs enrichment, we summarized CTCs number (Figure 4B) and positive rate (Table 2). The number of isolated CTCs ranged from 0 to 7 per 2 mL, with an average of 2±2 (mean ± SD). Statistical analyses were used to illustrate the relationship between CTCs and clinical characteristics of HCC patients. The positive rate of CTCs in patients with portal vein tumor thrombus (100%) was significantly higher than that in patients without portal vein tumor thrombus (48.5%; p=0.006) (Figure 4C), and the number of CTCs detected in patients with portal vein tumor thrombus (4±2) was significantly higher than that in patients without portal vein tumor thrombus (1±1; p< 0.001). Spearman correlation analysis showed that there was a high correlation between the CTCs' positive rate and tumor size (r=0.363, p=0.018) and between the number of CTCs and tumor size (r=0.424, p=0.005) (Figure 4D). The positive rate of CTCs was highly correlated with TNM staging from 66% in stage I to 100% in stage IV (r=0.436, p=0.004), and also the number of CTCs was highly correlated with TNM staging from 1±1 in stage I to 4±2 in stage IV (r=0.599, p< 0.001) (Figure 4E). Neither the positivity rate of CTCs nor the number of CTCs was correlated with age, sex, etiology, Child-Pugh class, AFP level or tumor number (Table 3).
Discussion
As demonstrated in Figures 1-3, sLeX-AP coated CTC-BioTChip showed sufficient capture efficiency for HCC CTCs; both sLex-AP and HA/CTS are integral components of the platform. CTC-BioTChip supplied the platform with excellent biocompatibility and transparency, this biomimetic surface achieved enhanced topographic interactions with captured CTCs. Recent study by other group also have confirmed the utility of HA/CTS (modified by PEG with specific capture agents) for CTCs capture [46]. Moreover, parts of live CTCs can be isolated from cancer patients and cultured [47, 48], and our pervious study have already shown that cancer cells could be cultured on this substrate [25], thus we believed that in future our CTC-BioTChip could be used for in situ ex-vivo culture of patients' CTCs for further biological molecular analysis. Another important part of the platform is sLeX-AP; its use significantly improved the capture efficiency of HCC CTCs through this platform. Notablely, our group [19] as well as other groups [23, 37-41] previously applied aptamers as functional molecules to achieve excellent CTCs detection in many types of cancers, which indicated that aptamers were applicable for rare CTCs detection. Especially, epithelial-mesenchymal transition (EMT) have been observed in CTCs, the main characteristic of EMT-CTCs was that the epithelial markers (EpCAM, E-cadherin etc.) downregulated and mesenchymal markers (Vimentin, N-cadherin etc.) accordingly upregulated [49], thus using anti-EpCAM as functional molecules could miss this valuable part of CTCs. It was proposed that EMT may promote CTCs migration and invasion during tumor metastasis [50], moreover several studies showed that CTCs' migration and invasion was partly affected by sLeX/E-selectin way [51-53].
Clinical Characteristics of 42 HCC patients.
| Variable | Number of patient | % |
|---|---|---|
| Age | ||
| <60 | 24 | 57.1 |
| ≥60 | 18 | 42.9 |
| Sex | ||
| Male | 35 | 83.3 |
| Female | 7 | 16.7 |
| Etiology | ||
| HBV | 34 | 81.0 |
| HCV | 1 | 2.4 |
| HBV and HCV | 0 | 0.0 |
| Non-HBV, non-HCV | 7 | 16.7 |
| Child-Pugh class | ||
| Child A | 34 | 81.0 |
| Child B | 8 | 19.0 |
| Child C | 0 | 0.0 |
| AFP | ||
| <20 | 17 | 40.5 |
| 20-100 | 5 | 11.9 |
| 100-400 | 5 | 11.9 |
| ≥400 | 15 | 35.7 |
| Tumor number | ||
| Single | 25 | 59.5 |
| Multi | 17 | 40.5 |
| Tumor size (cm) | ||
| <3 | 5 | 11.9 |
| 3-5 | 9 | 21.4 |
| ≥5 | 28 | 66.7 |
| Portal vein thrombus | ||
| With | 9 | 21.4 |
| Without | 33 | 78.6 |
| TNM stagea | ||
| Stage I | 16 | 38.1 |
| Stage II | 11 | 26.2 |
| Stage III | 12 | 28.6 |
| Stage IV | 3 | 7.1 |
| Total | 42 | 100.0 |
Abbreviations: HBV, hepatitis B virus; HCV, hepatitis C virus.
a. Sixth edition of UICC TNM staging system of HCC (2002).
Number of Detected CTCs from 2 ml peripheral blood.
| Patient Group | Patients (n) | CTCs+ Patients (n) | CTC Mean±SD | Positive rate |
|---|---|---|---|---|
| AFP | ||||
| <20 | 17 | 8 | 1±2 | 47.1% |
| 20-100 | 5 | 3 | 1±1 | 60.0% |
| 100-400 | 5 | 2 | 1±1 | 40.0% |
| ≥400 | 15 | 12 | 3±2 | 80.0% |
| Tumor number | ||||
| Single | 25 | 15 | 1±2 | 60.0% |
| Multi | 17 | 12 | 2±2 | 70.6% |
| Tumor size (cm) | ||||
| <3 | 5 | 1 | 0±1 | 20.0% |
| 3-5 | 9 | 4 | 1±1 | 44.4% |
| ≥5 | 28 | 22 | 2±2 | 78.6% |
| Portal vein thrombus | ||||
| With | 9 | 9 | 4±2 | 100.0% |
| Without | 33 | 18 | 1±1 | 54.5% |
| TNM stagea | ||||
| Stage I | 16 | 6 | 1±1 | 37.5% |
| Stage II | 11 | 6 | 1±1 | 54.5% |
| Stage III | 12 | 10 | 3±2 | 83.3% |
| Stage IV | 3 | 3 | 4±2 | 100.0% |
| Total | 42 | 25 | 2±2 | 59.5% |
a. Sixth edition of UICC TNM staging system of HCC (2002).
The relation between CTCs and Clinicopathological variables of HCC patients.
| Variable | P value | |
|---|---|---|
| CTCs' Positivity Rate | CTCs number | |
| Age | 0.348a | 0.074b |
| Sex | 1.000a | 0.974b |
| Etiology | 0.796c | 0.903c |
| Child-Pugh class | 0.334c | 0.340c |
| AFP | 0.079c | 0.309d |
| Tumor number | 0.339a | 0.098b |
| Portal vein thrombus | 0.006a | 0.000b |
| Tumor size (cm) | 0.018c | 0.005c |
| TNM stagee | 0.004c | 0.000c |
Abbreviation: NS, not significant.
a P values from Fisher's exact test.
b P values from the Mann-Whitney test
c P values from Spearman rank correlation analysis.
d P values from Kruskal-Wallis test
e Sixth edition of UICC TNM staging system of HCC (2002).
Consequentially, sLeX-AP coated CTC-BioTChip was designed to mimic vascular endothelium to interact with CTCs then trapped them; through the rolling (sLeX-AP) and stationary binding (sLeX-ap and CTC-BioTChip) steps, enhanced CTCs capture sensitivity achieved. Besides, during our experiment, we noticed that sLex-AP coated slides showed higher capture efficiency than anti-EpCAM coated slides for HepG2 cells, but showed lower capture efficiency than anti-EpCAM coated slides for MCF-7, HCT116 and MGC803 cells. The reason may be that Sialyl Lewis X had different expression level or surface density in different cancer, which could be a factor to affect the capture efficiency in different cell lines using slex-Ap [43] [54]. In spite of sufficient efficiency for our platform in capturing HCC CTCs, we still noticed certain degree of WBC noise. CTCs and WBCs, which were identified by three-color immunocytochemistry method from patient blood samples, had a ratio (CTC/background cell) ranged from 1/60~1/200 for one random microscope fields (200X). Although nonspecific absorbed cell could be partly washed away during HCC CTCs capture, our platform still showed suboptimal capture purity so far. Our future efforts will be focused on integrating our platform with microfluidic technology such as chaotic mixers, or with WBC negative depletion methods to improve both capture yield and purity for HCC CTCs.
CTC-BioTChip showed clinical usefulness in detecting CTCs from peripheral blood samples collected from 42 HCC patients at various stages (described in Table 1-3). Enrolled HCC patients were classified according to the sixth edition of UICC TNM staging system (42). Statistical analysis showed that CTCs could be detected in 25 HCC patients (59.5%), even in patients at early stage (37.5%) or in patients with tumor size less than 3 cm (20%). Both the positivity rate and the CTCs count were positively correlated with the disease extent as classified by the TNM stage, this correlation implied that CTCs could be used as supplementary to evaluate the degree of disease progression. The correlation between portal vein tumor thrombus and CTCs detection was another part of the results need to be discussed. Portal vein tumor thrombus was an important prognostic factor, and it was previously thought to be the main cause of intrahepatic metastases [55, 56]. However, in our study, data showed that both the positivity rate and the number of CTCs in patients with portal vein tumor thrombus were higher than those in patients without portal vein tumor thrombus, this correlation implied that portal vein tumor thrombus may also be a source of systemic spread of CTCs, and it may contribute to distant metastasis. Tumor size was another prognostic factor for HCC patients, our results showed that tumor size was positively correlated with both the positivity rate and the number of CTCs, this correlation indicated that larger HCC had the ability to release more CTCs to increase the risk of tumor metastasis. These clinical correlations not only suggested that sLex-AP coated CTC-BioTChip have sufficient specificity and sensitivity, also exhibited that CTCs detection by this platform might provide useful information for clinical practice.
However, our current study has the limitations of a small sample size, and data from a single study center. Thus, the correlation between CTCs and HCC clinicopathological characteristics should be interpreted with cautious. A rigorous prospective, multicenter, and comprehensive randomized clinical trial are needed to evaluate the clinical usefulness of our platform for CTC detection in patients with HCC.
Conclusion
Together, in this study, we have successfully developed our CTC-BioTChip for sensitive and specific CTCs detection in HCC patients with lower expense, and smaller blood volume. Our results indicate that CTC detection in HCC patients by this platform might be clinically useful. More importantly, through CTCs' enumeration, we expect our platform might play an important role in treatment responses monitor and patient prognosis indication, and then realize personalized therapy indication for HCC patients in the future.
Supplementary Material
Supplementary tables and figures.
Acknowledgements
The work was supported by grants from National High Technology Research and Development Program of China (Grant No. 2012AA02A502, 2012AA02A506), the National Natural Science Foundation of China (Grant No. 81572874).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559-64
2. Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679-95
3. Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12:895-904
4. Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8:329-40
5. Pantel K, Alix-Panabieres C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med. 2010;16:398-406
6. Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418:823
7. Dotan E, Cohen SJ, Alpaugh KR. et al. Circulating tumor cells: evolving evidence and future challenges. Oncologist. 2009;14:1070-82
8. Pantel K, Alix-Panabieres C. Real-time liquid biopsy in cancer patients: fact or fiction? Cancer Res. 2013;73:6384-8
9. Braun S, Vogl FD, Naume B. et al. A pooled analysis of bone marrow micrometastasis in breast cancer. N Engl J Med. 2005;353:793-802
10. Rahbari NN, Aigner M, Thorlund K. et al. Meta-analysis shows that detection of circulating tumor cells indicates poor prognosis in patients with colorectal cancer. Gastroenterology. 2010;138:1714-26
11. Wang S, Zheng G, Cheng B. et al. Circulating tumor cells (CTCs) detected by RT-PCR and its prognostic role in gastric cancer: a meta-analysis of published literature. PLoS One. 2014;9:e99259
12. Taback B, Chan AD, Kuo CT. et al. Detection of occult metastatic breast cancer cells in blood by a multimolecular marker assay: correlation with clinical stage of disease. Cancer Res. 2001;61:8845-50
13. Keilholz U, Goldin-Lang P, Bechrakis NE. et al. Quantitative detection of circulating tumor cells in cutaneous and ocular melanoma and quality assessment by real-time reverse transcriptase-polymerase chain reaction. Clin Cancer Res. 2004;10:1605-12
14. Liu Z, Jiang M, Zhao J. et al. Circulating tumor cells in perioperative esophageal cancer patients: quantitative assay system and potential clinical utility. Clin Cancer Res. 2007;13:2992-7
15. Schuster R, Bechrakis NE, Stroux A. et al. Circulating tumor cells as prognostic factor for distant metastases and survival in patients with primary uveal melanoma. Clin Cancer Res. 2007;13:1171-8
16. Zieglschmid V, Hollmann C, Bocher O. Detection of disseminated tumor cells in peripheral blood. Crit Rev Clin Lab Sci. 2005;42:155-96
17. Yoon HJ, Kozminsky M, Nagrath S. Emerging role of nanomaterials in circulating tumor cell isolation and analysis. ACS Nano. 2014;8:1995-2017
18. Lee SK, Kim DJ, Lee G. et al. Specific rare cell capture using micro-patterned silicon nanowire platform. Biosens Bioelectron. 2014;54:181-8
19. Shen Q, Xu L, Zhao L. et al. Specific capture and release of circulating tumor cells using aptamer-modified nanosubstrates. Adv Mater. 2013;25:2368-73
20. Wang S, Liu K, Liu J. et al. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew Chem Int Ed Engl. 2011;50:3084-8
21. Wang S, Wang H, Jiao J. et al. Three-dimensional nanostructured substrates toward efficient capture of circulating tumor cells. Angew Chem Int Ed Engl. 2009;48:8970-3
22. Jeon S, Hong W, Lee ES. et al. High-purity isolation and recovery of circulating tumor cells using conducting polymer-deposited microfluidic device. Theranostics. 2014;4:1123-32
23. Sheng W, Chen T, Tan W. et al. Multivalent DNA nanospheres for enhanced capture of cancer cells in microfluidic devices. ACS Nano. 2013;7:7067-76
24. He R, Zhao L, Liu Y. et al. Biocompatible TiO2 nanoparticle-based cell immunoassay for circulating tumor cells capture and identification from cancer patients. Biomed Microdevices. 2013;15:617-26
25. Cheng B, He Z, Zhao L. et al. Transparent, biocompatible nanostructured surfaces for cancer cell capture and culture. Int J Nanomedicine. 2014;9:2569-80
26. Zhao L, Lu YT, Li F. et al. High-purity prostate circulating tumor cell isolation by a polymer nanofiber-embedded microchip for whole exome sequencing. Adv Mater. 2013;25:2897-902
27. Zhang N, Deng Y, Tai Q. et al. Electrospun TiO2 nanofiber-based cell capture assay for detecting circulating tumor cells from colorectal and gastric cancer patients. Adv Mater. 2012;24:2756-60
28. Chen W, Weng S, Zhang F. et al. Nanoroughened surfaces for efficient capture of circulating tumor cells without using capture antibodies. ACS Nano. 2013;7:566-75
29. Cheng B, Song H, Wang S. et al. Quantification of rare cancer cells in patients with gastrointestinal cancer by nanostructured substrate. Transl Oncol. 2014;7:720-5
30. Porcell AI, De Young BR, Proca DM. et al. Immunohistochemical analysis of hepatocellular and adenocarcinoma in the liver: MOC31 compares favorably with other putative markers. Mod Pathol. 2000;13:773-8
31. Went PT, Lugli A, Meier S. et al. Frequent EpCam protein expression in human carcinomas. Hum Pathol. 2004;35:122-8
32. Schulze K, Gasch C, Staufer K. et al. Presence of EpCAM-positive circulating tumor cells as biomarker for systemic disease strongly correlates to survival in patients with hepatocellular carcinoma. Int J Cancer. 2013;133:2165-71
33. Sun YF, Xu Y, Yang XR. et al. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology. 2013;57:1458-68
34. Zhang Y, Li J, Cao L. et al. Circulating tumor cells in hepatocellular carcinoma: detection techniques, clinical implications, and future perspectives. Semin Oncol. 2012;39:449-60
35. Lin M, Chen JF, Lu YT. et al. Nanostructure embedded microchips for detection, isolation, and characterization of circulating tumor cells. Acc Chem Res. 2014;47:2941-50
36. Hong B, Zu Y. Detecting circulating tumor cells: current challenges and new trends. Theranostics. 2013;3:377-94
37. Shangguan D, Li Y, Tang Z. et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci U S A. 2006;103:11838-43
38. Sheng W, Chen T, Kamath R. et al. Aptamer-enabled efficient isolation of cancer cells from whole blood using a microfluidic device. Anal Chem. 2012;84:4199-206
39. Chen L, Liu X, Su B. et al. Aptamer-mediated efficient capture and release of T lymphocytes on nanostructured surfaces. Adv Mater. 2011;23:4376-80
40. Nair SV, Witek MA, Jackson JM. et al. Enzymatic cleavage of uracil-containing single-stranded DNA linkers for the efficient release of affinity-selected circulating tumor cells. Chem Commun (Camb). 2015;51:3266-9
41. Yu X, Wang B, Zhang N. et al. Capture and Release of Cancer Cells by Combining On-Chip Purification and Off-Chip Enzymatic Treatment. ACS Appl Mater Interfaces. 2015;7:24001-7
42. Schultz MJ, Swindall AF, Bellis SL. Regulation of the metastatic cell phenotype by sialylated glycans. Cancer Metastasis Rev. 2012;31:501-18
43. Takada A, Ohmori K, Yoneda T. et al. Contribution of carbohydrate antigens sialyl Lewis A and sialyl Lewis X to adhesion of human cancer cells to vascular endothelium. Cancer Res. 1993;53:354-61
44. Xie J, Zhao R, Gu S. et al. The architecture and biological function of dual antibody-coated dendrimers: enhanced control of circulating tumor cells and their hetero-adhesion to endothelial cells for metastasis prevention. Theranostics. 2014;4:1250-63
45. Greene F, Page D, Fleming I. et al. Liver including intrahepatic bile ducts. AJCC Cancer Staging Handbook. 2002:131-44
46. Sun N, Wang J, Ji L. et al. A Cellular Compatible Chitosan Nanoparticle Surface for Isolation and In Situ Culture of Rare Number CTCs. Small. 2015;11:5444-51
47. Yu M, Bardia A, Aceto N. et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216-20
48. Zhang L, Ridgway LD, Wetzel MD. et al. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci Transl Med. 2013;5:180ra48
49. Krebs MG, Metcalf RL, Carter L. et al. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat Rev Clin Oncol. 2014;11:129-44
50. Thiery JP, Lim CT. Tumor dissemination: an EMT affair. Cancer Cell. 2013;23:272-3
51. Burdick MM, McCarty OJ, Jadhav S. et al. Cell-cell interactions in inflammation and cancer metastasis. IEEE Eng Med Biol Mag. 2001;20:86-91
52. Barthel SR, Gavino JD, Descheny L. et al. Targeting selectins and selectin ligands in inflammation and cancer. Expert Opin Ther Targets. 2007;11:1473-91
53. Kannagi R. Molecular mechanism for cancer-associated induction of sialyl Lewis X and sialyl Lewis A expression-The Warburg effect revisited. Glycoconj J. 2004;20:353-64
54. Christiansen MN, Chik J, Lee L. et al. Cell surface protein glycosylation in cancer. Proteomics. 2014;14:525-46
55. Toyosaka A, Okamoto E, Mitsunobu M. et al. Intrahepatic metastases in hepatocellular carcinoma: evidence for spread via the portal vein as an efferent vessel. Am J Gastroenterol. 1996;91:1610-5
56. Mitsunobu M, Toyosaka A, Oriyama T. et al. Intrahepatic metastases in hepatocellular carcinoma: the role of the portal vein as an efferent vessel. Clin Exp Metastasis. 1996;14:520-9
Author contact
![]() Corresponding author: Bin Xiong, Dept. Of Oncology, Zhongnan Hospital of Wuhan University; Hubei Key Laboratory of Tumor Biological Behaviors & Hubei Cancer Clinical Study Center, Wuhan 430071, P.R. China. E-mail: Binxiong1961edu.cn.
Corresponding author: Bin Xiong, Dept. Of Oncology, Zhongnan Hospital of Wuhan University; Hubei Key Laboratory of Tumor Biological Behaviors & Hubei Cancer Clinical Study Center, Wuhan 430071, P.R. China. E-mail: Binxiong1961edu.cn.
 Global reach, higher impact
Global reach, higher impact