13.3
Impact Factor
Theranostics 2023; 13(14):4905-4918. doi:10.7150/thno.86921 This issue Cite
Research Paper
Detection of lineage-reprogramming efficiency of tumor cells in a 3D-printed liver-on-a-chip model
1. Department of Orthopaedic Surgery, Shanghai Key Laboratory of Orthopaedic Implants, Shanghai Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, 200011, China.
2. Wake Forest Institute for Regenerative Medicine, Wake Forest University Health Sciences, Winston-Salem, NC, USA.
3. Department of Cancer Biology, Wake Forest School of Medicine, Winston-Salem, NC, USA.
4. Department of Biomedical Engineering, The Ohio State University, Columbus, OH, USA.
5. Department of Surgery, Division of Surgical Oncology, Wake Forest Baptist Health, Winston‑Salem, NC, USA.
† These authors contributed equally to this work.
Abstract
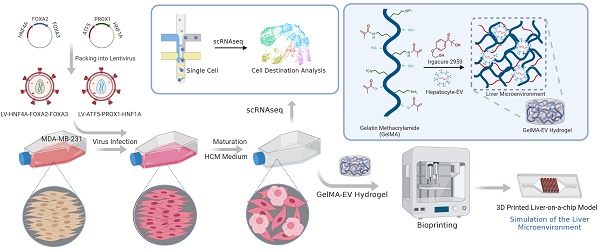
Background: The liver metastasis accompanied with the loss of liver function is one of the most common complications in patients with triple-negative breast cancers (TNBC). Lineage reprogramming, as a technique direct inducing the functional cell types from one lineage to another lineage without passing through an intermediate pluripotent stage, is promising in changing cell fates and overcoming the limitations of primary cells. However, most reprogramming techniques are derived from human fibroblasts, and whether cancer cells can be reversed into hepatocytes remains elusive.
Methods: Herein, we simplify preparation of reprogramming reagents by expressing six transcriptional factors (HNF4A, FOXA2, FOXA3, ATF5, PROX1, and HNF1) from two lentiviral vectors, each expressing three factors. Then the virus was transduced into MDA-MB-231 cells to generated human induced hepatocyte-like cells (hiHeps) and single-cell sequencing was used to analyze the fate for the cells after reprogramming. Furthermore, we constructed a Liver-on-a-chip (LOC) model by bioprinting the Gelatin Methacryloyl hydrogel loaded with hepatocyte extracellular vesicles (GelMA-EV) bioink onto the microfluidic chip to assess the metastasis behavior of the reprogrammed TNBC cells under the 3D liver microenvironment in vitro.
Results: The combination of the genes HNF4A, FOXA2, FOXA3, ATF5, PROX1 and HNF1A could reprogram MDA-MB-231 tumor cells into human-induced hepatocytes (hiHeps), limiting metastasis of these cells. Single-cell sequencing analysis showed that the oncogenes were significantly inhibited while the liver-specific genes were activated after lineage reprogramming. Finally, the constructed LOC model showed that the hepatic phenotypes of the reprogrammed cells could be observed, and the metastasis of embedded cancer cells could be inhibited under the liver microenvironment.
Conclusion: Our findings demonstrate that reprogramming could be a promising method to produce hepatocytes and treat TNBC liver metastasis. And the LOC model could intimate the 3D liver microenvironment and assess the behavior of the reprogrammed TNBC cells.
Keywords: Direct reprogramming, Induced hepatocytes, Triple-negative breast cancers, 3D printing, Organ-on-a-chip
 Global reach, higher impact
Global reach, higher impact