13.3
Impact Factor
Theranostics 2021; 11(9):4549-4566. doi:10.7150/thno.54967 This issue Cite
Review
The crosstalk between m6A RNA methylation and other epigenetic regulators: a novel perspective in epigenetic remodeling
1. Department of Pathology, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou, 310000, China.
2. Division of Hepatobiliary and Pancreatic Surgery, Department of Surgery, the First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, 310000, China.
Received 2020-10-25; Accepted 2021-2-7; Published 2021-3-4
Abstract
Epigenetic regulation involves a range of sophisticated processes which contribute to heritable alterations in gene expression without altering DNA sequence. Regulatory events predominantly include DNA methylation, chromatin remodeling, histone modifications, non-coding RNAs (ncRNAs), and RNA modification. As the most prevalent RNA modification in eukaryotic cells, N6-methyladenosine (m6A) RNA methylation actively participates in the modulation of RNA metabolism. Notably, accumulating evidence has revealed complicated interrelations occurring between m6A and other well-known epigenetic modifications. Their crosstalk conspicuously triggers epigenetic remodeling, further yielding profound impacts on a variety of physiological and pathological processes, especially tumorigenesis. Herein, we provide an up-to-date review of this emerging hot area of biological research, summarizing the interplay between m6A RNA methylation and other epigenetic regulators, and highlighting their underlying functions in epigenetic reprogramming.
Keywords: N6-methyladenosine (m6A), DNA methylation, chromatin remodeling, histone modification, non-coding RNA (ncRNA), RNA modification
Introduction
Epigenetics, which represents the modulation of heritable phenotypes without any alterations in DNA sequences, has become a significant regulatory mechanism of diverse physiological or pathological processes. The scope of epigenetics is extensive, typically including DNA methylation, chromatin remodeling, histone modification, non-coding RNAs (ncRNAs) and RNA modification [1]. The first three members are superstars in epigenetics, and have been studied extensively so far. ncRNAs, mainly comprised of microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) [2], have provoked accumulating interests nowadays. In addition, there are more than 100 categories of RNA chemical modifications, and the common types include N6-methyladenosine (m6A), pseudouridine (ψ), 2'-O-methylation (Nm), m1A, 5-methylcytosine (m5C), adenosine-to-inosine (A-to-I), and N6, 2'-O-dimethyladenosine (m6Am) [3-7]. Notably, m6A RNA methylation is the most abundant internal mRNA modification in mammals [8]. With the rapid development of detection methodologies and high-throughput sequencing, the genome-wide features of m6A are being uncovered, which have increasingly attracted the attention of bioscience researchers.
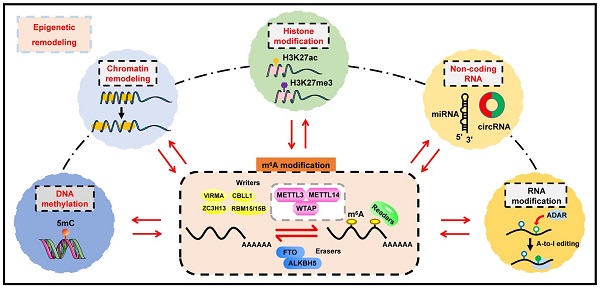
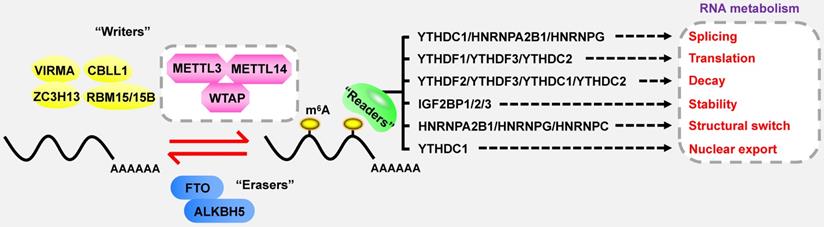
In the case of total RNA, m6A methylation occurs in approximately 0.1-0.4% of adenosines [9], predominantly located at 3' untranslated regions (3'UTRs), near stop codons and within the long internal exon [10, 11]. DRACH sequences are verified as the consensus motif of m6A (D = G/A/U; R = G/A; H = U/A/C) [12]. Strikingly, m6A modification is a reversible and dynamic process, which is deposited by methyltransferases (also called “writers”), and removed by demethylases (also called “erasers”) (Figure 1) [13, 14]. Subsequently, m6A-binding proteins (also called “readers” or “effectors”) recognize and bind to the m6A marks of targeted RNAs to influence their RNA metabolism, including stability, translation, alternative splicing and transport [15-18]. Furthermore, m6A plays a key role in far-ranging biological processes, such as cell differentiation, tissue development, environmental stress response, spermatogenesis, immune homeostasis and tumorigenesis [13].
Remarkably, it is commonly acknowledged that epigenetic regulations are intricate due to the interactions among epigenetic modifiers [19, 20]. As a research frontier, m6A is just like a storm center to frequently interact with its peripheral partners, the other epigenetic modulators. These partners can be modified and regulated by m6A modification, while m6A methylation may also be efficiently controlled by these regulators [20-22]. The coordinated relationships between m6A machinery and any other epigenetic counterparts elicit the epigenetic remodeling, which accounts for the perplexing modulations of various bioprocesses. Herein, we summarize the up-to-date findings about the interplay of m6A RNA methylation and other epigenetic modifications (Tables 1-3), and demonstrate how these associations impact biological functions, particularly in oncogenesis and tumor progression, highlighting the potential of m6A as a therapeutic target in the clinical practice.
The genealogy of m6A modification
m6A writers
The installation of m6A methylation is manipulated by the methyltransferase complex (MTC), which largely comprises of methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14), and Wilms tumor 1-associated protein (WTAP) [23]. METTL3 functions as a key catalytic element to facilitate the formation of m6A, while METTL14 acts as an RNA-binding scaffold to promote the enzymatic activity of METTL3 [24, 25]. WTAP is responsible for the stabilization of the METTL3-METTL14 heterodimer and ensuring their accurate localization to nuclear speckles [26]. Moreover, there are other co-factors involved in the conformation of MTC, including vir-like m6A methyltransferase associated (VIRMA, also known as KIAA1429) [27], Cbl proto-oncogene like 1 (CBLL1, also known as HAKAI), RNA-binding motif protein 15 (RBM15) with its paralogue RBM15B [28], and zinc finger CCCH domain-containing protein 13 (ZC3H13) [29, 30]. Notably, METTL16 is another m6A methyltransferase, which dominates cellular SAM levels and mediates m6A modification of U6 small nuclear RNAs (snRNAs), pre-mRNAs or certain types of lncRNAs [31, 32]. Additionally, METTL5 and ZCCHC4 have been identified as m6A methyltransferases for 18S rRNA and 28S rRNA, respectively [33, 34].
The complicated interactions between m6A and other epigenetic modifications
| Categories of epigenetics | Related components | m6A regulators | Mechanisms | References |
|---|---|---|---|---|
| DNA methylation | SlDML2 | SlALKBH2 | SlDML2-induced DNA methylation regulates the m6A demethylase SlALKBH2, while SlALKBH2-guided m6A demethylation strengthens the stability of 5mC demethylase SlDML2 in turn. | [62] |
| DNMT1, DNMT3a | METTL3 | The binding of DNMT1 and DNMT3a to METTL3 promoter is reduced by cigarette smoke condensate (CSC), leading to the hypomethylation of METTL3 and facilitating its expression. | [64] | |
| / | ALKBH5 | The CpG island of ALKBH5 is hypomethylated by CSC, which increases ALKBH5 expression. | [65] | |
| Chromatin remodeling | BAF155 | RBM15 | RBM15 accelerates the decay of chromatin remodeling factor BAF155 via the m6A methylation machinery. | [67] |
| carRNAs | METTL3 YTHDC1 | METTL3 promotes m6A methylation of chromosome-associated regulatory RNAs (carRNAs), while YTHDC1 mediates their degradation. | [22] | |
| Histone modification | H3K27ac, H3K27me3, CBP, p300 | METTL14 | METTL14 not only alters H3K27me3 modification, but also regulates H3K27ac modification by destabilizing CBP and p300 mRNAs. | [21] |
| H3K27me3, Ezh2 | METTL3 | METTL3 deposits m6A modification on histone methyltransferase Ezh2, which increases the level of H3K27me3. | [70] | |
| H3K4me3 | METTL3, METTL14, WTAP | The m6A modification catalyzed by METTL3/METTL14/WTAP complex substantially strengthens H3K4me3 modification. | [71] | |
| JMJD6 | hnRNPA2B1 | Arginine demethylase JMJD6 activates hnRNPA2B1 through facilitating its demethylation at Arg226. | [72] | |
| H3K27ac | METTL3 | H3K27ac modification on the promoter of METTL3 triggers its transcription. | [73] | |
| H3K4me3, KDM5C | METTL14 | KDM5C-mediated demethylation of H3K4me3 suppresses METTL14 transcription. | [74] | |
| H3K36me3 | METTL14 | H3K36me3 mark recognized by METTL14 promotes the binding of m6A methyltransferase complex to adjacent RNA polymerase II, depositing m6A co-transcriptionally. | [75] | |
| H3K9me2, KDM3B | YTHDC1 | YTHDC1 induces the H3K9me2 demethylation via recruiting KDM3B to the m6A-marked chromatin regions. | [76] | |
| RNA modification | m1A | FTO | FTO mediates demethylation of m1A in tRNA. | [37] |
| YTHDF1-3, YTHDC1 | m6A-binding proteins YTHDF1-3 and YTHDC1 are capable of directly binding to the m1A sites. | [80] | ||
| YTHDF2 | YTHDF2 recognizes m1A-modified transcripts and mediates their decay. | [81] | ||
| m5C | YTHDF2 | m6A reader YTHDF2 and m5C writer NSUN2 cooperatively facilitate murine leukemia virus (MLV) replication. | [88] | |
| METTL3, METTL14 | METTL3/METTL14-mediated m6A methylation and NSUN2-mediated m5C methylation collaborate with each other to strengthen the expression of p21 mRNA. | [89] | ||
| YTHDF2 | YTHDF2 binds to m5C in rRNA with the Trp432 residue, remarkably decreasing the m5C level. | [90] | ||
| A-to-I | / | Loss of m6A modification contributes to the elevated level of A-to-I editing via the favorable association of ADAR with m6A-depleted transcripts. | [4] | |
| Pseudogene | METTL3 | METTL3 elevates the expression of lncRNA pseudogene Olfr29-ps1 and facilitates its sponge to miR-214-3p. | [94] | |
| / | m6A modification and pseudouridine (Ψ) collaboratively weaken the binding of RBP hPUM2 to its targeted RNAs. | [95] | ||
| m6Am | METTL3, WTAP, ALKBH5 | The m6Am signal can be detected in METTL3, WTAP and ALKBH5, while the m6A signal is found in m6Am writer PCIF1. | [105] | |
| FTO | FTO is responsible for the demethylation of both m6A and m6Am modifications. | [25, 37] |
m6A erasers
Fat mass and obesity-associated (FTO) and alkB homolog 5 (ALKBH5) are the only two known m6A demethylases to date. Although both demethylases belong to the AlkB family of dioxygenases, they eliminate m6A through different mechanisms. As the first identified m6A demethylase, FTO induces demethylation activity depending on the oxidative function, which requires iron (II) and α-KG [35]. Specifically, FTO initially oxidizes m6A to form intermediate products, including N6-hydroxymethyladenosine (hm6A) and N6-formyladenosine (f6A), and subsequently hydrolyzes the products into adenosine, which is a sequential and multi-step procedure. However, the catalytic process mediated by ALKBH5 is a one-step reaction process, in which ALKBH5 directly abrogates m6A in an oxidative-dependent manner [36]. Furthermore, a discrepancy has been observed in the recognition of substrates between ALKBH5 and FTO. ALKBH5 acts as an m6A-specific demethylase, while FTO can demethylate a variety of RNA modifications, such as m6A, m6Am and m1A [37].
The specific molecular mechanisms and biological functions of m6A modification on ncRNAs
| Categories | m6A-related enzymes | Non-coding RNAs | Mechanisms | Biological functions | References |
|---|---|---|---|---|---|
| m6A-miRNA | METTL3 | miR-221/222 | Promoting miR-221/222 maturation. | Accelerating cell proliferation of bladder cancer. | [110] |
| METTL3 | miR-1246 | Facilitating miR-1246 maturation. | Promoting the metastasis of colorectal cancer. | [111] | |
| METTL3 | miR-873-5p | Strengthening miR-873-5p maturation. | Blocking oxidative stress and apoptosis in colistin-evoked nephrotoxicity. | [112] | |
| METTL3 | miR-143-3p | Enhancing miR-143-3p maturation. | Facilitating angiogenesis and brain metastasis of lung cancer. | [113] | |
| METTL3 | miR-320 | Increasing the m6A level of pre-miR-320. | Driving osteogenic differentiation of bone marrow-derived mesenchymal stem cells. | [114] | |
| METTL3 | miR-7212-5p | Mediating miR-7212‐5p maturation. | Inhibiting osteoblast differentiation and fracture healing. | [115] | |
| METTL3, NKAP | miR-25-3p | Accelerating miR-25-3p maturation. | Promoting the progression of pancreatic cancer. | [66] | |
| METTL14 | miR-126 | Inducing miR-126 maturation. | Suppressing hepatocellular carcinoma metastasis. | [117] | |
| m6A-lncRNA | METTL3, YTHDF3 | MALAT1 | Enhancing the stability of MALAT1. | Inducing drug resistance and metastasis of non-small cell lung cancer. | [128] |
| WTAP | XIST | Co-localizing with XIST. | Participating in XIST-mediated silencing. | [130] | |
| METTL3, WTAP, RBM15/15B, YTHDC1 | XIST | Promoting XIST-mediated transcriptional repression. | / | [28] | |
| METTL14, YTHDF2 | XIST | Abolishing the stability of XIST. | Suppressing proliferation and metastasis of colorectal cancer. | [133] | |
| METTL3 | LINC00958 | Promoting the stability of LINC00958. | Increasing the lipogenesis of hepatocellular carcinoma. | [135] | |
| METTL3, METTL14 | LNCAROD | Up-regulating the expression of LNCAROD. | Facilitating the progression of head and neck squamous cell carcinoma. | [136] | |
| VIRMA | CCA1/2 | Increasing the m6A level of CCA1/2. | Inducing aggressive phenotype of prostate cancer. | [137] | |
| IGF2BP2 | DANCR | Strengthening the stability of DANCR. | Enhancing stemness-like properties of pancreatic cancer. | [52] | |
| ALKBH5, YTHDF2 | PVT1 | Elevating PVT1 expression. | Promoting the tumorigenesis of osteosarcoma. | [138] | |
| METTL3 | RHPN1-AS1 | Enhancing RHPN1-AS1 expression. | Accelerating the proliferation and metastasis of epithelial ovarian cancer. | [139] | |
| METTL3, ALKBH5, hnRNPA2B1 | RP11 | Increasing the expression of RP11. | Triggering the metastasis of colorectal cancer. | [140] | |
| YTHDF3 | GAS5 | Promoting the decay of GAS5. | Inhibiting the tumorigenesis of colorectal cancer | [141] | |
| METTL3, METTL14, WTAP, ALKBH5, YTHDF1 | LINC00278 | Modulating the m6A modification of LINC00278 and then affecting YY1BM translation. | Regulating the progression of cigarette smoking-related esophageal squamous cell carcinoma. | [65] | |
| METTL3, YTHDC1 | pncRNA-D | Methylating pncRNA-D and inhibiting its binding to TLS. | Modulating cell cycle. | [142] | |
| METTL3 | linc1281 | Sustaining the interaction of linc1281 and pluripotency-related miRNAs. | Affecting the differentiation potential of embryonic stem cells. | [143] | |
| YTHDF2 | lnc-Dpf3 | Inducing the degradation of lnc-Dpf3. | Controlling the migration of dendritic cells. | [144] | |
| m6A-circRNA | YTHDC1 | circNSUN2 | Recognizing circNSUN2 to enhance its cytoplasmic transport. | Facilitating colorectal carcinoma liver metastasis. | [156] |
| METTL3, YTHDC1, YTHDF3 | circ-ZNF609 | Promoting circZNF609 translation. | / | [158] |
m6A readers
The m6A readers primarily consist of YT521-B homology (YTH) domain family proteins (YTHDF1/2/3), YTH domain containing proteins (YTHDC1/2) [15-17, 38, 39], insulin-like growth factor 2 mRNA-binding proteins (IGF2BP1/2/3) [40], and heterogeneous nuclear ribonucleoprotein (HNRNP) family (HNRNPA2B1, HNRNPC and HNRNPG) [41-43], which exert a great influence on the destiny of targeted RNAs.
The underlying molecular mechanisms and biological functions of ncRNAs on m6A modification
| Categories | Non-coding RNAs | m6A-related enzymes | Mechanisms | Biological functions | References |
|---|---|---|---|---|---|
| miRNA-m6A | miR-145 | YTHDF2 | Inhibiting the expression of YTHDF2. | / | [121] |
| miR-186 | METTL3 | Suppressing METTL3 expression. | Inhibiting the growth and metastasis of hepatoblastoma | [122] | |
| let-7g | METTL3 | Attenuating the expression of METTL3. | Accelerating the progression of breast cancer. | [123] | |
| lncRNA-m6A | LINRIS | IGF2BP2 | Maintaining the stability of IGF2BP2. | Promoting the aerobic glycolysis in colorectal cancer. | [145] |
| ARHGAP5-AS1 | METTL3 | Recruiting METTL3 to methylate and stabilize ARHGAP5. | Strengthening the chemoresistance of gastric cancer. | [146] | |
| GAS5-AS1 | ALKBH5 | Interacting with ALKBH5 to demethylate and stabilize GAS5. | Suppressing the growth and metastasis of cervical cancer. | [147] | |
| FOXM1-AS | ALKBH5 | Increasing the binding of ALKBH5 to FOXM1 pre-mRNA. | Facilitating the tumorigenicity of glioblastoma stem-like cells. | [116] | |
| GATA3-AS | KIAA1429 | Enhancing the interaction between KIAA1429 and GATA3 pre-mRNA. | Accelerating hepatocellular carcinoma progression. | [149] | |
| LINC00266-1 | IGF2BP1 | Promoting the recognition of IGF2BP1 upon m6A-modified RNAs like c-Myc. | Strengthening tumorigenesis of colorectal cancer. | [150] | |
| circRNA-m6A | circSTAG1 | ALKBH5 | Capturing ALKBH5 and reducing its intranuclear translocation. | Attenuating the depressive-like behaviors. | [163] |
As the first and most extensively investigated m6A reader, YTHDF2 can bind to the m6A residues in 3'UTR and facilitate RNA degradation [16, 44]. Unlike YTHDF2, YTHDF1 selectively recognizes m6A marks in 5'UTR and near the stop codon, and boots the translation efficiency of targeted genes via the interaction with eukaryotic initiation factor 3 (eIF3) [15]. Interestingly, YTHDF3 performs the dual functions of facilitating translation and inducing degradation of targeted transcripts [38]. However, a recent research carried out by Zaccara et al. challenges the conventional views and reveals that there is no evidence to demonstrate the direct role of YTHDF proteins in promoting RNA translation [45]. They also put forward an unified model of m6A function in which m6A modification predominantly affects mRNA degradation through the combined action of three redundant YTHDF proteins. These controversial viewpoints show the complex roles of YTHDFs in the m6A-based regulation, which require further discussion and verification. YTHDC1 is an m6A reader which not only regulates alternative splicing and nuclear export, but also accelerates mRNA degradation [17, 18, 22]. YTHDC2 can induce the translation elongation of m6A-modified mRNAs, but also reduce the stability of certain targeted mRNAs [39, 46, 47]. Furthermore, IGF2BPs are another cluster of readers whose K homology (KH) domains are required for m6A recognition. Generally, IGF2BPs can enhance the stability and translation of m6A-containing mRNAs [40, 48-52].
The binding of HNRNPA2B1 and m6A is mediated by a mechanism called “m6A switch”, in which alteration in the structure of targeted RNA caused by m6A methylation enhances the combination of m6A and HNRNPA2B1 [41, 43]. HNRNPA2B1 not only recognizes nuclear m6A-bearing transcripts to promote alternative splicing, but also strengthens primary miRNA processing [53]. HNRNPG is capable of regulating alternative splicing or gene expression [42]. Furthermore, HNRNPC may participate in the RNA processing of mRNAs or lncRNAs depending on m6A modification [41].
Strikingly, some reader-like effectors are also crucial for m6A regulation. For example, eIF3 can facilitate m6A-mediated translation [54]. METTL3 has the capacity to promote translation of several mRNAs independent of its methyltransferase activity and other m6A-binding proteins [55]. In addition, HuR is recognized to be involved in m6A-related events [56]. However, the regulatory modes of HuR are currently controversial.
The interplay between m6A and other epigenetic modifications
m6A and DNA methylation
DNA methylation is a well-known and crucial epigenetic modification [57]. Studies have revealed that DNA 5mC and 6mA methylations are the most common types of DNA modifications in eukaryotes and prokaryotes, respectively. Specifically, 5mC is generated by DNA methyltransferase 3A (DNMT3A) and DNMT3B [58], while demethylated either actively via ten-eleven translocation (TET) or passively by diluting DNA methylation labels during DNA replication [59, 60]. In addition, N6AMT1 and ALKBH1 have been characterized as methyltransferase and demethylase of 6mA modification, respectively [61].
Notably, an RNA methylome manifests the crosstalk between RNA and DNA methylation in fruit ripening [62]. The m6A demethylase SlALKBH2, which is responsible for the decreased m6A levels of fruit-ripening genes, is modulated by SlDML2-contained DNA methylation. In turn, the stability of 5mC demethylase SlDML2 is strengthened by SlALKBH2-guided m6A demethylation (Figure 2A). Most recently, a comprehensive interplay between 5mC and m6A regulators across 33 cancer species based on bioinformatics analyses has been reported [63]. The two types of methylations are functionally correlated with significant co-occurrences of genetic mutations. Some of the pivotal m6A/5mC genes are combined to establish an epigenetic module eigengene (EME). Interestingly, an elevated EME implies a strongly proliferative and aggressive cellular status, low inflammatory and immune infiltration, and enhanced enrichment of stromal signatures. Furthermore, EME level is useful to predict prognosis of cancer patients.
The dynamic and reversible processes of m6A modification. “Writers” deposit m6A methylation on RNAs, while “erasers” remove the m6A marks. Then “readers” are responsible for regulating the fate of targeted RNAs.
The complex interplay between m6A modification and other epigenetic regulators including DNA methylation, chromatin remodeling, histone modification and RNA modification. (A) The regulatory circuit of m6A modification and DNA methylation. (B) The loss of 5mC DNA methylation on METTL3 promotes its expression. (C) m6A methylation affects the chromatin state by regulating the expression of carRNAs. (D) m6A impacts histone modification through modulating the level of histone-associated enzymes. (E) JMJD6 mediates the demethylation of hnRNPA2B1, impelling its translocation to cytoplasm. (F) Histone acetylation facilitates METTL3 expression. (G) m6A methylation and m5C modification cooperatively promote the translation process. (H) Deficiency of m6A methylation leads to the enhanced level of A-to-I editing. (I) m6A methylation regulates the expression of pseudogene and impacts its sponge to miRNA.
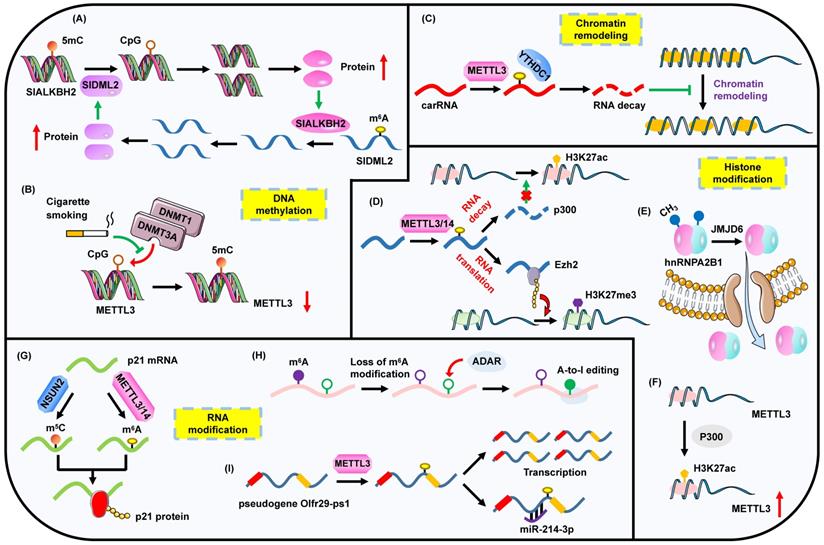
In pancreatic cancer, cigarette smoke condensate (CSC) is able to induce the hypomethylation of METTL3 through attenuating the bindings of DNMT1 and DNMT3a to the METTL3 promoter, which leads to the up-regulation of METTL3 and the following increased m6A levels of pri-miR-25 (Figure 2B) [64]. However, CSC can also contribute to a diminished m6A abundance through triggering the hypomethylation of ALKBH5 CpG island in esophageal squamous cell carcinoma [65]. Thus CSC seems to be a powerful factor to indirectly influence m6A modification via the straightforward impact on DNA methylation. Moreover, m6A profiling based on human fetal tissues reveals a preferential occupation of m6A on CpG-rich promoters. CpG-related promoters are capable of modulating m6A levels [66]. These results may suggest the co-transcriptional process of m6A biogenesis and DNA methylation.
m6A and chromatin remodeling
Chromatin remodeling is the rearrangement of chromatin state. An open or condensed state determines the accessible or unapproachable access for DNA binding proteins. Nowadays, several studies have demonstrated the crosstalk between chromain remodeling and m6A modification. BAF155 is a chromatin remodeling factor. RBM15 can negatively regulate the expression of BAF155 mRNA by decreasing its stability and promote its decay in an m6A-dependent manner [67]. Notably, the regulation capacity of RBM15 on BAF155 requires the activity of METTL3. Furthermore, a reverse correlation between METTL3/YTHDC1 and chromatin accessibility in mouse embryonic stem cells (ESCs) is observed [22]. Specifically, METTL3 promotes the m6A methylation of chromosome-associated regulatory RNAs (carRNAs), while YTHDC1 participates in the degradation of these m6A-marked RNAs. Thus METTL3/YTHDC1-guided m6A modification regulates the chromatin state and subsequent transcription by governing the expression of carRNAs (Figure 2C).
Remarkably, a study has revealed the deposition of m6A in chromatin-associated nascent pre-mRNAs from Hela cells. This m6A methylation, which is mainly present in exons and rarely in introns, is accomplished when mRNA is released into nucleoplasm. Surprisingly, m6A modification is required for the cytoplasmic mRNA stability of nascent transcripts, but not for the majority of splicing events [68].
m6A and histone modification
Histone modification is a significant participant of post-translational regulations, which is involved in chromatin structure modulation, nucleosome dynamics and gene transcription. It primarily contains histone methylation, acetylation and ubiquitination. Interestingly, some modifications lead to the repression of transcription like H3K9me2/3 and H3K27me3, while others are associated with the activation of transcription including H3K4me1-3, H3K27me1, H3K36me1-3 and H3K27ac [69].
During cell development, METTL14 plays a vital role in the proliferation and differentiation of neural stem cells (NSCs). Surprisingly, the increased levels of H3K27ac, H3K4me3 and H3K27me3 modifications are observed when METTL14 is deleted. MTT assays demonstrate that m6A modulates the proliferation of NSCs partially via regulating H3K27ac and H3K27me3. Mechanistically, METTL14-mediated m6A methylation suppresses the stability of both CREB binding protein (CBP) and p300 transcripts which are the crucial modifiers of H3K27ac (Figure 2D) [21]. In addition, METTL3-mediated m6A modification is necessary for neuronal development and neurogenesis. METTL3 regulates the m6A-modified histone methyltransferase Ezh2, which further advances the level of H3K27me3 (Figure 2D) [70]. For erythropoiesis, m6A enzymes facilitate the translation of erythroid genes, especially those encoding SETD histone methyltransferases. The impairment of m6A leads to a substantial inhibition of H3K4me3 modification which is responsible for KLF1-centered transcriptional program required for erythropoiesis, heme synthesis or hemoglobin assembly [71]. These studies suggest the divergent impacts on histone methylation induced by m6A, which may indicate that m6A-mediated histone regulation is cell-type-specific.
Apart from m6A-modulated histone modifications, histone modifiers also intimately participate in m6A rearrangement. The m6A reader hnRNPA2B1 is implicated in the immune response to DNA viruses. Herpes simplex virus-1 (HSV-1) infection induces the dimerization of hnRNPA2B1, which guides its nucleo-cytoplasmic translocation. Simultaneously, the arginine demethylase JMJD6 promotes the demethylation of hnRNPA2B1 at Arg226 and activates its translocation to cytoplasm, which further magnifies the expression of IFN-β (Figure 2E) [72]. In gastric cancer, the promoter of METTL3 is marked by p300-regulated H3K27ac modification, which triggers the transcription of METTL3 and then leads to an elevated m6A level of HDGF (Figure 2F) [73]. Furthermore, KDM5C-guided demethylation of H3K4me3 modification suppresses the transcription of METTL14 which can restrain the metastasis of colorectal cancer (CRC) via promoting the m6A level of SOX4 mRNA [74].
Moreover, two studies have afforded systematic evidence for the precise and dynamical deposition of m6A and histone modification. Huang et al. find that m6A peaks associated with H3K36me3 marks mainly locate near stop codons, while those H3K36me3 loci not modified by m6A are enriched in the coding sequence (CDS) or intron [75]. The correlated positions imply their intertwined relationships. Intriguingly, although H3K36me3 cannot impact the expression of m6A key enzymes, it may affect the interaction between m6A enzymes and their targets. In other words, H3K36me3 is able to recruit m6A complex to deposit m6A imprinting. The fundamental element for the binding of m6A complex and H3K36me3 is METTL14 which is further identified to recognize H3K36me3 marks via a Pol II-independent pattern during transcription elongation [75]. Additionally, Li et al. clarify that METTL3/METTL14-mediated m6A methylation modulates the levels of H3K9me2 [76]. The genome-wide correlation between m6A and KDM3B (H3K9me2 demethylase) is identified. To be specific, YTHDC1 recruits KDM3B to m6A-marked chromatin regions, triggering H3K9me2 demethylation and subsequent activation of gene expression. Conservatively, the co-occurrence of H3K36me2 and m6A is found in plants as well [77]. All these investigations reveal the co-transcriptional interplay or even co-occupancy between m6A and histone modification.
m6A and other RNA modifications
m6A and m1A
Currently, m1A is considered as a reversible modification in tRNAs, rRNAs, and mRNAs, which is methylated and demethylated by TRMTs and ALKBH1/3, respectively [78, 79]. Remarkably, increasing evidence indicates a close link between m1A and m6A. Wei et al. discover that FTO has the ability to mediate both nuclear and cytoplasmic demethylation of m1A in tRNAs, and to subsequently suppress the RNA translation process [37]. The special structure of FTO is analogous to the tRNA m5C methyltransferase NSUN6, which explains why another m6A demethylase ALKBH5 cannot recognize m1A at tRNAs as a substrate.
The m6A-binding proteins YTHDF1-3 and YTHDC1 are capable of directly binding to m1A sites. YTHDF2 accomplishes the recognition of m6A and m1A depending on its conserved residue Trp432 [80]. Functionally, YTHDF2 facilitates the degradation of m1A-modified transcripts [81].
Fortunately, two approaches including DART-seq and m1A-IP-seq/m1Aquant-seq, have been used to achieve genome-wide mapping of m6A and m1A with a single-base resolution, respectively [82, 83]. However, further research should be conducted to explore the mechanisms between the two types of modifications via using the novel tools.
m6A and m5C
The m5C modification, which is the methylation of cytosine at carbon 5, is catalyzed by NSUN proteins and DNMT2 [84, 85], and primarily occurs in tRNAs, rRNAs, and mRNAs [86]. Previous studies have reported that m5C methylation is of great significance in the RNA stability, export and transcription [87].
Remarkably, there is a subtle relationship between m5C and m6A modifications. Courtney et al. demonstrate that murine leukemia virus (MLV) transcripts exhibit high levels of m6A and m5C modifications, which lead to a high level of viral replication. Mechanistically, the ectopic expression of YTHDF2 facilitates MLV replication, while the inhibition of m5C writer NSUN2 hinders MLV replication [88], which suggests that m6A may cooperate with m5C to engage in some biological events. Coincidently, a direct synergistic effect of m6A and m5C has been reported [89]. METTL3/METTL14-catalyzed m6A methylation and NSUN2-induced m5C methylation can jointly enhance the expression of p21 mRNA in response to oxidative stress-triggered cellular senescence in tumor cells (Figure 2G). In addition to the cooperative relationship, an interaction between m6A and m5C has been observed. Specifically, METTL3/METTL14-mediated m6A modification can promote NSUN2-mediated m5C modification, and vice versa.
In addition, m6A reader YTHDF2 is capable of recognizing and binding to m5C in RNA [90]. Deletion of YTHDF2 results in a remarkably expanded m5C level in rRNA. Interestingly, YTHDF2 participates in the regulation of pre-rRNA processing, which may be achieved via its modulation of m5C level.
m6A and A-to-I
The transition of A-to-I is processed by adenosine deaminases acting on RNA (ADAR) enzymes, which is a principal form of RNA editing [91]. It is reported that A-to-I is a key factor influencing RNA metabolism, such as miRNA processing [92].
A reverse correlation between m6A and A-to-I has been identified using genomic analyses (Figure 2H). Loss of m6A modification contributes to the elevated level of A-to-I editing via a favorable association of ADAR with m6A-depleted transcripts. However, the underlying mechanism has not been fully elucidated. One possible reason for the occurrence is that the alteration of m6A-induced RNA structure may mediate the binding of ADAR and targeted genes. The occupation of m6A enzymes on RNAs may interfere with the localization of ADAR [4]. However, whether A-to-I is capable of modulating m6A level remains indeterminate.
m6A and pseudogene
Pseudogene is a type of genomic element, which is partially homologous to corresponding functional genes, although lacks protein-coding capability due to mutations. Pseudogene widely participates in gene regulation [93].
Studies have revealed that there is a potential association between m6A and pseudogenes. Olfr29-ps1 is a lncRNA pseudogene, which is stimulated by cytokine IL-6 in myeloid-derived suppressor cells (MDSCs). METTL3-mediated m6A methylation facilitates the expression of Olfr29-ps1, and simultaneously enhances its sponge to miR-214-3p (Figure 2I). Then MyD88, which is suppressed by miR-214-3p, is up-regulated to amplify the differentiation and immunosuppressive effects of MDSCs [94]. Moreover, it is reported that m6A and pseudouridine (ψ) can collaboratively disrupt the binding of hPUM2 to its targeted RNAs [95].
In addition, m6A and ψ play a crucial role in immunity. Durbin et al. apply a well-accepted RIG-I-related platform to examine the immunosuppressive potential of various RNA modifications [96]. The results reveal that either m6A or ψ negatively correlates with the alleviated innate immune signaling. Specifically, m6A-modified RNAs may poorly bind to RIG-I. Although ψ-containing RNAs can intimately interact with RIG-I, they are unable to initiate the canonical RIG-I antiviral signaling.
m6A and m6Am
When the transcription initiation nucleoside of mRNA is 2-O-methyladeonisine (Am), m6Am methyltransferase PCIF1 is capable of catalyzing methylation on its N6 position to further generate m6Am, which is dependent on the structure of 7-methylguanosine (m7G) cap [97-101]. Studies have revealed that m6Am can reinforce the stability of transcripts [102], while the findings about its effects on translation are inconsistently identified. Akichika et al. illustrate that m6Am enhances the translation of capped mRNAs [97]. However, another study suggests that m6Am may impede cap-dependent translation [98].
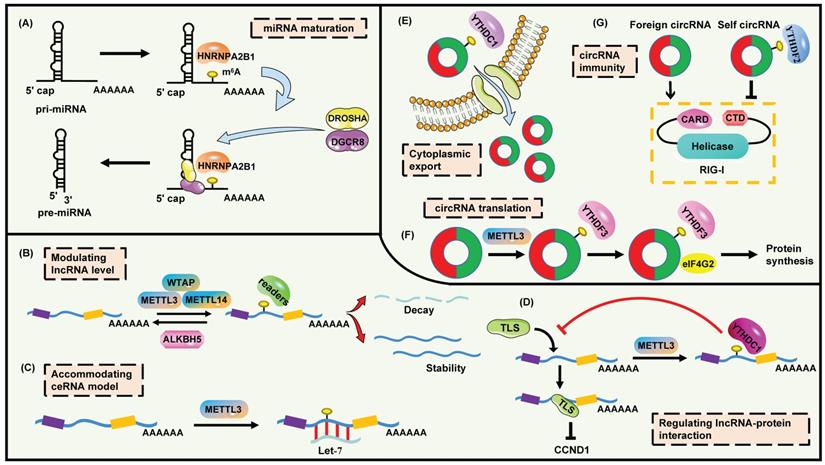
The functions and mechanisms of m6A modification on ncRNAs. (A) m6A promotes the maturation of miRNA. (B) m6A modulates lncRNA level. (C) m6A facilitates lncRNA to combine with miRNA. (D) m6A interferes the binding of lncRNA to proteins. (E) m6A mediates the cytoplasmic export of circRNA. (F) m6A regulates circRNA translation. (G) m6A assists the innate immune system to recognize self circRNA.
Several studies demonstrating the genome-wide landscape of m6A and m6Am have been conducted, which provide reliable evidence for their relationship [103-105]. The conserved m6Am signals can be detected in WTAP and ALKBH5, while the non-conserved m6Am signals can be identified in METTL3. Additionally, the non-conserved m6A signals can be found in PCIF1 [105]. Furthermore, FTO has been demonstrated to target m6Am. Functionally, FTO is responsible for the demethylation of m6Am in snRNA [37]. Nevertheless, additional functional relevance of m6A and m6Am remains to be explored.
m6A and ncRNAs
m6A modification exists in almost all types of ncRNAs, especially in miRNAs, lncRNAs and circRNAs. They are all vigorous performers participating in extensive biological processes, particularly in tumor malignancy. The crosstalk of m6A and ncRNAs is pervasive and inspiring, extending the scope of epigenetics.
m6A-miRNA
miRNA is a short non-coding RNA (no more than 22 nucleotides), and links to a variety of biological processes such as tumor growth, drug resistance, cell differentiation, and cellular senescence [106]. Initially, primary miRNA (pri-miRNA) is cleaved into precursor miRNA (pre-miRNA) by the microprocessor complex comprising of endonuclease Drosha and DGCR8 protein. After being transported to cytoplasm by exportin 5, pre-miRNA is further cleaved by Dicer to release the double-strands RNAs, which are then loaded onto an AGO protein constituting the RNA-induced silencing complex (RISC) [107].
Intriguingly, m6A is the mark for advancing the processing of pri-miRNAs [108]. METTL3 is sufficient to methylate massive pri-miRNAs to reinforce miRNA maturation through recruiting DGCR8 and m6A reader HNRNPA2B1 (Figure 3A). Moreover, HNRNPA2B1 interacts with DGCR8 to promote its binding to pri-miRNAs, which enhances the continuous generation of pri-miRNAs [53, 108]. There are plenty of illustrations about this regulatory pattern. In bladder cancer, METTL3 accelerates cell proliferation by promoting the maturation of pri-miR221/222 which targets at PTEN [109]. In CRC, METTL3 accounts for the aberrant m6A modification and boosts the production of mature miR-1246, which suppresses the SPRED/MAPK signaling [110]. Wang et al. clarify that up-regulation of METTL3 blocks oxidative stress and apoptosis in colistin-evoked nephrotoxicity via the promotion of miR-873-5p mature process and the regulation of Keap1-Nrf2 pathway [111]. Besides, mimicking the function of Dicer, METTL3-mediated m6A methylation leads to the splicing of pre-miR-143-3p which impairs VASH1 expression to facilitate angiogenesis and metastasis of lung cancers [112]. Moreover, METTL3 increases the m6A modification of pre-miR-320 and drives osteogenic differentiation of bone marrow-derived mesenchymal stem cells [113]. Another study demonstrates that the maturation of miR-7212-5p is impelled by METTL3-mediated m6A modification, while the miR-7212-5p/FGFR3 axis accounts for the regulation of osteoblast differentiation and fracture healing [114]. In addition, an interesting study shows that CSC activates the excessive miR-25-3p maturation dependent on m6A mechanism in pancreatic cancer [64]. The enhancement of METTL3 triggered by CSC contributes to the up-regulation of m6A level on pri-miR-25. Then NKAP serves as not only the m6A reader but also a splicing factor to stimulate the processing of pri-miR-25. Accumulating miR-25-3p suppresses PHLPP2, leading to the activation of AKT-p70S6K signaling [64]. Except for METTL3, another m6A writer METTL14 also modulates the maturation of miRNAs analogously. As a suppressor in hepatocellular carcinoma (HCC), METTL14 interacts with DGCR8 to promote the processing of pri-miR-126 via an m6A-dependent pattern, triggering the enhanced level of miR-126 which represses the tumor metastasis [115].
Now that m6A is frequently enriched in 3' UTRs (near stop codons), and miRNA binding sites on mRNA are also commonly observed within 3' UTRs, the relationship between m6A and miRNA binding is discussed. However, an inverse localization pattern is identified [10]. One reasonable explanation is that moderate spatial distance may be beneficial for mutual effects between m6A and miRNA. Actually, deficiency of m6A caused by loss of METTL3 or METTL14 restrains the miRNA-mRNA interaction as well as boosts HuR-mRNA interaction, which finally stabilizes the corresponding transcript [116]. A more vivid example is provided by Zhang et al. [117]. The m6A residue is found in the 3'UTR of YAP (353-357), and this modification is crucial for the conjugation of miR-582-3p and YAP. Hence, m6A modification may trigger the binding of miRNAs and targeted genes.
Additionally, AGO2 mRNA is highly methylated and positively modulated by m6A methyltransferases in human diploid fibroblasts. The miRNA abundance is controlled by m6A level based on the stability of AGO2 [118]. Knuckles et al. propose a model to delineate the RNA fate determined by m6A and microprocessor [119]. In normal temperature, METTL3-centered complex deposits the m6A labels to massive RNAs containing mRNAs, pri-miRNAs, lncRNAs and snoRNAs, followed by the induction of their degradation mediated by DGCR8. However, acute heat stress leads to the re-localization of the m6A complex and DGCR8 at heat-shock genes to facilitate their decay. Meanwhile, those transcripts previously modulated by METTL3 and DGCR8 accumulate. This is an indirect fashion of m6A to control the degradation of miRNAs or other ncRNAs.
miRNA-m6A
A bidirectional relationship exists between miRNAs and m6A because miRNAs can regulate m6A-related events as well. Dicer, but not AGO protein, mediates the formation of m6A without altering the amount of methyltransferases or demethylases, and it may modulate nuclear speckle localization of METTL3 [120]. miRNAs are able to trigger de novo m6A methylation through a sequence pairing pattern. Moreover, miRNAs are responsible for the manipulation of the binding of METTL3 to miRNA site-containing mRNAs to affect m6A abundance, which is tightly associated with cell reprogramming to pluripotency [120]. In HCC, miR-145 governs m6A level by inhibiting the expression of YTHDF2 [121]. METTL3 is targeted by miR-186, and it activates Wnt/β-catenin signaling in hepatoblastoma [122]. miRNA let-7g which is inhibited by HBXIP, attenuates the expression of METTL3. Simultaneously, HBXIP is activated by METTL3 in an m6A-dependent manner [123]. The positive feedback loop elaborates the complicated connection between miRNA and m6A.
m6A-lncRNA
LncRNAs are a group of non-coding transcripts longer than 200 nucleotides. The functions of lncRNAs are diverse, including regulating chromatin topology, serving as scaffolding for proteins or RNAs, governing RNA stabilization and transcription, or even producing peptides [124]. LncRNAs can be modulated via multiple levels containing transcriptional regulation, post-transcriptional processing and degradation control [125]. Importantly, the interaction between lncRNAs and m6A modification is a novel annotation (Figure 3B-D). Xiao et al. have generated the whole-transcriptome m6A landscape of human fetal tissues. Numerous lncRNAs are methylated by m6A especially in kidney, placenta and brain. Enhancer lncRNAs (originated from enhancers) have a higher enrichment in m6A modification compared with other lncRNAs. The distribution of m6A on lncRNAs is nearly balanced among 5'UTR, CDS and 3'UTR, which is different from the distribution on mRNA. Meanwhile, the proportion of m6A methylation on lncRNAs is lower than on mRNAs [66].
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a highly conserved nuclear lncRNA which is closely related to the metastasis of tumors [126]. As an abundant and essential transcript, MALAT1 is a paradigm to describe m6A-participated modification of lncRNAs. It is reported that m6A-modified MALAT1 (at A2577) is adequate for the binding of HNRNPC, one of the m6A readers which is essential for pre-mRNA processing [41]. Similarly, m6A methylation (at A2515) increases the accessibility of MALAT1 for RNA-binding protein (RBP) through exposing its purine-rich sequences. Then HNRNPG, which governs gene expression and alternative splicing, binds to MALAT1 using its low-complexity region [42]. In addition, the putative m6A writer METTL16 can interact with the U6 snRNA, pre-mRNAs and lncRNAs, such as MALAT1 [31]. To be specific, the 3' triple helix domain of MALAT1 is the binding site of METTL16 [127]. Jin et al. demonstrate that METTL3-guided m6A modulation contributes to the elevated expression of MALAT1 with the support of YTHDF3 in non-small cell lung cancer (NSCLC) [128]. Then MALAT1 sponges miR-1914-3p to increase YAP activity and strengthen the metastatic potential of NSCLC.
XIST is another well-characterized mammalian lncRNA. It is the master regulator of X-chromosome inactivation (XCI), a dosage compensation process to balance X-linked gene expression via the suppression of transcription [129]. Moindrot et al. employ a pooled shRNA screen to reveal that WTAP is one of key factors for XIST-mediated silencing, which co-localizes with XIST RNA in nuclear perichromatin spaces [130]. Moreover, a following study demonstrates that XIST is heavily m6A methylated, and highlights the role of m6A modification in XIST-dependent transcriptional silencing [28]. WTAP and METTL3 can be recruited by RBM15/15B to achieve the m6A modification on XIST. Then m6A-marked XIST is recognized by the reader YTHDC1, which promotes XIST-mediated gene inhibition [28]. In addition, SPEN is a vital orchestrator of XCI by binding to XIST. The SPOC domain of SPEN is clarified to be involved in the recruitment of m6A machinery to XIST [131]. Nevertheless, Nesterova et al. have conducted the systematic allelic analysis of XIST-mediated suppression in two interspecific mice models, and put forward the viewpoint that RBM15-centered m6A complex may provide minor contribution to this type of gene silencing [132]. The possible causes for different consequences might rely on the redundancy of m6A modification and the various approaches to assay transcriptional inhibition. Although whether m6A can indeed control XIST-guided silencing is controversial, METTL14/YTHDF2 axis is feasible for regulating the stability of XIST [133]. Therefore, the sophisticated m6A-XIST interaction deserves further explorations.
Actually, there are many other m6A-bearing lncRNAs which have been reported in multiple tumors with the major mechanisms of RNA stability regulation. The lncRNA FAM225A is overexpressed in nasopharyngeal carcinoma, and m6A modification is identified on FAM225A enhancing its RNA stability [134]. In HCC, METTL3-mediated modulation contributes to the stabilization of LINC00958 which intensifies the HCC lipogenesis [135]. Similarly, LNCAROD is up-regulated by METTL3/METTL14 in head and neck squamous cell carcinoma (HNSCC) and impels its tumorigenicity through preventing YBX1 from degradation [136]. VIRMA induces aggressive phenotype of prostate cancer through sustaining m6A levels and abundance of the oncogenic lncRNAs CCAT1/2 [137]. DANCR, initially recognized as an anti-differentiation lncRNA, is strengthened by IGF2BP2 based on an m6A-modified site, and boosts stemness like properties of pancreatic cancer [52]. ALKBH5-guided m6A demethylation enhances the expression of PVT1 with the assistance of YTHDF2 in osteosarcoma [138]. In epithelial ovarian cancer, METTL3 increases the level of RHPN1-AS1 [139]. Besides, there are several studies about m6A-related lncRNAs in CRC. RP11 promotes the dissemination of CRC cells by regulating the epithelial mesenchymal transition (EMT). Specifically, the expression of RP11 is collaboratively regulated by METTL3 and ALKBH5. Elevated RP11 recruits the hnRNPA2B1 which recognizes and stabilizes the mRNAs of Siah1 and Fbxo45, thereby preventing the Siah1 and Fbxo45-dependent proteasomal degradation of Zeb1 [140]. Moreover, a study demonstrates a regulatory loop between lncRNA and m6A. LncRNA GAS5 combines with the WW domain of YAP and promotes YAP degradation via modulating its nucleo-cytoplasmic translocation. During CRC tumorigenesis, the m6A reader YTHDF3 interacts with m6A-modified GAS5 and promotes its decay, thus inhibiting the degradation of YAP. Accumulating YAP further activates the transcription of YTHDF3. This is a complicated but intriguing negative feedback loop between m6A and lncRNA [141].
Furthermore, other regulatory layers contain the m6A-mediated translation and the RBP-binding of lncRNA. LINC00278 is an m6A-methylated transcript regulated by METTL3/METTL14/WTAP and ALKBH5. Studies have revealed that LINC00278 encodes a tumor-suppressing micropeptide called YY1BM, which suppresses the combination of YY1 and androgen receptor, rendering ESCC cells more sensitive to nutrient deprivation. Mechanistically, m6A modification promotes the translation of YY1BM via a YTHDF1-dependent manner [65]. pncRNA-D is an irradiation-triggered lncRNA which interacts with RBP TLS/FUS. The interaction of pncRNA-D with TLS is associated with CCND1 inhibition. METTL3 is responsible for the half-time of m6A-methylated pncRNA-D. YTHDC1 competitively inhibits the binding of pncRNA-D to TLS, thereby alleviating TLS-mediated suppression of CCND1. The decrease of m6A modification leads to a G0/G1 arrest in the cell cycle relying on CCND1 [142].
In addition to tumorigenesis, m6A-lncRNA interaction is involved in other cellular procedures as well. Yang et al. find that linc1281 is indispensable for appropriate mouse ESC differentiation. The METTL3-dependent m6A mark in the last exon of linc1281 is responsible for not only its functional roles, but also the interactions with pluripotency-related miRNAs [143]. For immune homeostasis regulation, m6A-modified lnc-Dpf3 controls the migration of dendritic cell (DC). It is well-accepted that although rapid DC migration is vital for initiation of immune defense, timely cessation of its trafficking is also indispensable for the avoidance of excessive inflammation. In the early stage, CCR7-mediated DC migration accelerates in response to CCL19/CCL21. However, during the late stage, CCR7 stimulation triggers the expression of lnc-Dpf3 by removing its m6A methylation and protecting it from YTHDF2-mediated degradation. Then lnc-Dpf3 binds to HIF-1α to suppress the HIF1α-dependent glycolysis and the migratory capacity of DC [144].
lncRNA-m6A
Apart from the m6A-lncRNA interaction, lncRNA is able to impact the m6A methylation as well. In CRC, LINRIS maintains the stability of the m6A reader IGF2BP2 through blocking its ubiquitination/autophagy-lysosome pathway, which facilitates MYC-mediated glycolysis [145]. Moreover, the antisense lncRNA may reinforce the interaction of parent transcripts (mature or nascent) with m6A enzymes to control gene expression. For example, the up-regulation of ARHGAP5 is associated with chemoresistance in gastric cancer. In the nucleus, ARHGAP5-AS1 enhances the transcription of ARHGAP5 by binding to its promoter. Furthermore, ARHGAP5-AS1 can recruit METTL3 in the nucleus to induce the elevated m6A modification on ARHGAP5 mRNA, eventually facilitating the stability of ARHGAP5 [146]. Similarly, GAS5-AS1 enhances the stability of GAS5 by interacting with ALKBH5 which eliminates m6A modification in cervical cancer [147]. In addition, FOXM1-AS increases the binding of ALKBH5 to FOXM1 pre-mRNA in glioblastoma. ALKBH5-triggered demethylation impels the effects of RNA-binding protein HuR, contributing to the elevated level of FOXM1 [148]. GATA3-AS promotes the interaction of KIAA1429 with GATA3 pre-mRNA in HCC [149]. Recently, a study by Zhu et al. reveals another interesting regulatory mode. LncRNA LINC00266-1 can encode a small peptide which tightly interacts with IGF2BP1. The binding of peptide strengthens the recognition of IGF2BP1 on m6A-modified RNAs like c-Myc, further enhancing the stability of targets which are closely associated with CRC tumorigenesis [150].
m6A-circRNA
CircRNA is a species of covalently closed and evolutionally conservative circular transcript, mainly deriving from back-splicing of exons [151]. The structure of circRNA is quite stable. It is broadly expressed in various kinds of specimens via a cell or tissue-specific manner. CircRNA is extensively involved in biological processes, such as developmental modulation, pathogenesis of heart diseases, chemoresistance and tumorigenesis [152]. It primarily functions as the sponge of miRNAs (ceRNA), as well as participates in the interaction with protein, transcription, splicing regulation, and even the non-canonical translation [153].
The information of m6A-modified circRNAs is finite but attractive. Zhou et al. have established a genome-wide map of m6A-circRNAs in hESCs and Hela cells, and revealed the cell-type-specific patterns of m6A modification on circRNAs [153]. There are several features about m6A-circRNAs. For example, circRNAs containing long single exons instead of multi-exons are more likely to be modified by m6A. m6A-circRNAs are commonly generated from those exons without m6A peaks in mRNAs. Like mRNAs, circRNAs are methylated by METTL3 and recognized by YTHDF1/YTHDF2 [154]. Park et al. prove that both linear and circular m6A-marked RNAs can be edited by the YTHDF2-HRSP12-RNase P/MRP axis [155]. CircNSUN2 is an m6A-methylated circRNA which promotes the liver metastasis of CRC patients. The m6A motif “GAACU” on circNSUN2 is recognized by YTHDC1, which enhances circNSUN2 export from nucleus to cytoplasm (Figure 3E) [156].
It is inspiring to observe that circRNAs possess widespread m6A modification, which is adequate to drive protein synthesis with even a single m6A site. This cap-independent translation requires the assistance of eIF4G2 and YTHDF3 [157, 158]. As expected, the translation can be abolished by FTO, while enhanced by METTL3 or METTL14 [157]. Besides, circE7 is identified as an m6A-marked, cytoplasmatic and polysomes-associated circRNA. E7 oncoprotein is produced from the translation of circE7 human papillomavirus, while the mutation of possible m6A motifs strongly suppresses E7 protein expression [159]. Timoteo et al. reveal that METTL3 regulates the m6A levels while YTHDC1 impacts the back-splicing of circRNAs. The cooperation of METTL3 and YTHDC1 regulates the biogenesis of various circRNAs including circ-ZNF609 which is translatable. Moreover, YTHDF3 and eIF4G2 recognize circ-ZNF609 to regulate its translation (Figure 3F) [158]. Tang et al. identify that approximately half of spermiogenesis-related circRNAs are created via the back-splicing at m6A-enriched sites in linear mRNAs where start and stop codons are usually located. The outcome is that these circRNAs embrace m6A-accociated open reading frames (ORFs) in their junctions, which reveals the novel role of m6A in coding-circRNAs biogenesis [160]. These results enrich the m6A-based non-canonical functions of circRNAs.
In addition, m6A-circRNAs are also involved in the immunoregulation and environmental stress response (Figure 3G). Foreign circRNAs, instead of self-counterparts, are efficient to trigger T cell activation and antitumor immunity in vivo. The m6A methylation patterns of exogenous and endogenous circRNAs are quite distinct. Mechanistically, unmodified foreign circRNAs heavily stimulate MAVS polymerization and interferon production after the RIG-I recognition. Nevertheless, m6A modification impairs activation of immune genes induced by endogenous circRNAs to prevent aberrant responses, which means that m6A can be the identity for self circRNAs. YTHDF2 is required for the suppression of circRNA-mediated innate immune signaling [161]. Intriguingly, a transcriptome-wide profiling of m6A-circRNAs is revealed based on the hypoxia mediated pulmonary hypertension (HPH) model. The m6A abundance of circRNAs is diminished but its expression is increased in hypoxia. m6A-circRNAs are predominantly derived from encoding transcripts spanned single exons. Furthermore, the network of circRNA/miRNA/mRNA is also regulated by m6A in HPH. CircXpo6 and circTmtc3 are both m6A-modified and then down-regulated in HPH [162].
circRNA-m6A
However, studies about the functions of circRNAs on m6A modifications are rare. Recently, a study has revealed the role of circRNA-modulated m6A machinery in major depressive disorder (MDD) [163]. CircSTAG1 is down-regulated in MDD animal models or patients with MDD. CircSTAG1 has the capacity to capture ALKBH5 to reduce its translocation into the nucleus. Then m6A modification is enhanced, which results in an increased degradation of fatty acid amide hydrolase (FAAH) mRNA and a subsequent decrease in depressive-like behaviors, as well as astrocyte loss. In short, circSTAG1 ameliorates MDD through inhibiting the translocation of ALKBH5 and then augmenting m6A levels of FAAH mRNA. Further researches should be conducted to elucidate the complex interactions between circRNA and m6A.
The potential clinical values of m6A-centered epigenetic modifications
Nowadays, it is generally believed that epigenetic regulations exert a crucial role in the pathogenesis of various diseases. Therefore, exploring the possible pharmaceutical agents targeting epigenetic modifications seems to be a promising therapeutic strategy. For example, it is reported that DNA methyltransferase inhibitor (DNMTi), 5-Aza-2'-deoxycytidine, is able to enhance immunotherapy in esophageal carcinoma by promoting the expression of MAGE-A11 [164]. Moreover, histone deacetylase inhibitor (HDACi) MPT0B291 is capable of suppressing glioma growth partially via facilitating the acetylation of p53 [165]. Interestingly, the synergistic effects on treatment by combining multiple types of epigenetic inhibitors are widely reported [166-168].
In addition, inhibitors based on m6A-related enzymes have been actively investigated. However, current studies mostly focus on FTO, instead of m6A methyltransferases or m6A-binding proteins. As a highly selective inhibitor of FTO, meclofenamic acid 2 (MA2) can dramatically suppress the growth and self-renewal of GSC [169, 170]. Chen et al. reveal that R-2HG can inhibit FTO and lead to the decreased stability of MYC and CEBPA, thereby impairing the proliferation of leukemia cells [171]. There are other small molecule drugs targeting FTO that exerted substantial inhibitory effects in tumors, such as FTO-04 [172] and FB23-2 [173]. Moreover, FTO inhibitors participate in the immunotherapy as well. In melanoma, FTO repression promotes tumor growth and increases the response of cancer to anti-PD-1 blockade [174]. Analogously, the freshly recognized inhibitor of ALKBH5, ALK-04, is capable of reinforcing the efficacy of anti-PD-1 therapy [175]. Furthermore, two series of adenine derivatives is identified as the selective inhibitors of METTL3, in spite of their elusive roles in clinical applications [176].
Notably, the intricate crosstalk between m6A and other epigenetic modifiers is tightly involved in tumor progression as mentioned above. Therefore, abolishing these interplay in human cancers may be the meaningful therapeutic perspective. For example, in gastric cancer, p300-guided H3K27ac modification can trigger the transcription of METTL3, eventually facilitating the malignancy of tumor [73]. Perhaps, combination of HDACi and METTL3 inhibitors may become the feasible approach to interrupt the progression of gastric cancer. In addition, METTL3 mediates the m6A level of MALAT1 to increase its stability, which results in the drug resistance and metastasis of NSCLC [64]. Possibly, suppressing the activity of METTL3 to regulate lncRNA levels may enhance the sensitive of NSCLC to cisplatin. Moreover, the diminished DNA methylation triggers the enhancement of METTL3, which further induces the maturation of pri-miR-25, promoting the development of pancreatic cancer [128]. It is inspiring to try the combined treatment of DNMTi and METTL3 inhibitors to collapse the vicious axis of DNA methylation/m6A/ncRNA in this terrible cancer.
Nevertheless, all these ideas remain theoretical owing that investigations about drugs targeting at the crosstalk of m6A and other modifications are quite rare. The potential clinical values remind us that further explorations are urgently required.
Conclusion
m6A RNA methylation, which is a new trajectory of epigenetic modification, has increasingly attracted the attention of researchers over the last few years. Studies have revealed that m6A plays a crucial role in RNA metabolism, such as degradation, alternative splicing, and translation. In addition, the interactions of m6A and targeted RNAs exert great influence on various biological processes, particularly in tumorigenesis. Meanwhile, accumulating evidence has deciphered the interplay between m6A and other epigenetic modulators (DNA methylation, chromatin remodeling, histone modification, RNA modification and ncRNAs), further unveiling the mysteries of epigenetic reprogramming.
Briefly, m6A and DNA methylation may exhibit a cooperative relationship, which relies on the interaction between m6A demethylase and DNA demethylase. In chromatin remodeling, m6A writers or readers are able to regulate the expression of chromatin-related RNAs, thus accommodating the chromatin state. However, there is still a dearth of information regarding the two crosstalk. For example, whether the regulatory loop between m6A and 6mA DNA methylation is available deserves further explorations. Notably, the complicated links between m6A and histone modification gradually emerge. m6A methylation modulates the status of histone methylation or acetylation, while histone modification also intends to affect the expression of m6A-related genes. The co-transcriptional regulation expounds the accurate deposition of m6A and histone modification, which determines the precise control of bioprocesses. Furthermore, m6A not only controls the level of other RNA modifications such as m5C and A-to-I editing, but also collaborates with them to govern multiple physiological processes. These shed light on the reciprocal associations of m6A and other RNA modifications and pave the way to further comprehend other types of RNA modifications. There is also a close relationship between m6A modification and ncRNAs, including miRNAs, lncRNAs and circRNAs. In most cancers, m6A machinery plays a promoting or suppressive role through altering the expression of targeted ncRNAs. In turn, ncRNAs regulate the stability and expression of m6A-assciated enzymes. It breaks the stereotype of ncRNAs and opens up a new paradigm for exploring the potential roles of ncRNAs. Nevertheless, the crosstalk between m6A methylation and circRNAs has not been clearly elucidated, particularly the function of circRNAs on m6A regulation.
Generally, interactions between m6A modification and other epigenetic members actively participate in the progression of tumors. These crosstalk can not only serve as the essential biomarkers for cancers, but also provide insightful mechanisms to develop the promising therapeutic strategies. Admittedly, these findings are only the tip of the iceberg. In the future, firstly, abundant efforts are still required to uncover more underlying roles of the interplay among these epigenetic modifiers and reach the deeper understanding of epigenetics in cancers. Secondly, it is imperative to explore potential remedies targeting at these interactions to reverse the erroneous epigenetic remodeling and reshape the balance. To be specific, perhaps the combination of m6A enzymes inhibitors and other modifiers inhibitors (DNMTi, HDACi, etc.) deserve validations in multiple tumors. It may be more attracting to directly target at the crosstalk instead of the modification itself. Moreover, it is noteworthy that the associations between FTO and other modifications are poorly investigated. FTO is the most unambiguous drug target with several selective inhibitors. Clarifying the mystery of crosstalk between FTO and other epigenetic members might guide to improve treatment efficiency of cancers.
Abbreviations
ncRNA: non-coding RNA; miRNA: microRNA; lncRNA: long non-coding RNA; circRNA: circular RNA; m6A: N6-methyladenosine; m5C: 5-methylcytosine; A-to-I: adenosine into inosine; m6Am: N6, 2'-O-dimethyladenosine; METTL3: methyltransferase-like 3; METTL14: methyltransferase-like 14; WTAP: Wilms tumor 1-associated protein; VIRMA: vir-like m6A methyltransferase associated; CBLL1: Cbl proto-oncogene like 1; RBM15: RNA-binding motif protein 15; ZC3H13: zinc finger CCCH domain-containing protein 13; FTO: fat mass and obesity-associated; ALKBH5: alkB homolog 5; YTH: YT521-B homology; YTHDFs: YTH domain family proteins; YTHDCs: YTH domain containing proteins; IGF2BPs: insulin-like growth factor 2 mRNA-binding proteins; HNRNPs: heterogeneous nuclear ribonucleoproteins; carRNA: chromosome-associated regulatory RNA; CRC: colorectal cancer; HCC: hepatocellular carcinoma; NSCLC: non-small cell lung cancer.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Katada S, Imhof A, Sassone-Corsi P. Connecting threads: epigenetics and metabolism. Cell. 2012;148:24-8
2. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339-46
3. Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science. 2018;361:1346-9
4. Xiang JF, Yang Q, Liu CX, Wu M, Chen LL, Yang L. N6-methyladenosines modulate A-to-I RNA editing. Mol Cell. 2018;69:126-35
5. Li X, Zhu P, Ma S, Song J, Bai J, Sun F. et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol. 2015;11:592-7
6. Legrand C, Tuorto F, Hartmann M, Liebers R, Jacob D, Helm M. et al. Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res. 2017;27:1589-96
7. Ayadi L, Galvanin A, Pichot F, Marchand V, Motorin Y. RNA ribose methylation (2'-O-methylation): Occurrence, biosynthesis and biological functions. Biochim Biophys Acta Gene Regul Mech. 2019;1862:253-69
8. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974;71:3971-5
9. Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5' terminus of HeLa cell messenger RNA. Cell. 1975;4:379-86
10. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149:1635-46
11. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201-6
12. Kane SE, Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol Cell Biol. 1985;5:2298-306
13. Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N6-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28:507-17
14. Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18:31-42
15. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, at al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388-99
16. Zhou J, wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591-4
17. Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF. et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016;61:507-19
18. Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y. et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife. 2017;6:e31311
19. Atlasi Y, Stunnenberg HG. The interplay of epigenetic marks during stem cell differentiation and development. Nat Rev Genet. 2017;18:643-58
20. Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G. et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019;12:121
21. Wang Y, Li Y, Yue M, Wang J, Kumar S, Wechsler-Reya RJ. et al. N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat Neurosci. 2018;21:195-206
22. Liu J, Do X, Chen C, Chen C, Liu C, Xu MM. et al. N6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science. 2020;367:580-6
23. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93-5
24. Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z. et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575-8
25. Sledz P, Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife. 2016;5:e18434
26. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177-89
27. Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z. et al. VIRMA mediates preferential m6A mRNA methylation in 3' UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10
28. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M. et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369-73
29. Wen J, Lv R, Ma H, Shen H, He C, Wang J. et al. Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol Cell. 2018;69:1028-38
30. Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH. et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32:415-29
31. Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Hoebartner C. et al. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18:2004-14
32. Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP. et al. The U6 snRNA m6A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell. 2017;169:824-35
33. Nhan Van T, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P. et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019;47:7719-33
34. Ma H, Wang X, Cai J, Dai Q, Natchiar SK, Lv R. et al. N6-Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol. 2019;15:88-94
35. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y. et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885-7
36. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang C-M, Li CJ. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18-29
37. Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC. et al. Differential m6A, m6A(m), and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol Cell. 2018;71:973-85
38. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ. et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315-28
39. Mao Y, Dong L, Liu XM, Guo J, Ma H, Shen B. et al. m6A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat Commun. 2019;10:5332
40. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H. et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285-95
41. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560-4
42. Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45:6051-63
43. Wu B, Su S, Patil DP, Liu H, Gan J, Jaffrey SR. et al. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat Commun. 2018;9:420
44. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M. et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626
45. Zaccara S, Jaffrey SR. A Unified Model for the Function of YTHDF Proteins in Regulating m6A-Modified mRNA. Cell. 2020;181:1582-95.e18
46. Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y. et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115-27
47. Zhou B, Liu C, Xu L, Yuan Y, Zhao J, Zhao W. et al. N6-methyladenosine reader protein Ythdc2 suppresses liver steatosis via regulation of mRNA stability of lipogenic genes. Hepatology. 2020 https://doi.org/10.1002/hep.31220. [Online ahead of print]
48. Muller S, Glass M, Singh AK, Haase J, Bley N, Fuchs T. et al. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res. 2019;47:375-90
49. Wang S, Chim B, Su Y, Khil P, Wong M, Wang X. et al. Enhancement of LIN28B-induced hematopoietic reprogramming by IGF2BP3. Genes Dev. 2019;33:1048-68
50. Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN. et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18:112
51. Elcheva IA, Wood T, Chiarolanzio K, Chim B, Wong M, Singh V. et al. RNA-binding protein IGF2BP1 maintains leukemia stem cell properties by regulating HOXB4, MYB, and ALDH1A1. Leukemia. 2020;34:1354-63
52. Hu X, Peng WX, Zhou H, Jiang J, Zhou X, Huang D. et al. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2020;27:1782-94
53. Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162:1299-308
54. Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O. et al. 5' UTR m(6)A promotes cap-independent translation. Cell. 2015;163:999-1010
55. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell. 2016;62:335-45
56. Chen K, Lu Z, Wang X, Fu Y, Luo G-Z, Liu N. et al. High-resolution N(6)-methyladenosine (m(6)A) map using photo-crosslinking-assisted m(6)A sequencing. Angew Chem Int Ed Engl. 2015;54:1587-90
57. Greenberg MVC, Bourc'his D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. 2019;20:590-607
58. Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068-70
59. Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930-5
60. Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300-3
61. Xiao CL, Zhu S, He M, Chen D, Zhang Q, Chen Y. et al. N6-methyladenine DNA modification in the human genome. Mol Cell. 2018;71:306-18
62. Zhou L, Tian S, Qin G. RNA methylomes reveal the m6A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol. 2019;20:156
63. Chen YT, Shen JY, Chen DP, Wu CF, Guo R, Zhang PP. et al. Identification of cross-talk between m6A and 5mC regulators associated with onco-immunogenic features and prognosis across 33 cancer types. J Hematol Oncol. 2020;13:22
64. Zhang J, Bai R, Li M, Ye H, Wu C, Wang C. et al. Excessive miR-25-3p maturation via N6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 2019;10:1858
65. Wu S, Zhang L, Deng J, Guo B, Li F, Wang Y. et al. A novel micropeptide encoded by Y-linked LINC00278 links cigarette smoking and AR signaling in male esophageal squamous cell carcinoma. Cancer Res. 2020;80:2790-803
66. Xiao S, Cao S, Huang Q, Xia L, Deng M, Yang M. et al. The RNA N6-methyladenosine modification landscape of human fetal tissues. Nat Cell Biol. 2019;21:651-61
67. Xie Y, Castro-Hernandez R, Sokpor G, Linh P, Narayanan R, Rosenbusch J. et al. RBM15 modulates the function of chromatin remodeling factor BAF155 through RNA methylation in developing cortex. Mol Neurobiol. 2019;56:7305-20
68. Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vagbo CB, Geula S. et al. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 2017;31:990-1006
69. Zhao Z, Shilatifard A. Epigenetic modifications of histones in cancer. Genome Biol. 2019;20:245
70. Chen J, Zhang YC, Huang C, Shen H, Sun B, Cheng X. et al. m6A regulates neurogenesis and neuronal development by modulating histone methyltransferase Ezh2. Genomics Proteomics Bioinformatics. 2019;17:154-68
71. Kuppers DA, Arora S, Lim Y, Lim AR, Carter LM, Corrin PD. et al. N6-methyladenosine mRNA marking promotes selective translation of regulons required for human erythropoiesis. Nat Commun. 2019;10:4596
72. Wang L, Wen M, Cao X. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science. 2019;365:eaav0758
73. Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J. et al. METTL3-mediated m6A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69:1193-205
74. Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B. et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer. 2020;19:106
75. Huang H, Weng H, Zhou K, Wu T, Zhao BS, Sun M. et al. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature. 2019;567:414-9
76. Li Y, Xia L, Tan K, Ye X, Zuo Z, Li M. et al. N6-Methyladenosine co-transcriptionally directs the demethylation of histone H3K9me2. Nat Genet. 2020;52:870-7
77. Shim S, Lee HG, Lee H, Seo PJ. H3K36me2 is highly correlated with m6A modifications in plants. J Integr Plant Biol. 2020;62:1455-60
78. Zhang C, Jia G. Reversible RNA modification N1-methyladenosine (m1A) in mRNA and tRNA. Genomics Proteomics Bioinformatics. 2018;16:155-61
79. Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J. et al. ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell. 2016;167:816-28.e16
80. Dai X, Wang T, Gonzalez G, Wang Y. Identification of YTH domain-containing proteins as the readers for N1-methyladenosine in RNA. Anal Chem. 2018;90:6380-4
81. Seo KW, Kleiner RE. YTHDF2 recognition of N1-Methyladenosine (m1A)-modified RNA is associated with transcript destabilization. ACS Chem Biol. 2020;15:132-9
82. Meyer KD. DART-seq: an antibody-free method for global m6A detection. Nat Methods. 2019;16:1275-80
83. Zhou H, Rauch S, Dai Q, Cui X, Zhang Z, Nachtergaele S. et al. Evolution of a reverse transcriptase to map N1-methyladenosine in human messenger RNA. Nat Methods. 2019;16:1281-8
84. Reid R, Greene PJ, Santi DV. Exposition of a family of RNA m(5)C methyltransferases from searching genomic and proteomic sequences. Nucleic Acids Res. 1999;27:3138-45
85. Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X. et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395-8
86. Trixl L, Lusser A. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip Rev RNA. 2019;10:e1510
87. Bohnsack KE, Höbartner C, Bohnsack MT. Eukaryotic 5-methylcytosine (m5C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease. Genes (Basel). 2019;10:102
88. Courtney DG, Chalem A, Bogerd HP, Law BA, Kennedy EM, Holley CL. et al. Extensive epitranscriptomic methylation of A and C residues on murine leukemia virus transcripts enhances viral gene expression. mBio. 2019;10:e01209-19
89. Li Q, Li X, Tang H, Jiang B, Dou Y, Gorospe M. et al. NSUN2-mediated m5C methylation and METTL3/METTL14-mediated m6A methylation cooperatively enhance p21 translation. J Cell Biochem. 2017;118:2587-98
90. Dai X, Gonzalez G, Li L, Li J, You C, Miao W. et al. YTHDF2 binds to 5-methylcytosine in RNA and modulates the maturation of ribosomal RNA. Anal Chem. 2020;92:1346-54
91. Barbon A, Magri C. RNA Editing and Modifications in Mood Disorders. Genes (Basel). 2020;11:872
92. Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol. 2016;17:83-96
93. Chen X, Wan L, Wang W, Xi WJ, Yang AG, Wang T. Re-recognition of pseudogenes: From molecular to clinical applications. Theranostics. 2020;10:1479-99
94. Shang W, Gao Y, Tang Z, Zhang Y, Yang R. The pseudogene Olfr29-ps1 promotes the suppressive function and differentiation of monocytic MDSCs. Cancer Immunol Res. 2019;7:813-27
95. Durbin AF, Wang C, Marcotrigiano J, Gehrke L. RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. mBio. 2016;7:e00833-16
96. Vaidyanathan PP, Alsadhan I, Merriman DK, Al-Hashimi HM, Herschlag D. Pseudouridine and N6-methyladenosine modifications weaken PUF protein/RNA interactions. RNA. 2017;23:611-8
97. Akichika S, Hirano S, Shichino Y, Suzuki T, Nishimasu H, Ishitani R. et al. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science. 2019;363:eaav0080
98. Sendinc E, Valle-Garcia D, Dhall A, Chen H, Henriques T, Navarrete-Perea J. et al. PCIF1 catalyzes m6Am mRNA methylation to regulate gene expression. Mol Cell. 2019;75:620-30
99. Sun H, Zhang M, Li K, Bai D, Yi C. Cap-specific, terminal N6-methylation by a mammalian m6Am methyltransferase. Cell Res. 2019;29:80-82
100. Boulias K, Toczydłowska-Socha D, Hawley BR, Liberman N, Takashima K, Zaccara S. et al. Identification of the m(6)Am Methyltransferase PCIF1 Reveals the Location and Functions of m(6)Am in the Transcriptome. Mol Cell. 2019;75:631-43.e8
101. Pandey RR, Delfino E, Homolka D, Roithova A, Chen KM, Li L. et al. The Mammalian Cap-Specific m(6)Am RNA Methyltransferase PCIF1 Regulates Transcript Levels in Mouse Tissues. Cell Rep. 2020;32:108038
102. Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP. et al. Reversible methylation of m6Am in the 5' cap controls mRNA stability. Nature. 2017;541:371-5
103. Engel M, Eggert C, Kaplick PM, Eder M, Roeh S, Tietze L. et al. The role of m6A/m-RNA methylation in stress response regulation. Neuron. 2018;99:389-403
104. Tan B, Liu H, Zhang S, da Silva SR, Zhang L, Meng J. et al. Viral and cellular N6-methyladenosine and N6, 2'-O-dimethyladenosine epitranscriptomes in the KSHV life cycle. Nat Microbiol. 2018;3:108-20
105. Liu Je, Li K, Cai J, Zhang M, Zhang X, Xiong X. et al. Landscape and regulation of m6A and m6Am methylome across human and mouse tissues. Mol Cell. 2020;77:426-40
106. Wang Y, Wang L, Chen C, Chu X. New insights into the regulatory role of microRNA in tumor angiogenesis and clinical implications. Mol Cancer. 2018;17:22
107. Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20:21-37
108. Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482-5
109. Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC. et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18:110
110. Peng W, Li J, Chen R, Gu Q, Yang P, Qian W. et al. Upregulated METTL3 promotes metastasis of colorectal cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019;38:393
111. Wang J, Ishfaq M, Xu L, Xia C, Chen C, Li J. METTL3/m6A/miRNA-873-5p attenuated oxidative stress and apoptosis in colistin-induced kidney injury by modulating Keap1/Nrf2 pathway. Front Pharmacol. 2019;10:517
112. Wang H, Deng Q, Lv Z, Ling Y, Hou X, Chen Z. et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer. 2019;18:181
113. Yan G, Yuan Y, He M, Gong R, Lei H, Zhou H. et al. m6A methylation of precursor-miR-320/RUNX2 controls osteogenic potential of bone marrow-derived mesenchymal stem cells. Mol Ther Nucleic Acids. 2020;19:421-36
114. Mi B, Xiong Y, Yan C, Chen L, Xue H, Panayi AC. et al. Methyltransferase-like 3-mediated N6-methyladenosine modification of miR-7212-5p drives osteoblast differentiation and fracture healing. J Cell Mol Med. 2020;24:6385-96
115. Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F. et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6-methyladenosine-dependent primary microRNA processing. Hepatology. 2017;65:529-43
116. Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191-8
117. Zhang X, Xu Y, Qian Z, Zheng W, Wu Q, Chen Y. et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. 2018;9:1091
118. Min KW, Zealy RW, Davila S, Fomin M, Cummings JC, Makowsky D. et al. Profiling of m6A RNA modifications identified an age-associated regulation of AGO2 mRNA stability. Aging Cell. 2018;17:e12753
119. Knuckles P, Carl SH, Musheev M, Niehrs C, Wenger A, Buehler M. RNA fate determination through cotranscriptional adenosine methylation and microprocessor binding. Nat Struct Mol Biol. 2017;24:561-9
120. Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W. et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:289-301
121. Yang Z, Li J, Feng G, Gao S, Wang Y, Zhang S. et al. MicroRNA-145 modulates N6-methyladenosine levels by targeting the 3'-untranslated mRNA region of the N6-methyladenosine binding YTH domain family 2 protein. J Biol Chem. 2017;292:3614-23
122. Cui X, Wang Z, Li J, Zhu J, Ren Z, Zhang D. et al. Cross talk between RNA N6-methyladenosine methyltransferase-like 3 and miR-186 regulates hepatoblastoma progression through Wnt/β-catenin signalling pathway. Cell Prolif. 2020;53:e12768
123. Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang Z. et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11-9
124. Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19:143-57
125. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47-62
126. Ji P, Diederichs S, Wang WB, Boing S, Metzger R, Schneider PM. et al. MALAT-1, a novel noncoding RNA, and thymosin beta 4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031-41
127. Brown JA, Kinzig CG, DeGregorio SJ, Steitz JA. Methyltransferase-like protein 16 binds the 3'-terminal triple helix of MALAT1 long noncoding RNA. Proc Natl Acad Sci U S A. 2016;113:14013-8
128. Jin D, Guo J, Wu Y, Du J, Yang L, Wang X. et al. m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2019;12:135
129. Gendrel AV, Heard E. Noncoding RNAs and epigenetic mechanisms during X-chromosome inactivation. Annu Rev Cell Dev Biol. 2014;30:561-80
130. Moindrot B, Cerase A, Coker H, Masui O, Grijzenhout A, Pintacuda G. et al. A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Rep. 2015;12:562-72
131. Dossin F, Pinheiro I, Zylicz JJ, Roensch J, Collombet S, Le Saux A. et al. SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature. 2020;578:455-60
132. Nesterova TB, Wei G, Coker H, Pintacuda G, Bowness JS, Zhang T. et al. Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat Commun. 2019;10:3129
133. Yang X, Zhang S, He C, Xue P, Zhang L, He Z. et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19:46
134. Zheng ZQ, Li ZX, Zhou GQ, Lin L, Zhang LL, Lv JW. et al. Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cancer Res. 2019;79:4612-26
135. Zuo X, Chen Z, Gao W, Zhang Y, Wang J, Wang J. et al. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol. 2020;13:5
136. Ban Y, Tan P, Cai J, Li J, Hu M, Zhou Y. et al. LNCAROD is stabilized by m6A methylation and promotes cancer progression via forming a ternary complex with HSPA1A and YBX1 in head and neck squamous cell carcinoma. Mol Oncol. 2020;14:1282-96
137. Barros-Silva D, Lobo J, Guimaraes-Teixeira C, Carneiro I, Oliveira J, Martens-Uzunova ES. et al. VIRMA-dependent N6-methyladenosine modifications regulate the expression of long non-coding RNAs CCAT1 and CCAT2 in prostate cancer. Cancers. 2020;12:771
138. Chen S, Zhou L, Wang Y. ALKBH5-mediated m6A demethylation of lncRNA PVT1 plays an oncogenic role in osteosarcoma. Cancer Cell Int. 2020;20:34
139. Wang J, Ding W, Xu Y, Tao E, Mo M, Xu W. et al. Long non-coding RNA RHPN1-AS1 promotes tumorigenesis and metastasis of ovarian cancer by acting as a ceRNA against miR-596 and upregulating LETM1. Aging. 2020;12:4558-72
140. Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F. et al. m6A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer. 2019;18:87
141. Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A. et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol Cancer. 2019;18:143
142. Yoneda R, Ueda N, Uranishi K, Hirasaki M, Kurokawa R. Long noncoding RNA pncRNA-D reduces cyclin D1 gene expression and arrests cell cycle through RNA m6A modification. J Biol Chem. 2020;295:5626-39
143. Yang D, Qiao J, Wang G, Lan Y, Li G, Guo X. et al. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018;46:3906-20
144. Liu J, Zhang X, Chen K, Cheng Y, Liu S, Xia M. et al. CCR7 chemokine receptor-inducible lnc-Dpf3 restrains dendritic cell migration by inhibiting HIF-1α-mediated glycolysis. Immunity. 2019;50:600-15
145. Wang Y, Lu JH, Wu QN, Jin Y, Wang DS, Chen YX. et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer. 2019;18:174
146. Zhu L, Zhu Y, Han S, Chen M, Song P, Dai D. et al. Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis. 2019;10:383
147. Wang X, Zhang J, Wang Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am J Transl Res. 2019;11:4909-21
148. Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z. et al. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591-606
149. Lan T, Li H, Zhang D, Xu L, Liu H, Hao X. et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol Cancer. 2019;18:186
150. Zhu S, Wang JZ, Chen D, He YT, Meng N, Chen M. et al. An oncopeptide regulates m6A recognition by the m6A reader IGF2BP1 and tumorigenesis. Nat Commun. 2020;11:1685
151. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205-11
152. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428-42
153. Su M, Xiao Y, Ma J, Tang Y, Tian B, Zhang Y. et al. Circular RNAs in cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer. 2019;18:90
154. Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N. et al. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 2017;20:2262-76
155. Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK. et al. Endoribonucleolytic Cleavage of m6A-Containing RNAs by RNase P/MRP Complex. Mol Cell. 2019;74:494-507
156. Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD. et al. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695
157. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y. et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626-41
158. Di Timoteo G, Dattilo D, Centron-Broco A, Colantoni A, Guarnacci M, Rossi F. et al. Modulation of circRNA metabolism by m6A modification. Cell Rep. 2020;31:107641
159. Zhao J, Lee EE, Kim J, Yang R, Chamseddin B, Ni C. et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat Commun. 2019;10:2300
160. Tang C, Xie Y, Yu T, Liu N, Wang Z, Woolsey RJ. et al. m6A-dependent biogenesis of circular RNAs in male germ cells. Cell Res. 2020;30:211-28
161. Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L. et al. N6-methyladenosine modification controls circular RNA immunity. Mol Cell. 2019;76:96-109
162. Su H, Wang G, Wu L, Ma X, Ying K, Zhang R. Transcriptome-wide map of m6A circRNAs identified in a rat model of hypoxia mediated pulmonary hypertension. BMC Genomics. 2020;21:39
163. Huang R, Zhang Y, Bai Y, Han B, Ju M, Chen B. et al. N6-methyladenosine modification of fatty acid amide hydrolase messenger RNA in circular RNA STAG1-regulated astrocyte dysfunction and depressive-like behaviors. Biol psychiatry. 2020;88:392-404
164. Wu Y, Sang M, Liu F, Zhang J, Li W, Li Z. et al. Epigenetic modulation combined with PD-1/PD-L1 blockade enhances immunotherapy based on MAGE-A11 antigen-specific CD8+T cells against esophageal carcinoma. Carcinogenesis. 2020;41:894-903
165. Buyandelger B, Bar EE, Hung KS, Hung KS, Chen RM, Chiang YH. et al. Histone deacetylase inhibitor MPT0B291 suppresses Glioma Growth in vitro and in vivo partially through acetylation of p53. Int J Biol Sci. 2020;16:3184-99
166. Wang C, Hamacher A, Petzsch P, Köhrer K, Niegisch G, Hoffmann MJ. et al. Combination of Decitabine and Entinostat Synergistically Inhibits Urothelial Bladder Cancer Cells via Activation of FoxO1. Cancers (Basel). 2020;12:337
167. Blagitko-Dorfs N, Schlosser P, Greve G, Pfeifer D, Meier R, Baude A. et al. Combination treatment of acute myeloid leukemia cells with DNMT and HDAC inhibitors: predominant synergistic gene downregulation associated with gene body demethylation. Leukemia. 2019;33:945-56
168. Su Y, Hopfinger NR, Nguyen TD, Pogash TJ, Santucci-Pereira J, Russo J. Epigenetic reprogramming of epithelial mesenchymal transition in triple negative breast cancer cells with DNA methyltransferase and histone deacetylase inhibitors. J Exp Clin Cancer Res. 2018;37:314
169. Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H. et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43:373-84
170. Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G. et al. m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017;18:2622-34
171. Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y. et al. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell. 2018;172:90-105.e23
172. Huff S, Tiwari SK, Gonzalez GM, Wang YS, Rana TM. m(6)A-RNA Demethylase FTO Inhibitors Impair Self-Renewal in Glioblastoma Stem Cells. ACS Chem Biol. 2021 doi: 10.1021/acschembio.0c00841. [Online ahead of print]
173. Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H. et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell. 2019;35:677-91.e10
174. Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y. et al. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019;10:2782
175. Li N, Kang Y, Wang L, Huff S, Tang R, Hui H. et al. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc Natl Acad Sci U S A. 2020;117:20159-70
176. Bedi RK, Huang D, Eberle SA, Wiedmer L, Śledź P, Caflisch A. Small-molecule inhibitors of METTL3, the major human epitranscriptomic writer. ChemMedChem. 2020;15:744-8
Author contact
![]() Corresponding authors: Mei Jin, E-mail: 3193013edu.cn; Jin Wang, E-mail: 3197039edu.cn.
Corresponding authors: Mei Jin, E-mail: 3193013edu.cn; Jin Wang, E-mail: 3197039edu.cn.
 Global reach, higher impact
Global reach, higher impact