13.3
Impact Factor
Theranostics 2019; 9(14):4101-4114. doi:10.7150/thno.35186 This issue Cite
Research Paper
Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer
1. Department of Colorectal Surgery, Fudan University Shanghai Cancer Center, Shanghai, 200032, China
2. Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, 200032, China
3. Department of GI Surgery, Shanghai Tenth People's Hospital Affiliated to Tongji University, Shanghai, 200072, China
4. Center for Precision Medicine, Department of Internal Medicine, Section of Molecular Medicine, Medical Center Boulevard, Winston-Salem, NC 27157, USA.
5. Department of Pathology, Shanghai Tenth People's Hospital Affiliated to Tongji University, Shanghai 200072, China
6. Department of Clinical Laboratory, Yangpu Hospital, Tongji University School of Medicine, Shanghai, China
7. Center for Translational Medicine, Yangpu Hospital, Tongji University School of Medicine, Shanghai, China
8. Center for Gastrointestinal Research, Center for Epigenetics, Cancer Prevention and Cancer Genomics, Baylor Scott &White Research Institute and Charles A. Sammons Cancer Center, Texas, USA
#Equal contribution
Abstract
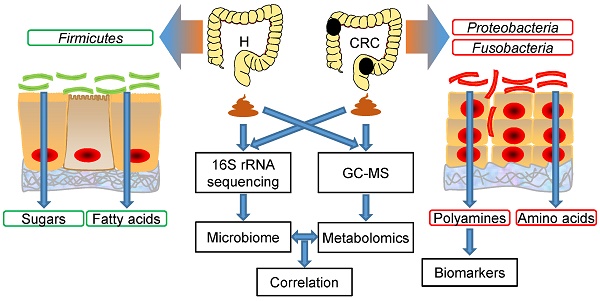
Rationale: Colorectal cancer (CRC) is a malignant tumor with the third highest morbidity rate among all cancers. Driven by the host's genetic makeup and environmental exposures, the gut microbiome and its metabolites have been implicated as the causes and regulators of CRC pathogenesis. We assessed human fecal samples as noninvasive and unbiased surrogates to catalog the gut microbiota and metabolome in patients with CRC.
Methods: Fecal samples collected from CRC patients (CRC group, n = 50) and healthy volunteers (H group, n = 50) were subjected to microbiome (16S rRNA gene sequencing) and metabolome (gas chromatography-mass spectrometry, GC-MS) analyses. The datasets were analyzed individually and integrated for combined analysis using various bioinformatics approaches.
Results: Fecal metabolomic analysis led to the identification of 164 metabolites spread across 40 metabolic pathways in both groups. In addition, there were 42 and 17 metabolites specific to the H and CRC groups, respectively. Sequencing of microbial diversity revealed 1084 operational taxonomic units (OTUs) across the two groups, and there was less species diversity in the CRC group than in the H group. Seventy-six discriminatory OTUs were identified for the microbiota of H volunteers and CRC patients. Integrated analysis correlated CRC-associated microbes with metabolites, such as polyamines (cadaverine and putrescine).
Conclusions: Our results provide substantial evidence of a novel interplay between the gut microbiome and metabolome (i.e., polyamines), which is drastically perturbed in CRC. Microbe-associated metabolites can be used as diagnostic biomarkers in therapeutic explorations.
Keywords: colorectal cancer, gut, microbiome, metabolomics, biomarkers
 Global reach, higher impact
Global reach, higher impact