13.3
Impact Factor
Theranostics 2017; 7(17):4183-4191. doi:10.7150/thno.21299 This issue Cite
Review
Identifying and Characterizing circRNA-Protein Interaction
1. Sunnybrook Research Institute, Sunnybrook Health Sciences Centre, Toronto;
2. Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto;
3. State Key Laboratory of Applied Microbiology Southern China, Guangdong Provincial Key Laboratory of Microbial Culture Collection and Application, Guangdong Institute of Microbiology, Guangzhou, 510070, China;
4. Yuewei Edible Fungi Technology Co. Ltd., Guangzhou, 510070, China;
5. Atta-ur-Rahman School of Applied Biosciences (ASAB), National University of Sciences and Technology (NUST), H-12 Islamabad, Pakistan;
6. Institute of Medical Science, University of Toronto, Toronto, Canada.
Received 2017-6-2; Accepted 2017-8-4; Published 2017-9-26
Abstract
Circular RNAs have been identified as naturally occurring RNAs that are highly represented in the eukaryotic transcriptome. Although a large number of circRNAs have been reported, circRNA functions remain largely unknown. CircRNAs can function as miRNA sponges, thereby reducing their ability to target mRNAs. We hypothesize that circRNAs may bind, store, sort, and sequester proteins to particular subcellular locations, and act as dynamic scaffolding molecules that modulate protein-protein interactions. Here, we review the biological implication and function of circRNA-protein interaction, and reveal a dynamic model of the interaction in various tissues, development stages and physiological conditions. Improved techniques to identify and characterize the dynamic RNA-protein interactions may elucidate the molecular mechanisms associated with the expression and functional diversity of circRNAs.
Functional Significance of circRNAs
Circular RNAs are a novel class of endogenous RNAs with covalently closed loop structures. They are generated during RNA splicing and arise from exons (exonic circular RNAs or circRNAs), introns (intronic circular RNAs or ciRNAs), or a combination of both (EIciRNAs)1. ciRNAs are end-joined in a different way than circRNAs and EIciRNAs. Although circular RNAs were first identified over twenty years ago, they were generally considered to represent splicing errors and thought to be present in low abundance. With the advent of high-throughput sequencing and novel computational approaches for non-polyadenylated RNA transcripts, thousands of circular RNAs have been successfully identified in various species.2 CircRNAs from back-spliced exons have been identified as naturally occurring RNAs that are highly represented in the eukaryotic transcriptome3. Although most circular RNA transcripts are generally expressed at low levels relative to their respective linear RNAs, some circRNAs have been reported to be expressed more abundantly than their linear counterparts.4 CircRNAs are found mostly in the cytoplasm and have been identified in exosome of culture media.5, 6 Thus far, although some circular RNAs may function as ceRNAs in regulating the activity of the corresponding linear mRNAs by binding miRNAs, no specific association in terms of functionality has been identified between a circular RNA and its corresponding linear transcripts. Furthermore, circRNAs have been implicated in tissue and organ development and may play a role in various disease processes, such as neuro-degeneration and cancer development. 7, 8
The function of circRNAs remains largely unknown. A handful of circRNAs have been shown to be involved in post-transcriptional regulation by functioning as "sponges" of miRNAs, reducing their ability to target mRNAs.9,10 Specifically, the circRNA ciRS-7/CDR1as has been found to harbour over 70 conventional miR-7 binding sites and functions as a specific miRNA sponge, repressing the function of miR-7. High-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) suggests that the circRNA CDR1as is densely bound by the miRNA effector protein Ago2 and miR-7.9,10 Similarly, the circular transcript of the Sry gene has 16 binding sites for miR-138 and was pulled-down by an antibody against AGO2 when miR-138 was overexpressed. miR-138 mediated mRNA targeting was then shown to be attenuated with circular Sry overexpression.9 Initially, two circRNAs, ciRS-7/CDR1as and Sry were reported to contain multiple miRNA binding sites.11,12 Later on, a couple other circular RNAs have been reported to function as sponges of miRNAs, enhancing targeting gene translation13,14. We have found that circ-Foxo3 can also function as an miRNA sponge, regulating cell cycle progression 15. To function as an miRNA sponge, circular RNAs need to either harbor many miRNA binding sites or be expressed at very high levels in the cytosol 16.
Another study on circRNAs in neural tissue used a bioinformatics approach to estimate the potential of the brain-derived circRNAs to serve as miRNA sponges. This study revealed that as a general class, these circRNAs did not exhibit a greater capacity to serve as miRNA sponges than their corresponding linear mRNAs. 17 It is debated whether or not miRNA inhibition is a general feature of circRNAs. Although some reports have indicated that certain circRNAs function as miRNA sponges, several studies have showed that the majority of circRNAs do not act as miRNA sponges.11, 12, 17-19
It has been proposed that some circRNAs may “sponge” other factors, such as RNA binding proteins. 20 circRNAs may be used to bind, store, or sequester molecules such as transcription factors, to particular subcellular locations. In this way, circRNAs may act as dynamic scaffolds for the assembly of other components. However, some studies indicated that based on nucleotide sequence alone, circRNAs, as a group, are not more likely to bind to RNA binding proteins (RBPs) than linear mRNAs. 17
Abundantly expressed circular RNAs may have other unknown regulatory functions. EIciRNAs, a subclass of circular RNAs, have been shown to promote transcription of the gene from which they were derived, through interactions with host U1 snRNP and RNA Polymerase II.1 circ-RasGEF1B, a stress related circRNA, conserved in humans and mice, was shown to be up-regulated by the NF-κB pathway. This circRNA positively regulates the expression of ICAM-1 by enhancing the stability of mature ICAM-1 mRNA. 21 Bioinformatics analyses revealed no shared miRNA binding site between circ-RasGEF1B and the ICAM-1 transcript, ruling out the possibility that circ-RasGEF1B acts directly via miRNA-based regulation. The mechanism by which circ-RasGEF1B regulates ICAM-1 mRNA stability remains unknown. Although many circRNAs may possess biological functions, it remains possible that a large number of circRNAs may actually be inconsequential side-products of pre-mRNA splicing.12
Increased evidence indicates that circular RNAs may not be a real class of noncoding RNAs, at least some of them are translatable22-24. It is reported that a protein can be translated from circular RNAs in human cells driven by N6-methyladenosine22. The circular RNA Circ-ZNF609 contains an open reading frame spanning from the start codon, in common with the linear transcript, that is terminated at an in-frame stop codon, creating circularization. This circular RNA can be translated into a specific protein in a splicing-dependent and cap-independent manner, controlling myoblast proliferation 24.
Biological implications of circRNA-protein interaction
Many circRNAs are predicted to interact with RNA binding proteins (RBPs) 20, although bioinformatic analyses of circRNA sequences found very little enrichment in binding sites of RBPs compared with those of its corresponding linear mRNA.17 Due to the unique tertiary structure of circRNAs, the protein binding capacity of circRNA is likely to be more complex than previously thought. While emerging evidence indicates that some circRNA are translatable 22-24, the majority of circular RNAs are not translated as linear mRNAs are. Thus, RBPs bound to circRNAs are not displaced by ribosomes.11,12 The traditional nucleotide sequence-based approaches that are mainly used for assessing the binding of linear mRNAs with proteins may not work appropriately for analyzing circRNA-protein interaction.
It has been reported that some RNA-binding proteins (RBPs), such as Argonaute and MBL can bind to circRNAs.1, 9,25 ciR-7/CDR1as contains more than 70 selectively conserved miRNA target sites, and is widely associated with Argonaute (AGO) proteins in an miR-7-dependent manner.9 The circular MBL/MBNL1 RNA contains conserved muscleblind (MBL) binding sites, which are strongly and specifically bound by MBL. This study supported a direct role for MBL in circMbl biosynthesis. Modulation of MBL levels significantly affects circMbl biosynthesis, and this effect is dependent on MBL binding. The introns flanking circMbl have MBL binding sites, which will decide the circularization rates of bracketed exons, dependent on MBL levels. Expression of MBL may decrease the production of the parental mRNA by promoting circMbl production.25 The circular RNA circ-PABPN1 has been reported to bind to HuR, and extensive binding of HuR to circ-PABPN1 prevents its binding to PABPN1 mRNA and lowers PABPN1 translation 26. This study provides an example of competition between a circRNA and its cognate mRNA for an RBP that affects translation.
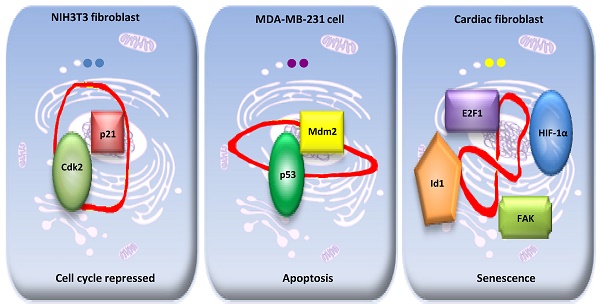
We have found that an endogenous cytosolic circRNA, a circular transcript from forkhead box O3 (circ-Foxo3) can facilitate, localize, and regulate the functions of proteins.27 Circ-Foxo3 displays high binding affinity to some senescence and stress related transcription factors, such as Id-1, E2F1, HIF-α, and FAK. Expression of circ-Foxo3 reduced nuclear translocation of Id-1, E2F1, and HIF-α, as well as mitochondrial translocation of FAK during cardiac stress, promoting cardiac senescence in vitro and in vivo.27 Holdt and co-workers recently found that a circRNA that is transcribed at a locus of atherosclerotic cardiovascular disease on chromosome 9p21 (circANRIL), was associated with reduced risk of atherosclerosis. CircANRIL does not have miRNA sponging activity and does not regulate the protein-coding genes, but does interact with the C-terminal lysine-rich domain of PES1, thereby competing with the rRNA for binding to protein PES1 and impairing ribosome biogenesis, inducing apoptosis. 28 In contrast, linear ANRIL, a known long noncoding RNA, was previously reported to have the opposite effect and promote atherosclerosis. 29 This confirms that circANRIL cannot cross-talk with the linear ANRIL by sponging miRNAs. Our further studies revealed that circ-Foxo3 is highly expressed in non-malignant cell lines and that its expression is correlated with physiological cell cycle progression.15 In the mouse fibroblast NIH3T3 cell line, circ-Foxo3 interacts with both p21 and CDK2, facilitating the inhibition of CDK2 by p21, and repressing cell cycle progression at the G1 stage.15 This study addressed for the first time how circRNA could act as a scaffold to modulate protein-protein interaction.
Furthermore, our study showed that circ-Foxo3 could regulate the expression of its binding proteins by modulating protein-protein interaction. Ectopic circ-Foxo3 enhanced breast cancer cell sensitivity to Doxorubicin and Cisplatin, potentiating cellular apoptosis following chemotherapeutic treatment.30 Mechanistically, circ-Foxo3 increased Foxo3 protein levels but repressed p53 expression. By binding to p53 and MDM2, circ-Foxo3 promoted MDM2-induced p53 ubiquitination and subsequent degradation, resulting in an overall decrease of p53 in p53 mutant breast cancer cell lines. The low binding affinity of Foxo3 protein to circ-Foxo3 prevented MDM2 from inducing Foxo3 ubiquitination and degradation, resulting in increased levels of Foxo3 protein. As a result, cell apoptosis was induced by up-regulation of Foxo3 and its down-stream target Puma. 30 Our previous study showed that circ-Foxo3 functioned as a sponge for miRNA binding and promoted Foxo3 expression, since circ-Foxo3 is highly expressed in cytosol. 31 Thus, circ-Foxo3 promotes Foxo3 expression by two different mechanisms: decreasing Foxo3 protein degradation and enhancing Foxo3 translation through sponging miRNAs.
Detecting circRNA-protein interactions
Global bioinformatic analyses of circRNA sequences have indicated that circRNAs show no enrichment in binding sites of RBPs compared with those of its linear mRNA. 17 However, tertiary structures of circRNAs may be more influential in determining protein binding capacity than the linear RNA sequences. To date, the interactions between circRNAs and proteins are mainly analyzed by RNA pull-down assay or RNA immunoprecipitation (RIP).
In the RNA pull-down assay, cell cultures are lysed in co-IP buffer, and incubated with biotinylated DNA oligo probes against putative binding circRNAs, for example, the circ-Foxo3 probe that is biotinylated. Streptavidin magnetic beads are added to each binding reaction and further incubation takes place allowing binding of the Streptavidin beads to the biotinylated circ-Foxo3 probe. The biotinylated probes will pull down the circular RNAs, for example circ-Foxo3, and proteins that bind to the circular RNAs. The beads are then washed, and bound proteins are processed by Western blotting or mass spectrometry to analyze circRNA binding proteins (Zeng et al., Theranostics. 2017, in press)15,27. Biotinylated random oligo pull-down samples can be used as a negative control for the experimental system. A biotinylated oligo probe against a specific circRNA can be applied to the pull-down assay. Since most circular RNA transcripts are expressed at low levels relative to their respective linear RNAs, overexpression of certain circRNAs in the samples may be a good approach for positive controls in the pull-down experiments. Mass spectrometry results can be further confirmed by RNA pull-down assays followed by Western blotting. Thus, in the RNA pull-down assay, the RNA is pulled-down by the probe, allowing analysis of associated proteins. The probe used in the back-splice junction is the only sequence element that is unique to the circRNA. Due to the sequence overlap with the often more abundant linear host RNA, the probe may pull down the host linear mRNA 32. To avoid this, pre-treatment with RNAse R would be necessary. The length of the probe can be around 30-40 nucleotides, which target the back-splicing junction region of the circular RNAs. Due to their complex tertiary structures, circRNAs could generally pull down more non-specific binding proteins in the RNA pull-down assay. This experiment should be carefully designed with sufficient control samples. Using circRNA overexpressing and silencing techniques will help to confirm RNA-protein interactions by quantitatively comparing pulled down circRNA with specific pulled down proteins.
RNA binding protein immunoprecipitation assay (RIP) followed by circRNA sequencing is another feasible strategy to analyze circRNA-protein interaction (Zeng et al., Theranostics. 2017, in press)15, 27, 33. In the circular RNA immunoprecipitation (RIP) assay, a protein is immunoprecipitated, allowing analysis of associated RNA. In brief, harvested cells are washed in ice-cold PBS, lysed in co-IP buffer, and incubated with a primary antibody. A 50% slurry of protein A-Sepharose is added to each sample, and incubated. Pellets are washed with PBS and resuspended in Tri Reagent. The eluted co-precipitated RNA in the aqueous solution is subject to circRNA sequencing. Recently, Schneider et al. (2016) performed protein immunoprecipitation assay with antibody against Insulin-like growth factor 2 binding protein 3 (IMP3, IGF2BP3) 33, followed by RNA sequence analysis of pulled down mixtures, and found 34 circRNAs to be associated with IMP3. IMP3 is an RBP, a known tumor marker involved in post-transcriptional regulation and a potential driver of tumor development.33 This study demonstrates that a protein may bind a set of different circRNAs. Generally, pulled down mixtures from anti-mouse or rabbit IG can be used as negative controls. Comparing the pulled down mixtures from the tested samples with positive and negative controls will correlate pulled down proteins and circRNAs, providing evidence of specific protein-circRNA interactions. The sequencing results may be confirmed by real-time PCR analysis to demonstrate the presence of the binding products using their respective primers. The pull-down mixture can also be processed by Western blotting to confirm the interaction of circular RNA with proteins 15. Immunoprecipitation assay of circRNAs and binding proteins using antibodies against the binding proteins followed by circRNA sequencing is another feasible strategy to analyze circRNA-protein interaction. If the fragment of circular RNA is long enough, it could be protected by the binding protein. In this way, it is similar to RNase protection assays (RPAs). However, in many circular RNA-protein interactions analyzed in our studies, we detected diverse distributions of the protein binding nucleotides in the circular RNAs. In some locations, only one or a few nucleotides are involved. RNase protection assay may not allow identification of such protein binding sites.
RNase protection assay (RPA) is a powerful method for detecting RNA and RNA fragments in cell extracts. RPA assays can also be used to map protein-RNA interactions. In such an assay, RNase H is used to cleave a target RNA molecule at a specific site hybridized with a DNA probe. If the oligonucleotides are able to anneal to the complementary sequence of the RNA, RNase H will cleave the RNA outside the double-stranded DNA/RNA region. However, if a protein is bound to the RNA at the target sequence, it will prevent cleavage by RNase H and indicate a site of interaction between protein and RNA. Using many small oligonucleotide probes allows the entire sequence of RNA to be mapped for sites of interaction. Since circRNA-protein interactions are more likely based on the tertiary structures of circRNA, the binding RNA fragments may be at multiple sites, and of varying nucleotides in length. In addition, the junctional region of circRNAs may not necessarily be part of the binding sites, further complicating the identification of circular transcript from linear RNAs. Thus, identifying the circular RNA sequence from the protected RNA fragments remains an intriguing field of research.
Analysis of circRNAs and protein co-localization by microscopy is another strategy that has been used to identify circRNA-protein interaction. Fluorescence in situ Hybridization (FISH/ISH) techniques can be used to detect the circular RNA transcript with a DNA oligo probe in the cells or tissue samples. Meanwhile, immunofluorescence staining can be applied to detect the position and abundance of proteins with specific antibodies. The read-out is a visual image and a co-localized signal for both RNA and protein of interest indicating complex formation. Our recent study reported that circ-Foxo3 could bind to stress related protein HIF-1α, and repressed its nuclear translocation to nuclei during stress.27 Fluorescence in situ hybridization and immunofluorescence staining showed that circ-Foxo3 is mainly expressed in the cytoplasm in fibroblasts, where it co-localized with stress related protein HIF-1α. The control cells displayed significant nuclear translocation of HIF-1α when treated with H2O2, while overexpression of circ-Foxo3 retained HIF-1α in the cytoplasm, interacting with circ-Foxo3 even after H2O2 treatment.
Dynamic circRNA-protein interactions
CircRNAs have unique covalently closed loop structures and specific tertiary structures, and exhibit tissue- and developmental-specific expression, playing crucial roles in multiple cellular processes. Circular RNAs may adopt tertiary structures distinct from related linear mRNAs 11, 34. Increasing evidence indicates that circRNA expression is dynamic, and allows for the maintenance of diverse spatial and temporal expression profiles.
We have found that circ-Foxo3 is expressed at significantly higher levels in non-tumor cell lines than in most cancer cell lines. 15 The expression levels of circ-Foxo3 in human breast cancer tumor specimen are significantly lower than in the adjacent benign tissues. Human and mouse heart tissues express high levels of circ-Foxo3 as they senesce, when compared to samples obtained from younger specimens. circ-Foxo3 expression levels increased in primary mouse cardiac fibroblasts, when the cells were maintained over confluence or under oxidative stress. circ-Foxo3 levels were also found to be increased during tumor cell apoptosis. 30 Most interestingly, circ-Foxo3 showed tissue specific biological functions in both in vitro and in vivo studies. circ-Foxo3 repressed cell cycle progression in non-cancer cell lines but enhanced breast cancer cell sensitivity to chemotherapeutic reagents via inducing cellular apoptosis, both of which are consistent with the functions of its parental gene Foxo3. 15, 27 On the other hand, circ-Foxo3 promotes cardiac senescence by sequestering stress and senescence related proteins involved in the stress responses. Overexpression of circ-Foxo3 promotes cardiac senescence in vitro and in vivo, and silencing circ-Foxo3 repressed cardiac senescence, indicating that this circRNA may not always function consistently with its parental gene. 30
Although circ-Foxo3 might sponge a handful of miRNAs that could modulate its parent gene expression at the post-transcriptional level, 31 the significant functions of circ-Foxo3 in tissue and developmental stage may be mainly related to its specific binding proteins. Our studies showed that circ-Foxo3 presumably exerted significant functional diversity in these types of cell lines as it bound to different binding proteins. In the non-cancer cell line NIH3T3 cells, circ-Foxo3 mainly interacted with p21 and CDK2, facilitating the inhibition of CDK2 by p21, and repressing cell cycle progression at the G1 stage.15 In the breast cancer cell line MDA-MB-231, most circ-Foxo3 bound to Mdm2 and p53, promoting Mdm2-induced p53 ubiquitination and subsequent degradation, resulting in an overall decrease of p53 in this p53 mutant cell line. 30
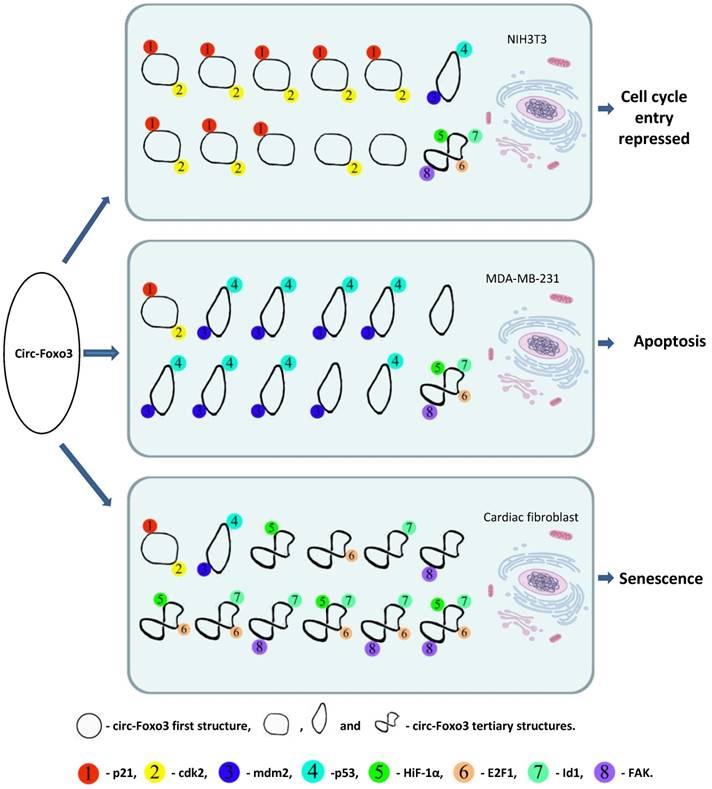
Usually RNAs interact with proteins through electrostatic interactions, hydrogen bonding, hydrophobic interactions and base stacking in a manner similar to DNA-protein interactions 35. However, RNA-protein interactions are significantly influenced by the tertiary structure of the RNA molecules 36. The tertiary structure of RNA molecules may also be modulated by their binding proteins 37. Our recent studies indicated that circRNAs are likely to form different tertiary models that prefer to bind specific proteins. Certain tertiary models may dynamically exist in certain tissues and cells resulting in different affinity with different binding proteins. Since the cellular environments, such as solvent and metal ions will have a strong influence on circRNA dynamic tertiary structure, the major circRNA tertiary structure may be different in various cell lines, tissues, and developmental stages. Thus, these circRNAs could display a variety of different functions in different tissues or development stages via binding different functional proteins. We applied in silico prediction and docking of tertiary structure of circ-Foxo3 with the above proteins 30, and found that the major tertiary structure models of circ-Foxo3 in tumor and non-tumor cells are different, which may be the main reason why this circRNA molecule showed functional diversity by binding different proteins in tumor and non-tumor cell lines (Figure 1). In our recent studies, we found that another circular RNA, circ-Amotl1, also showed significantly functional diversity with regard to its tissue specific binding proteins 38. Some proteins have been reported to bind a set of different circRNAs,33 whereas some circRNAs can also dynamically bind different proteins. 27, 30, 15 Both of these studies support a dynamic circRNA-protein interaction in various tissues, developmental stage, and cellular conditions (Figure 1).
RNA-protein interactions influencing dynamic expression of circRNAs and proteins
By binding to proteins, circRNAs may modulate various aspects of cell physiology. The rate of translation and stability of RNAs can also be influenced by protein-RNA interactions. circRNA-binding proteins play critical roles in regulating circRNA synthesis and degradation 19. It has been demonstrated that RNA circulation is facilitated by complementary sequences and is regulated by specific RNA-binding proteins.39 Thus far, RNA-binding proteins, such as Quaking (QKI) and muscleblind (MBL) have been reported to play a role in circRNA biogenesis of some circular RNAs.19,25 The splicing factors QKI and MBL are presumed to induce back-splicing, which can increase circRNA formation from genes containing protein binding motifs in flanking intronic regions. 9, 19, 25 RNA-protein interaction can also promote circRNA formation by stabilizing complementary sequences or by inhibiting canonical splicing 34. Most circRNAs are highly stable compared with their linear counterparts, at least partly because they are resistant to linear RNA decay mechanisms 40. With low basal transcription rates, the high biological stability of circRNA makes these molecules reach high levels in certain tissues 41. The dynamic expression of circRNA allows these molecules to function with diverse spatial and temporal biological profiles 41.
A dynamic circRNA-protein interaction model for circ-Foxo3 in different tissues. Our studies suggest that circFoxo3 displays a variety of tertiary structures in various cell/tissue environments. The major tertiary structure in mouse fibroblast NIH3T3 can bind p21 and CDK1, which represses cell cycle entry. The major tertiary structure of circ-Foxo3 in a breast cancer cell line allows this circular RNA to bind Mdm2 and p53, leading to induction of tumor cell apoptosis. The major tertiary structure of this circRNA could bind to stress related proteins in cardiomyocytes and cardiac fibroblasts, which promote cardiac senescence.
It has been reported that QKI regulates human circRNA biogenesis, and that circRNA abundance decreased upon QKI knockdown.19 Binding of QKI to motifs present in both of the intronic regions close to the circularized exons could facilitate RNA looping and back-splicing through protein-protein dimerization. Mutation of all QKI binding motifs repressed circRNA expression. In addition, insertion of QKI binding motifs in both the upstream and downstream introns flanking the central exon in an expression minigene was enough to produce circRNA from genes that did not otherwise give rise to circRNA. 19 In addition to QKI, the MBL protein has been shown to enhance circularization, giving rise to circMbl.25 Modulation of MBL levels significantly affects circMbl biosynthesis, and this effect is dependent on MBL binding. The introns flanking circMbl have MBL binding sites, which decide the circularization rates of bracketed exons dependent on MBL levels. Expression of MBL can repress the production of its own mRNA by promoting circMbl synthesis. 25
Thus, RBPs may serve as activators or inhibitors of the formation of circRNAs, and regulate circRNA expression levels. Given the high degree of stability of circRNAs, and their presumed resistance to exonucleolytic degradation, it is likely that cellular mechanisms exist for the control of circRNA levels. Extracellular vesicles such as exosomes, recovered from cell culture media have been found to contain high levels of certain circRNAs. 6 Of these circRNAs, most became enriched over their linear counterparts within the exosome. 6 Expulsion of circRNAs into the extracellular space by exosomal release can therefore be a mechanism by which cells clear and degrade circRNAs.5 Also, degradation pathways for at least some circRNAs may be mediated through endonuclease cleavage. Studies have shown that some circRNAs are densely bound by the miRNA effector protein Ago2, and the ciR-7/CDR1as is subject to the slicer activity of Ago2 in an miR-671 dependent manner.42
Our recent study showed that in the G1 restriction phase, NIH3T3 cells express higher levels of circ-Foxo3 relative to the S and G2 phases.15 Endogenous circ-Foxo3 expression decreased when the cells were treated with epidermal growth factor (EGF), and increased when cells were treated with the EGF inhibitor AG1478. Thus there was a correlation between circ-Foxo3 expression and cell cycle status. We further found that circ-Foxo3 could bind to both CDK2 and p21 protecting circ-Foxo3 from digestion by RNAse A. To corroborate our findings, we separately repressed both CDK2 and p21 using siRNA, and found reduced levels of endogenous circ-Foxo3 RNA. Conversely, circ-Foxo3 was found to be increased in the exosomal fractions of culture media, indicating that the binding of CDK2 and p21 to circ-Foxo3 may be critical to protecting circ-Foxo3 from degradation, by avoiding exosomal release into the extracellular space.
RNA-protein interactions have been reported to influence protein expression, function, and biogenesis. Some intronic circular RNAs and circRNAs with retained introns were also reported to function as positive regulators of RNA Pol II transcription in the nucleus. 1,43 In particular, EIciRNAs could promote their parental gene's transcription via interaction with host U1 snRNP and RNA Polymerase II. 1 The process of circRNA biogenesis may also regulate the formation of linear mRNAs through the competition between splicing and back-splicing. 44 Our recent studies showed that circ-Foxo3 could bind to the E3 ubiquitin-protein ligase Mdm2, and modulate its function in polyubiquitinating and degrading targeting proteins. In this manner, Foxo3, a protein target of Mdm2 with low binding affinity for circ-Foxo3, showed increased expression in circ-Foxo3-expressing breast cancer cells. Conversely, expression of p53, which had a high binding affinity to circ-Foxo3, was reduced. 30
CircRNAs for targeting therapeutics
In recent years, nanoparticles have emerged as a promising platform for theranostic application 45, 46. This approach is used to deliver nucleic acids such as small interfering RNA (siRNA), small hairpin RNA (shRNA), antisense oligonucleotides (AON), and plasmid DNA (pDNA), and have shown promise in pre-clinical treatment 47-50. Nanoparticles are linked to bioactive molecules, which may be able to direct the nanoparticles to specific sites within cells or the body. Nanoparticles can pass through cellular membranes but they may not be able to enter the nucleus, endoplasmic reticulum, or Golgi complex, although the mechanism is not clear 51. As such, nanoparticles can only be used to deliver molecules that function in the cytoplasm. There are many different types of nanoparticles and they function differently. Iron oxide nanoparticle (IONP)-linked immunosorbent assay (ILISA) can be used for disease screening and diagnosis 52. Systemic delivery of siRNA by nanoparticles can be used to deliver siRNAs for treating cancer and viral infections 53. mPEG conjugated gold nanoparticles were initially applied to deliver miRNAs or antisense in animal studies 54. This nucleic acid delivery technique was extended to deliver specific circular RNAs and siRNAs against circular RNAs in vivo in breast cancer, skin wound healing, and heart failure animal models 38-40.
The recent emerging circular RNAs may represent a great potential for theranostics. Circular RNAs play crucial roles in cell homeostasis and are involved in many disease phenotypes when there are deregulated. Due to their high stability, tissue and developmental stage related expression, as well as the potential binding of proteins they interact with, they are potentially promising targets for therapeutic approaches. Our recent studies suggest that conjugation of circular RNA expression plasmids with nanoparticles is a useful approach for delivery of circular RNAs. Though the delivery efficiency is not as high as application in siRNAs or AONs against circular RNAs, we have shown that the circular RNA circ-Foxo3 can be delivered in vivo to inhibit tumor progression by conjugating the expression plasmids of circ-Foxo3 with gold nanoparticles 30. This approach was also used to improve the healing process of skin wounds by delivering the expression plasmids of a circular RNA circ-Amotl1 conjugated with gold nanoparticles 39. Since nanoparticles are not able to enter the nucleus, the therapeutic approach can only focus on exonic circular RNAs, since this class of circular RNAs are mainly detected in the cytoplasm. With the advantage of high delivery efficiency, delivery of siRNAs or AONs against circular RNAs will be a valuable approach for future circular RNA studies. As the field continues to grow and resolve delivery challenges, new techniques may be developed to deliver circular RNAs to other organelles within the cell. It is expected that circular RNA gene therapy may become a new class of drugs for a wide range of different diseases owing to the development of non-toxic nanoparticles with specific tissue targeting.
Concluding remarks
Research interest in the physiologic and pathologic roles of circRNAs is growing. Although some individual circRNAs have been characterized, the functions of circRNAs remain largely unknown. The ability of circRNAs to interact with proteins, thereby binding, sequestering or translocating proteins to particular subcellular fractions, allows circRNAs to act as dynamic scaffolds that modulate protein-protein interaction. This provides a further layer of complexity to the biogenesis and degradation of both proteins and circRNAs, and may have further relevance in cell physiology. Advances in chemical and biological research techniques will improve current techniques to identify dynamic RNA-protein interactions, and further elucidate the molecular mechanisms associated with circular RNAs.
Abbreviations
FOXO3: forkhead box O3; CDK2: Cyclin-dependent kinase 2; circRNAs: exonic circular RNAs; ciRNAs: intronic circular RNAs; EIciRNAs: a circular RNA contains exon and intron; HITS-CLIP: High-throughput sequencing of RNA isolated by crosslinking immunoprecipitation; miRNA: microRNA; siRNA: small interfering RNA; shRNA: small hairpin RNA; AON: antisense oligonucleotides; pDNA: plasmid; RBP: RNA binding protein; AGO: Argonaute; MBL: muscleblind; QKI: Quaking; EGF: epidermal growth factor; IGF2BP3 or IMP3: Insulin-like growth factor 2 binding protein 3; RIP: RNA immunoprecipitation; RPA: RNase protection assay; FISH/ISH: Fluorescence in situ Hybridization.
Acknowledgements
This work was supported by grants from Canadian Institutes of Health Research (PJT-149083 and PJT-153105) and the High-level Leading Talent Introduction Program of GDAS (No.2016GDASRC-0102).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G. Exon-intron circular rnas regulate transcription in the nucleus. Nature structural & molecular biology. 2015;22:256-264
2. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134-147
3. Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell reports. 2015;10:103-111
4. Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular rna abundance with proliferation-exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Scientific reports. 2015;5:8057
5. Lasda E, Parker R. Circular rnas co-precipitate with extracellular vesicles: A possible mechanism for circrna clearance. PLoS One. 2016;11:e0148407
6. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular rna is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell research. 2015;25:981-984
7. Caiment F, Gaj S, Claessen S, Kleinjans J. High-throughput data integration of rna-mirna-circrna reveals novel insights into mechanisms of benzo[a]pyrene-induced carcinogenicity. Nucleic acids research. 2015;43:2525-2534
8. Lukiw WJ. Circular rna (circrna) in alzheimer's disease (ad). Frontiers in genetics. 2013;4:307
9. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural rna circles function as efficient microrna sponges. Nature. 2013;495:384-388
10. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular rnas are a large class of animal rnas with regulatory potency. Nature. 2013;495:333-338
11. Jeck WR, Sharpless NE. Detecting and characterizing circular rnas. Nature biotechnology. 2014;32:453-461
12. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular rnas. Genome Biol. 2014;15:409
13. Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, Li N, Zhou W, Yu Y, Cao X. Circular rna mto1 acts as the sponge of mir-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017
14. Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J, Ao Y. Circular rna related to the chondrocyte ecm regulates mmp13 expression by functioning as a mir-136 'sponge' in human cartilage degradation. Scientific reports. 2016;6:22572
15. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular rna retards cell cycle progression via forming ternary complexes with p21 and cdk2. Nucleic acids research. 2016;44:2846-2858
16. Li J, Yang J, Zhou P, Le Y, Zhou C, Wang S, Xu D, Lin HK, Gong Z. Circular rnas in cancer: Novel insights into origins, properties, functions and implications. American journal of cancer research. 2015;5:472-480
17. You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, Wang X, Hou J, Liu H, Sun W, Sambandan S, Chen T, Schuman EM, Chen W. Neural circular rnas are derived from synaptic genes and regulated by development and plasticity. Nature neuroscience. 2015;18:603-610
18. Gao W, Wang Z, Lv L, Yin D, Chen D, Han Z, Ma Y, Zhang M, Yang M, Gu Y. Photodynamic therapy induced enhancement of tumor vasculature permeability using an upconversion nanoconstruct for improved intratumoral nanoparticle delivery in deep tissues. Theranostics. 2016;6:1131-1144
19. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The rna binding protein quaking regulates formation of circrnas. Cell. 2015;160:1125-1134
20. Hentze MW, Preiss T. Circular rnas: Splicing's enigma variations. EMBO J. 2013;32:923-925
21. Xing L, Shi Q, Zheng K, Shen M, Ma J, Li F, Liu Y, Lin L, Tu W, Duan Y, Du L. Ultrasound-mediated microbubble destruction (ummd) facilitates the delivery of ca19-9 targeted and paclitaxel loaded mpeg-plga-pll nanoparticles in pancreatic cancer. Theranostics. 2016;6:1573-1587
22. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, Wong CC, Xiao X, Wang Z. Extensive translation of circular rnas driven by n6-methyladenosine. Cell research. 2017;27:626-641
23. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, Shenzis S, Samson M, Dittmar G, Landthaler M, Chekulaeva M, Rajewsky N, Kadener S. Translation of circrnas. Molecular cell. 2017;66:9-21 e27
24. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I. Circ-znf609 is a circular rna that can be translated and functions in myogenesis. Molecular cell. 2017;66:22-37 e29
25. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. Circrna biogenesis competes with pre-mrna splicing. Molecular cell. 2014;56:55-66
26. Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL, Gorospe M. Identification of hur target circular rnas uncovers suppression of pabpn1 translation by circpabpn1. RNA biology. 2017;14:361-369
27. Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, Li X, Yang BB. Foxo3 circular rna promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. European heart journal. 2017;38:1402-1412
28. Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, Gabel G, Beutner F, Scholz M, Thiery J, Musunuru K, Krohn K, Mann M, Teupser D. Circular non-coding rna anril modulates ribosomal rna maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429
29. Holdt LM, Sass K, Gabel G, Bergert H, Thiery J, Teupser D. Expression of chr9p21 genes cdkn2b (p15(ink4b)), cdkn2a (p16(ink4a), p14(arf)) and mtap in human atherosclerotic plaque. Atherosclerosis. 2011;214:264-270
30. Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, Yang BB. Induction of tumor apoptosis through a circular rna enhancing foxo3 activity. Cell death and differentiation. 2017;24:357-370
31. Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular rna and foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919-3931
32. McHugh CA, Russell P, Guttman M. Methods for comprehensive experimental identification of rna-protein interactions. Genome biology. 2014;15:203
33. Schneider T, Hung LH, Schreiner S, Starke S, Eckhof H, Rossbach O, Reich S, Medenbach J, Bindereif A. Circrna-protein complexes: Imp3 protein component defines subfamily of circrnps. Scientific reports. 2016;6:31313
34. Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular rnas. Wiley interdisciplinary reviews. RNA. 2015;6:563-579
35. Koh YY, Wang Y, Qiu C, Opperman L, Gross L, Tanaka Hall TM, Wickens M. Stacking interactions in puf-rna complexes. Rna. 2011;17:718-727
36. Lai D, Proctor JR, Meyer IM. On the importance of cotranscriptional rna structure formation. Rna. 2013;19:1461-1473
37. Sengupta A, Sung HL, Nesbitt DJ. Amino acid specific effects on rna tertiary interactions: Single-molecule kinetic and thermodynamic studies. The journal of physical chemistry. B. 2016
38. Yang ZG, Awan FM, Du WW, Zeng Y, Lyu J, Wu, Gupta S, Yang W, Yang BB. The circular rna interacts with stat3, increasing its nuclear translocation and wound repair by modulating dnmt3a and mir-17 function. Molecular therapy: the journal of the American Society of Gene Therapy. 2017
39. Chen LL. The biogenesis and emerging roles of circular rnas. Nature reviews. Molecular cell biology. 2016;17:205-211
40. Dang Y, Yan L, Hu B, Fan X, Ren Y, Li R, Lian Y, Yan J, Li Q, Zhang Y, Li M, Ren X, Huang J, Wu Y, Liu P, Wen L, Zhang C, Huang Y, Tang F, Qiao J. Tracing the expression of circular rnas in human pre-implantation embryos. Genome biology. 2016;17:130
41. Gruner H, Cortes-Lopez M, Cooper DA, Bauer M, Miura P. Circrna accumulation in the aging mouse brain. Scientific reports. 2016;6:38907
42. Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. Mirna-dependent gene silencing involving ago2-mediated cleavage of a circular antisense rna. The EMBO journal. 2011;30:4414-4422
43. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding rnas. Molecular cell. 2013;51:792-806
44. Lasda E, Parker R. Circular rnas: Diversity of form and function. RNA. 2014;20:1829-1842
45. Satterlee AB, Huang L. Current and future theranostic applications of the lipid-calcium-phosphate nanoparticle platform. Theranostics. 2016;6:918-929
46. Ho YJ, Chang YC, Yeh CK. Improving nanoparticle penetration in tumors by vascular disruption with acoustic droplet vaporization. Theranostics. 2016;6:392-403
47. Xiao B, Zhang Z, Viennois E, Kang Y, Zhang M, Han MK, Chen J, Merlin D. Combination therapy for ulcerative colitis: Orally targeted nanoparticles prevent mucosal damage and relieve inflammation. Theranostics. 2016;6:2250-2266
48. Zhang P, Hu C, Ran W, Meng J, Yin Q, Li Y. Recent progress in light-triggered nanotheranostics for cancer treatment. Theranostics. 2016;6:948-968
49. Jo SD, Ku SH, Won YY, Kim SH, Kwon IC. Targeted nanotheranostics for future personalized medicine: Recent progress in cancer therapy. Theranostics. 2016;6:1362-1377
50. Wang Y, Rajala A, Cao B, Ranjo-Bishop M, Agbaga MP, Mao C, Rajala RV. Cell-specific promoters enable lipid-based nanoparticles to deliver genes to specific cells of the retina in vivo. Theranostics. 2016;6:1514-1527
51. Greulich C, Diendorf J, Simon T, Eggeler G, Epple M, Koller M. Uptake and intracellular distribution of silver nanoparticles in human mesenchymal stem cells. Acta Biomater. 2011;7:347-354
52. Zhang L, Tong S, Zhou J, Bao G. Accurate quantification of disease markers in human serum using iron oxide nanoparticle-linked immunosorbent assay. Theranostics. 2016;6:1353-1361
53. Draz MS, Fang BA, Zhang P, Hu Z, Gu S, Weng KC, Gray JW, Chen FF. Nanoparticle-mediated systemic delivery of sirna for treatment of cancers and viral infections. Theranostics. 2014;4:872-892
54. Tan L, Peng J, Zhao Q, Zhang L, Tang X, Chen L, Lei M, Qian Z. A novel mpeg-pdlla-pll copolymer for docetaxel delivery in breast cancer therapy. Theranostics. 2017;7:2652-2672
Author contact
![]() Corresponding author: BB Yang, S110, S-Wing Research Building, Sunnybrook Health Sciences Centre, 2075 Bayview Ave, Toronto M4N 3M5 Canada, tel: (416) 480-5874 e-mail: byangutoronto.ca
Corresponding author: BB Yang, S110, S-Wing Research Building, Sunnybrook Health Sciences Centre, 2075 Bayview Ave, Toronto M4N 3M5 Canada, tel: (416) 480-5874 e-mail: byangutoronto.ca
 Global reach, higher impact
Global reach, higher impact