13.3
Impact Factor
Theranostics 2019; 9(2):540-553. doi:10.7150/thno.28374 This issue Cite
Research Paper
Comprehensive characterization of the mutational landscape in multiple myeloma cell lines reveals potential drivers and pathways associated with tumor progression and drug resistance
1. IGH, CNRS, Univ Montpellier, France
2. CHU Montpellier, Department of Biological Hematology, Montpellier, France
3. Univ Montpellier, UFR de Médecine, Montpellier, France
4. CHU Montpellier, Department of Clinical Hematology, Montpellier, France.
5. Univ Montpellier, UMR CNRS 5235, Montpellier, France.
6. Englander Institute for Precision Medicine, Institute for Computational Biomedicine, Weill Cornell Medical College, 1305 York Avenue, New York, NY 10021, USA;
*These authors contributed equally to this work and share last authorship.
Abstract
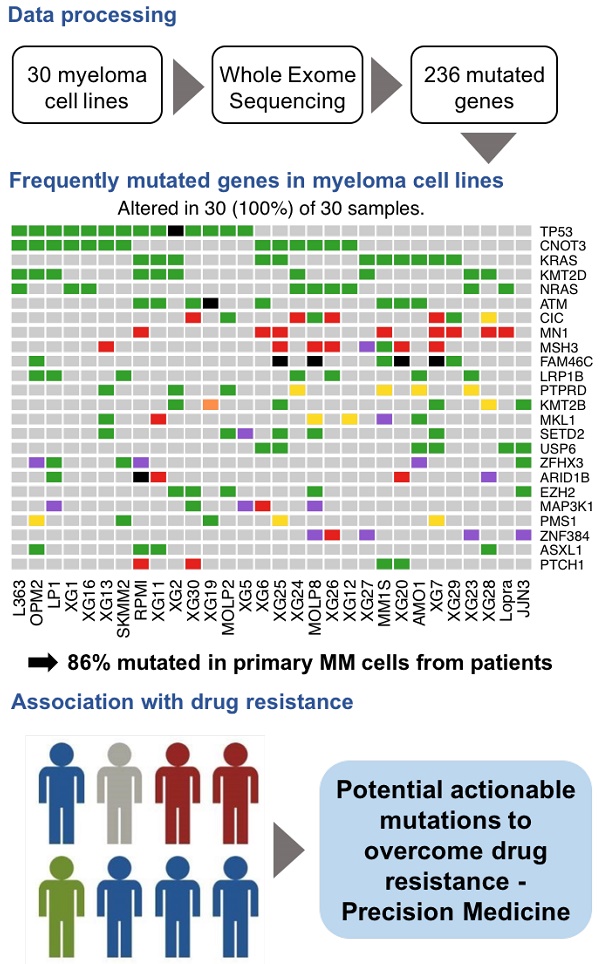
Human multiple myeloma tumor cell lines (HMCLs) have been a cornerstone of research in multiple myeloma (MM) and have helped to shape our understanding of molecular processes that drive tumor progression. A comprehensive characterization of genomic mutations in HMCLs will provide a basis for choosing relevant cell line models to study a particular aspect of myeloma biology, or to screen for an antagonist of certain cancer pathways.
Methods: We performed whole exome sequencing on a large cohort of 30 HMCLs, representative of a large molecular heterogeneity of MM, and 8 control samples (epstein-barr virus (EBV)-immortalized B-cells obtained from 8 different patients). We evaluated the sensitivity of HMCLs to ten drugs.
Results: We identified a high confidence list of 236 protein-coding genes with mutations affecting the structure of the encoded protein. Among the most frequently mutated genes, there were known MM drivers, such as TP53, KRAS, NRAS, ATM and FAM46C, as well as novel mutated genes, including CNOT3, KMT2D, MSH3 and PMS1. We next generated a comprehensive map of altered key pathways in HMCLs. These include cell growth pathways (MAPK, JAK-STAT, PI(3)K-AKT and TP53 / cell cycle pathway), DNA repair pathway and chromatin modifiers. Importantly, our analysis highlighted a significant association between the mutation of several genes and the response to conventional drugs used in MM as well as targeted inhibitors.
Conclusion: Taken together, this first comprehensive exome-wide analysis of the mutational landscape in HMCLs provides unique resources for further studies and identifies novel genes potentially associated with MM pathophysiology, some of which may be targets for future therapeutic intervention.
Keywords: human multiple myeloma cell lines, exome sequencing, mutational landscape
 Global reach, higher impact
Global reach, higher impact