13.3
Impact Factor
Theranostics 2024; 14(3):973-987. doi:10.7150/thno.90246 This issue Cite
Research Paper
Deep learning for automatic organ and tumor segmentation in nanomedicine pharmacokinetics
1. Princess Margaret Cancer Centre, University Health Network, 101 College Street, Toronto, M5G 1L7, Ontario, Canada.
2. Department of Medical Biophysics, University of Toronto, 101 College Street, Toronto, M5G 1L7, Ontario, Canada.
3. Department of Laboratory Medicine and Pathobiology, University of Toronto, 1 King's College Circle, Toronto, M5S 1A8, Ontario, Canada
4. Peter Munk Cardiac Centre, University Health Network, 190 Elizabeth St, Toronto, M5G 2C4, Ontario, Canada.
5. Department of Computer Science, University of Toronto, 101 College Street, Toronto, M5G 1L7, Ontario, Canada.
6. Institute of Biomedical Engineering, University of Toronto, 101 College Street, Toronto, M5G 1L7, Ontario, Canada.
7. Vector Institute for Artificial Intelligence, 661 University Avenue, Toronto, M4G 1M1, Ontario, Canada.
8. Techna Institute, University Health Network, 190 Elizabeth Street, Toronto, M5G 2C4, Ontario, Canada.
9. Department of Radiology, Stanford University, 1201 Welch Road, Stanford, 94305-5484, California, United States of America.
Abstract
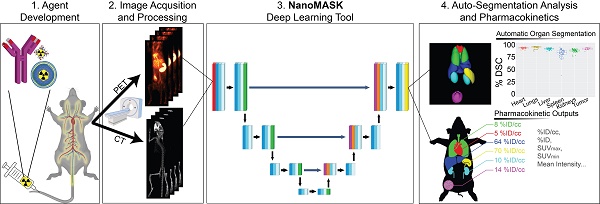
Rationale: Multimodal imaging provides important pharmacokinetic and dosimetry information during nanomedicine development and optimization. However, accurate quantitation is time-consuming, resource intensive, and requires anatomical expertise.
Methods: We present NanoMASK: a 3D U-Net adapted deep learning tool capable of rapid, automatic organ segmentation of multimodal imaging data that can output key clinical dosimetry metrics without manual intervention. This model was trained on 355 manually-contoured PET/CT data volumes of mice injected with a variety of nanomaterials and imaged over 48 hours.
Results: NanoMASK produced 3-dimensional contours of the heart, lungs, liver, spleen, kidneys, and tumor with high volumetric accuracy (pan-organ average %DSC of 92.5). Pharmacokinetic metrics including %ID/cc, %ID, and SUVmax achieved correlation coefficients exceeding R = 0.987 and relative mean errors below 0.2%. NanoMASK was applied to novel datasets of lipid nanoparticles and antibody-drug conjugates with a minimal drop in accuracy, illustrating its generalizability to different classes of nanomedicines. Furthermore, 20 additional auto-segmentation models were developed using training data subsets based on image modality, experimental imaging timepoint, and tumor status. These were used to explore the fundamental biases and dependencies of auto-segmentation models built on a 3D U-Net architecture, revealing significant differential impacts on organ segmentation accuracy.
Conclusions: NanoMASK is an easy-to-use, adaptable tool for improving accuracy and throughput in imaging-based pharmacokinetic studies of nanomedicine. It has been made publicly available to all readers for automatic segmentation and pharmacokinetic analysis across a diverse array of nanoparticles, expediting agent development.
Keywords: Deep Learning, Nanomedicine, Pharmacokinetics, Auto-Segmentation, Radioimaging, Functional Imaging, Multimodal, Multiparameter, Contouring, PET, CT
 Global reach, higher impact
Global reach, higher impact