13.3
Impact Factor
Theranostics 2012; 2(1):55-65. doi:10.7150/thno.3465 This issue Cite
Review
Magnetic Nanoparticles and microNMR for Diagnostic Applications
1. Center for Systems Biology, Massachusetts General Hospital, 185 Cambridge St, CPZN 5206, Boston, MA 02114;
2. Department of Systems Biology, Harvard Medical School, 200 Longwood Ave, Boston, MA 02115, USA
# Present address: Department of Applied Bioscience, CHA University, Seoul 135-081, Republic of Korea.
Received 2011-8-31; Accepted 2011-10-3; Published 2012-1-1
Abstract
Sensitive and quantitative measurements of clinically relevant protein biomarkers, pathogens and cells in biological samples would be invaluable for disease diagnosis, monitoring of malignancy, and for evaluating therapy efficacy. Biosensing strategies using magnetic nanoparticles (MNPs) have recently received considerable attention, since they offer unique advantages over traditional detection methods. Specifically, because biological samples have negligible magnetic background, MNPs can be used to obtain highly sensitive measurements in minimally processed samples. This review focuses on the use of MNPs for in vitro detection of cellular biomarkers based on nuclear magnetic resonance (NMR) effects. This detection platform, termed diagnostic magnetic resonance (DMR), exploits MNPs as proximity sensors to modulate the spin-spin relaxation time of water molecules surrounding the molecularly-targeted nanoparticles. With new developments such as more effective MNP biosensors, advanced conjugational strategies, and highly sensitive miniaturized NMR systems, the DMR detection capabilities have been considerably improved. These developments have also enabled parallel and rapid measurements from small sample volumes and on a wide range of targets, including whole cells, proteins, DNA/mRNA, metabolites, drugs, viruses and bacteria. The DMR platform thus makes a robust and easy-to-use sensor system with broad applications in biomedicine, as well as clinical utility in point-of-care settings.
Keywords: biosensor, diagnostics, magnetic nanoparticle, microfluidics, nuclear magnetic resonance.
INTRODUCTION
Rapid and sensitive characterization of rare biomarkers in easily accessible bodily sources (e.g., fine needle aspirates, biopsies, and whole blood) will have significant impact on life sciences and clinical practice [1, 2]. Such diagnostic platforms could be used for screening and early diagnosis, comprehensive disease characterization in patients, and targeted therapy based on personal responses to treatments [3, 4]. Clinical detection technologies ideally should 1) provide high sensitivity and specificity, 2) enable rapid measurements with minimal sample processing, and 3) allow for multiple biomarker detection in a single parent sample (multiplexed detection) for accurate diagnosis [5]. A number of sensors fulfilling some of these criteria have been developed based on optical [6, 7], electronic [8, 9] or magnetic detection [10, 11]. The clinical adaptation of these systems, however, is still limited, because they often require lengthy sample purification or long assay times.
Biosensing strategies based on magnetic nanoparticles (MNPs) have recently received considerable attention. Magnetic fields experience little interference from native biological samples as most biological entities have negligible magnetic susceptibilities, similar to that of water (~10-6). Even optically turbid samples thus will most often appear transparent to magnetic fields. However, when cells of interests are magnetically tagged, they will attain a high contrast against the biological background. To detect biomarkers using MNPs, various detection technologies have been developed [12]. These include techniques that use magnetometers, such as superconducting quantum interference device (SQUID) [13-15], magnetoresistive sensors [16-19], and Hall sensors [20], which directly measure the magnetic fields from magnetically-labeled biological targets.
We have recently developed a new magnetic sensing platform, diagnostic magnetic resonance (DMR) [21]. In contrast to direct detection of magnetic moments with magnetometers, which requires MNP-labeled targets to be closely positioned to the sensing elements, DMR exploits nuclear magnetic resonance (NMR) as the detection mechanism. When placed in NMR magnetic fields, MNPs create local magnetic fields, which change the relaxation rate of surrounding water molecules [22]. The mechanism simplifies and expedites the detection assays since the analytical signal is amplified and generated from the entire sample volume. By optimizing MNPs and miniaturizing NMR detectors, the DMR detection sensitivity for various target types have been considerably improved over the last few years. These developments nowadays enable rapid and multiplexed detection on a wide range of targets in microliter sample volumes, including nucleic acids [23], proteins [21], drugs, bacteria [24], and tumor cells [25-28]. With the recent integration of bioorthogonal targeting strategies [26, 29, 30] as well as accurate and real-time control of device temperature [31], the DMR platform has become more robust and sensitive, allowing operation in clinical settings [25]. This review will report on the latest developments of new MNPs and DMR systems for improving detection sensitivities. Specific biomedical and clinical DMR applications will also be discussed.
PRINCIPLE OF DMR ASSAY
Due to their small size, MNPs assume unique physical properties which are different from those of bulk materials. The most salient feature is the paramagnetic behavior of an ensemble of small MNPs, termed superparamagnetism [32]. For most magnetic materials (e.g., ferrite and iron), MNPs with diameter of < 20 nm have a single domain with magnetic moments confined in a particular direction defined by magnetic anisotropy. At sufficiently high temperature, thermal fluctuation can overcome the anisotropy barrier and spontaneously flip the magnetic moments of MNPs [33]. An ensemble of MNPs consequently displays negligible remnant magnetic moments in the absence of external magnetic fields, but the magnetic moments grow with increasing external magnetic fields. This superparamagnetic property ensures that MNPs do not spontaneously aggregate under physiological solutions. A MNP is typically comprised of an inorganic magnetic core and a biocompatible surface coating, which can be modified with functional ligands to confer the MNP with molecular specificity.
By producing local magnetic dipoles with strong spatial dependence, MNPs efficiently destroy the coherence in the spin-spin relaxation of water protons. The net effect is a change in magnetic resonance signal, which is measured as a shortening of the longitudinal (T1, spin-lattice) and transverse (T2, spin-spin) relaxation times. The capacities of MNPs to decrease T2 and T1 are respectively defined as the transverse (r2) and the longitudinal (r1) relaxivities. Typically, because the transverse relaxivities (r2) of MNPs are greater than their longitudinal relaxivities (r1), T2 is used for NMR-based biosensing applications. With a higher r2 relaxivity, fewer numbers of nanoparticles are required to produce detectable T2 changes.
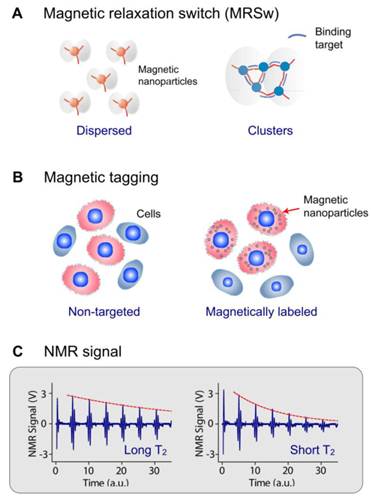
DMR detection exploits targeted MNPs to modulate the spin-spin T2 relaxation time of biological samples. Depending on the size of the target biomarker, DMR assays can take two forms. For the detection of small molecular analytes such as metabolites, oligonucloetides, and proteins, magnetic relaxation switching (MRSw) effect can be exploited. MRSw relies on the changes of organizational state of MNPs in solution. When MNPs cluster in solution, the aggregates will assume different r2 values, causing corresponding T2 changes. MRSw assays are performed without removing excess unbound MNPs (Fig. 1A), and thereby facilitate the detection of small molecules. For larger biological targets such as bacteria, mammalian cells or cellular components, cell biomarkers can be tagged with functional MNPs and unbound MNPs are removed (Fig. 1B). This gain of magnetic moment (change of 1/T2) is proportional to the number of MNPs bound, and also indicative of the abundance of relevant biomarkers (Fig. 1C).
MAGNETIC NANOPARTICLES AND THEIR RELAXATION PROPERTIES
Many different types of MNPs have been explored in an effort to maximize DMR detection sensitivity. In addition to having good stability in aqueous media, newer generations of MNPs are often engineered to have very high transverse relaxivity (r2) in order to induce pronounced T2 changes. According to the outer-sphere model, the transverse relaxivity of a MNP is proportional to τd ·M2, where τd is the residence time of water molecules around the particle and M is the particle magnetization [34]. The efforts to enhance r2 were thus focused on synthesizing larger MNPs using magnetically stronger materials.
DMR assay configurations. (A) Magnetic relaxation switching (MRSw) assays detect changes in the organizational states of magnetic nanoparticles (MNPs) in solution. MRSw assays can be designed to cause forward switching, whereby molecular targets cross-link MNPs to induce MNP clustering, thus effecting a corresponding decrease in T2. Alternatively, the assays can cause reverse switching, where enzymatic cleavage or competitive binding of molecular targets disassembles pre-formed clusters to cause an increase in T2. (B) Magnetic tagging assays detect the presence of bound MNPs on larger biological entities. Bound MNPs impart a magnetic moment to tagged cells, leading to a decrease in T2 relaxation time. Unbound MNPs must be removed to ensure detection sensitivity. (C) This gain of magnetic moment (change of 1/T2) is proportional to the number of MNPs bound; shorter T2 indicates higher expression of the relevant biomarkers. (Reproduced with permission from [23] [21]. Copyright 2002, 2008 Nature Publishing Group.)
Ferrite-based MNPs
Among different ferrite particles, cross-linked iron oxide (CLIO) nanoparticles have been widely used for DMR applications, notably because of their excellent stability and biocompatibility [35]. CLIO nanoparticles contain a superparamagnetic iron oxide core (3-5 nm monocrystalline iron oxide) composed of ferrimagnetic magnetite (Fe3O4) and/or maghemite (γFe2O3). The metallic core is coated with biocompatible dextran, before being cross-linked and functionalized with primary amine. Amine-terminated CLIO nanoparticles have an average hydrodynamic diameter of 25-40 nm, approximately 40-80 amines per nanoparticle for bioconjugation, and a r2 of ~50 s-1.mM-1 [Fe] [23, 36].
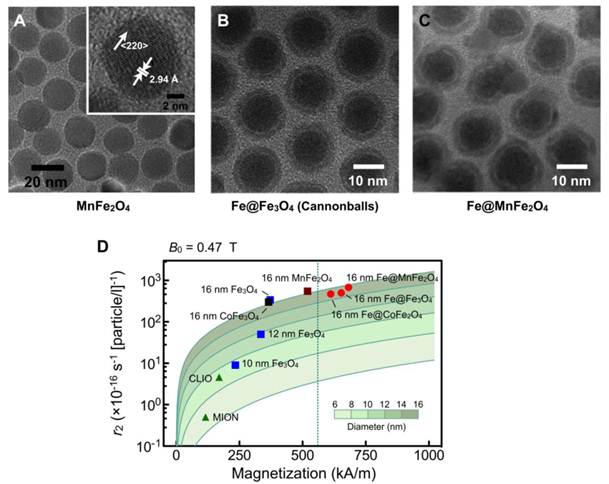
To enhance the magnetization of ferrite nanoparticles, two main strategies have been employed, namely magnetic doping and nanoparticle sizing. Doping of ferrite with ferromagnetic elements such as manganese (Mn), cobalt (Co) or nickel (Ni) [24, 37, 38] has been shown to improve the MNP magnetization. Among the singly-doped ferrite MNPs, MnFe2O4 nanoparticles have the highest magnetization and r2 value, on account of their electron spin configurations, followed by FeFe2O4, CoFe2O4, and NiFe2O4. More recently, it has been demonstrated that magnetization can be further enhanced via additional Zn2+ doping in MnFe2O4 nanoparticles [39]. In addition, nanoparticle magnetization is known to increase with particle size [40]. Ideally, each magnetic spin within a bulk magnetic material would align parallel to the external magnetic field. However, in the nanoscale regime, surface spins tend to be tilted (spin canting), a feature that decreases the overall magnetic moment. By increasing the MNP size, this surface effect is reduced to increase the overall magnetization; the increase in particle size further enhances the particle r2.
Both magnetic doping and sizing strategies were employed by our laboratory to produce MnFe2O4 nanoparticles with superior r2 relaxivity [28]. These particles were synthesized by reacting iron (III) acetylacetonate [Fe(acac)3], manganese (II) acetylacetonate [Mn(acac)2] and 1,2-hexadecanediol at high temperature (300 °C). Through a seed-mediated growth approach, the magnetic core diameter was increased from 10 nm to 12, 16, or 22 nm. MnFe2O4 nanoparticles with diameter ≤ 16 nm were found to be highly monodisperse and superparamagnetic at 300 K (Fig. 2A). The MNPs were subsequently rendered water-soluble using the small molecule meso-2,3-dimercaptosuccinic acid (DMSA) [24, 40, 41]. Due to DMSA's small size, the hydrodynamic diameter of MnFe2O4 nanoparticles was found to be smaller than that of CLIO nanoparticles, despite their larger magnetic core. More importantly, these MnFe2O4 nanoparticles assumed high relaxivities with r2 values reaching 420 s-1.mM-1[metal] (equal to 6×10−14 L·s−1 per particle), more than 8 times greater than CLIO nanoparticles in metal basis (50 s-1.mM-1[metal] or 7×10−16 L·s−1 per particle) [28].
Fe-core MNPs
Ferromagnetic metals, rather than their corresponding oxides, have been suggested as an ideal constituent for MNPs for their superior magnetization. This motivates the creation of Fe-core MNPs to achieve high r2 relaxivities [42, 43]. However, because Fe cores are extremely reactive, monometallic MNPs typically require protective layers to prevent progressive oxidation.
Recently, a unique 16 nm Fe-core/ferrite shell MNP, known as “cannonball”, has been developed for DMR applications (Fig. 2B) [44]. These structures are unique in that they contain elemental iron as core (not iron oxide) and a protecting oxide shell. These cannonballs were synthesized by thermally decomposing iron (0) pentacarbonyl [Fe(CO)5] to form the Fe core. A protective ferrite shell was formed by controlled oxidation with oxygen gas; this resulted in a thinner shell and retained a larger Fe core. The cannonballs were then coated with DMSA as described above. Because of their large Fe core, superparamagnetic cannonballs showed high magnetization (763 kA·m-1). The relaxivity of cannonballs is similar to that of MnFe2O4 nanoparticles (6×10−14 L·s−1 per particle) (Fig. 2D).
Higher r2-relaxivity MNPs developed to enhance in vitro detection sensitivity. (A) Transmission electron micrograph (TEM) images of manganese-doped ferrite nanoparticles (MnFe2O4). These nanoparticles have narrow size distribution and high crystallinity, and were synthesized using a seed-growth method to produce 10, 12, 16, and 22 nm nanoparticles. (B) TEM image of elemental iron (Fe) core/ferrite shell magnetic nanoparticles (CB; cannonballs). These particles have a large Fe core (11 nm) passivated with a thin ferrite shell (2.5 nm) that is produced through controlled oxidation. (C) TEM image of Fe core/ferrite shell magnetic nanoparticles with tunable core size and shell composition. The Fe cores were enlarged into a thermally stable ferromagnetic state to increase the overall magnetization. Protective ferrite shells were then grown onto the cores, without oxidizing the cores, and metal-doped to further enhance magnetization. The resultant particles displayed hysteresis with negligible coercivity. (D) Comparison of size, r2 relaxivity, and saturated magnetization of various doped-ferrite and elemental Fe-based nanoparticles. CLIO, cross-linked iron oxide; MION, monocrystalline iron oxide. (Adapted with permission from [28] Copyright 2009 National Academy of Sciences, USA. Reproduced with permission from [44]. Copyright 2009 John Wiley and Sons, Inc. Reproduced with permission from [45] Copyright 2011 John Wiley and Sons, Inc.)
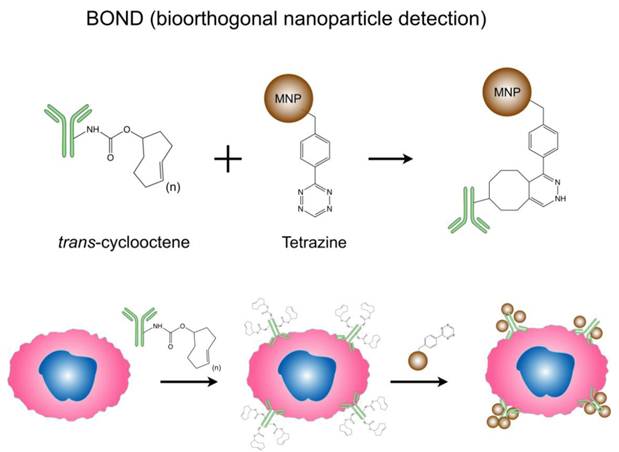
Bioorthogonal nanoparticle detection (BOND) strategy for DMR detection. Schematics showing the conjugation chemistry and coupling between antibody and nanoparticle. Antibodies against biomarkers were modified with trans-cyclooctene (TCO) and used as scaffolds to couple more tetrazine (Tz) modified nanoparticles onto live cells. The strategy is fast, catalyst-free, specific even in complex biological environment, and amplifies biomarker signals. (Reproduced with permission from [26]. Copyright 2010 Nature Publishing Group.)
We have further advanced Fe-core MNPs by developing a new synthetic approach for tunable core size and shell composition [45]. These new generation MNPs consist of an Fe core and an artificial ferrite shell (Fe@MFe2O4, M = Fe, Mn, Co). The Fe cores were enlarged into a thermally stable ferromagnetic state to increase the overall magnetization. Instead of traditionally oxidizing the Fe core to form the shell, protective ferrite shells were grown onto the cores and metal-doped to further enhance magnetization. The resultant particles displayed a unique magnetic feature, the presence of hysteresis with negligible coercivity. Further analysis revealed a novel magnetization process wherein the shell effectively reduces the coercivity of the ferromagnetic cores by leading the magnetization process at small magnetic fields. The resulting particles, especially Fe@MnFe2O4 MNPs (Fig. 2C) attained high saturation magnetization (796 kA·m-1) and r2 (7×10−14 L·s−1 per particle; 430 s-1·mM-1[metal]) (Fig. 2D) but with negligible remanence to prevent inter-particle aggregations. When applied for DMR assays, these Fe@MnFe2O4 MNPs achieved superior performance, capable of detecting picomolar avidin and single cancer cells in whole blood samples.
Bioorthogonal nanoparticle detection (BOND)
In addition to the previous strategies to enhance MNP relaxivities, surface modification of particles and novel targeting approach also improve DMR sensing capacities. Bioorthogonal cycloaddition between a 1,2,4,5-tetrazine (Tz) and a trans-cyclooctene (TCO) is a fast and chemoselective reaction that does not require a catalyst, and can be used for small molecule labeling in serum [46, 47]. Recently, this chemistry has been successfully adapted to MNP targeting, and has been shown to improve nanoparticle binding valency and detection sensitivity. Termed 'bioorthogonal nanoparticle detection' (BOND), this novel targeting platform employs Tz and TCO as the coupling agents [26]. Specific antibodies are first modified with TCO; once bound to cellular biomarkers of interest, the TCO-modified antibodies facilitate the coupling of Tz-modified nanoparticles (Fig. 3). Because of the small size of the coupling reagents, their high multiplicity on antibodies/nanoparticles resulted in higher nanoparticle binding to cells. This not only amplifies the biomarker signals but also significantly improves the detection sensitivity of DMR assays. Moreover, this platform is broadly-applicable for both extracellular and intracellular biomarker detection, and scalable for clinical uses [27, 48]. Already, BOND has been successfully adapted for DMR molecular profiling of experimental cellular samples [26] and clinical fine needle aspirate samples [25]. Recently, we have developed newer two-step detection schemes based on complementary oligonucleotide approaches [29], alternative cycloaddition chemistries [49] and cyclodextrin/adamantine supramolecular interactions [30].
MINIATURIZED NMR SYSTEMS
A key to sensitive detection in DMR is the miniaturized nuclear magnetic resonance (µNMR) systems (Fig. 4) [21]. The miniaturization of the NMR device brings distinctive advantages. First, it lowers the detection limit by reducing the sample volumes and hence effectively increasing analyte concentrations [50]. Second, miniaturized NMR probes (coils) produce much stronger radio-frequency (RF) magnetic fields per unit current, leading to higher signal-to-noise per unit sample volume [51]. Third, with smaller RF coils, the requirement for spatial homogeneity of static magnetic fields becomes less stringent, making it possible to use small, portable magnets [21].
The first prototype (µNMR-1; Fig. 4a) was designed to test the feasibility of miniaturization. The system consisted of a microfabricated NMR probe, a small permanent magnet (B = 0.5 T), custom-designed NMR electronics, and a microfluidic network [21]. The probe had multiple planar microcoils (a 2 × 4 array) to enable parallel detection. In this system, the coils excite biological samples by emitting RF pulses, and subsequently receive resulting NMR signals. The microfluidic networks provide efficient mixing between the samples and MNPs and guide the samples to eight different planar coils. To compensate for the magnetic field inhomogeneity from the small permanent magnet, Carr-Purcell-Meiboom-Gill (CPMG) spin-echo pulse sequences were used for transverse (T2) relaxation time measurements. To generate versatile pulse sequences while using minimal electronic parts, we devised a new circuit schematic for NMR electronics, that has served as a blueprint for subsequent NMR systems.
The second prototype (µNMR-2) was designed to improve the DMR detection sensitivity by optimizing the NMR probe (Fig. 4B) [28, 44]. We selected wire-wound solenoidal coils as the probe format, as they could produce more homogeneous RF magnetic fields and have less electrical resistance. The overall signal-to-noise ratio (SNR) was further enhanced by fully embedding the coils into the fluidic system. In brief, solenoidal coils were prepared by winding insulated wires around a polyethylene tube. The coil-tube assembly was placed on a template for microfluidic systems, and cast-molded by pouring polydimethylsiloxane (PDMS). After PDMS curing, the tube was extracted to open up the entire coil-bore for fluidic access for high filling factor (≈ 1). Combined with lower electrical noise of the coils, the µNMR-2 showed 10-fold higher mass detection sensitivity than µNMR-1, and enabled highly sensitive detection (single mammalian cells, a few bacteria). In parallel with the probe development, we also strategized to miniaturize the entire NMR system. Particularly, we custom-designed a CMOS (complementary-metal-oxide semiconductor) chip (1.4 × 1.4 mm2 ) that integrates all NMR electronics (Fig. 4C) [52, 53]. The whole NMR setup could be packaged as a mobile hand-held device, making µNMR-2 the world's smallest NMR system.
Most recently, we have developed a third generation µNMR-3 device optimized for routine clinical applications (Fig. 4D) [31]. A major challenge in moving µNMR from a basic science laboratory to a clinical environment has been the system's inherent sensitivity to temperature. The latter originates from the temperature-dependent fluctuation of the magnetic field generated by the permanent magnet. These fluctuations lead to drifts in the NMR frequency, resulting in an artifact in the measured T2 values. In a laboratory setting, the problem can be addressed by controlling the environmental and system temperatures. Such a solution, however, significantly increases the cost and size of the µNMR system, undermining its use for point-of-care applications. In the new µNMR-3, we solved this problem through an electronic approach: we implemented a feedback routine that automatically tracks temperature changes and correspondingly reconfigures the measurement settings. This solution guarantees reliable and robust DMR measurements. For example, when the tracking routine was turned off, T2 values fluctuated up to 200% of its initial value in a typical laboratory setting. With the tracking on, however, T2 variations considerably decreased (< 1%; Fig. 4D right).
Miniaturized devices developed for DMR biosensing. (A) The first-generation miniaturized device (µNMR-1) to measure relaxation times of biological samples consists of an array of microcoils for NMR measurements, microfluidic networks for sample handling and mixing, miniaturized NMR electronics and a portable permanent magnet to generate a polarizing magnetic field. (B) The second-generation (µNMR-2) consists of a solenoidal coil embedded in a microfluidic device. As compared to the previous generation, this improved device has a higher filling factor, better signal-to-noise ratio, and reduces sample volume requirement to ~1 μl. (C) 0.1 kg “palm” µNMR-2 system is 20× lighter and 30× smaller than previous generation. To achieve this significant size reduction, a small 0.56 T magnet was used. To compensate for the signal reduction from the smaller magnet, this device incorporates a new RF transceiver fully integrated in the 0.18-μm CMOS. (D) The latest generation of µNMR is equipped with the capability to automatically track and compensate for temperature changes, ensuring robust and reliable T2 measurements. With the temperature compensation turned on, the measured T2 values of the same sample showed <1% variation, whereas T2 values fluctuated >200% without such scheme (right). The new system communicates with mobile devices (smartphones, tablet computers) for easy system control and data logging. (Reproduced with permission from [24]. Copyright 2008 Nature Publishing Group. Reproduced with permission from [28]. Copyright 2009 National Academy of Sciences, USA. Reproduced with permission from [52]. Copyright 2010 IEEE. Reproduced with permission from [31]. Copyright 2011 RSC Publishing.)
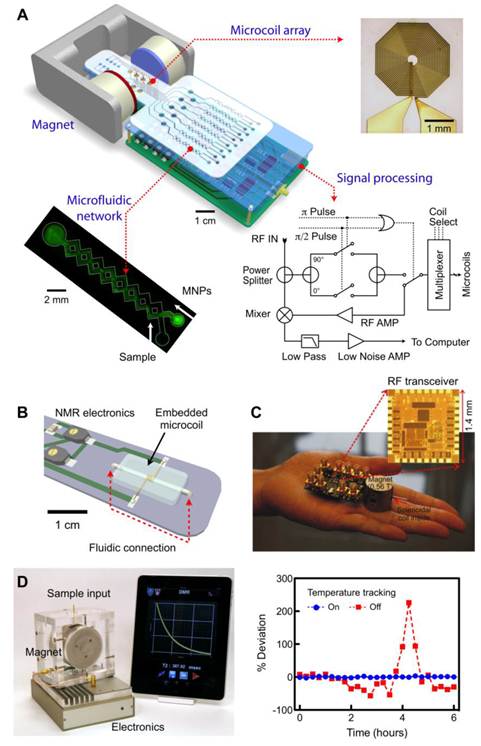
The µNMR-3 features other important aspects for practical use. Unlike the previous generations of µNMR, small-form factor NMR electronics (20 × 12 × 5 cm3) were assembled using off-the-shelf integrated circuit (IC) chips (e.g., microcontrollers, RF synthesizers), reducing the cost of the system (<$200) and enhancing the programmability. Moreover, the µNMR-3 interfaces with mobile devices (e.g., iPhone™, iPAD™) for its operation. This maximizes the portability of µNMR-3 and improves user-friendliness; medical personnel can obtain sample data promptly via customized DMR applications and achieve real-time data logging/sharing over an encrypted wireless network in a remote clinical site. We have recently employed this system in several clinical trials of cancer detection and profiling.
To date, the DMR platform has been successfully applied to sensitively quantify a wide range of biological targets including DNA/mRNA [23, 54], proteins [21, 23, 55], enzyme activities [23, 56, 57], small molecules/drugs [58, 59], bacteria [21, 44], viruses [60] and mammalian tumor cells [21, 25, 26, 28]. As described previously, the detection mode of DMR depends on the size of its targets. For small targets such as oligonucleotides, proteins and other small molecule metabolites, MRSw sensing has been used. In these examples, MNPs were conjugated with targeting ligands to function as multivalent sensors. In the presence of the analytes, MNP binding and subsequent clustering causes corresponding T2 changes to indicate the quantity of the targets. Moreover, such assay strategy can also be used to analyze enzymatic activities. For example, specific MNPs have been designed to assess human telomerase (hTERT) activity by hybridizing with the 30-base pair telomeric repeat sequences produced by hTERT activity [54]. More recently, myeloperoxidase (MPO) sensors were generated by attaching phenol-containing molecules, such as dopamine or serotonin, to CLIO nanoparticles [61]. In the presence of peroxidase activity, tyroxyl radicals were formed to cross-link the nanoparticles. Using this assay configuration, leukocyte-derived MPO has been shown to play a critical role in the pathogenesis of atherosclerotic plaques.
CANCER DETECTION AND PROFILING
Sensitive detection and rapid profiling of tumor cells in unprocessed biological samples will have significant impact on both biomedical research and clinical practice. DMR molecular profiling of cancer markers (HER2/neu, EGFR, and EpCAM) on mammalian cells was initially demonstrated using the first-generation DMR device (µNMR-1) [21]. In these early experiments, CLIO nanoparticles were directly conjugated to monoclonal antibodies. Using the second-generation μNMR-2 and the highly magnetic MnFe2O4 nanoparticles, cellular detection sensitivity was remarkably improved [28].
Tumor cell detection and profiling with DMR. (A) The detection sensitivity of DMR was approximately two cells (in 1 µl sample volume) using the improved µNMR-2 device (Figure 4B) and the highly magnetic MnFe2O4 nanoparticles (Figure 2A) , making this detection platform superior to current clinical methods (cytology and histology). (B and C) Different human breast cancer cells with varying expression of HER2/neu and control cells were labeled with MnFe2O4 nanoparticles. DMR measurements correlated well with standard molecular analyses such as flow cytometry, Western blot and immunofluorescence, but required substantially fewer cells. (Reproduced with permission from [28]. Copyright 2009 National Academy of Sciences, USA.)
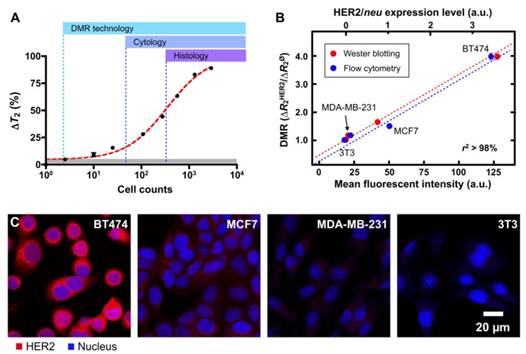
Notably, as shown in Fig. 5A, the detection threshold was reduced to approximately single-cell level, far surpassing the sensitivity of other conventional clinical methods. There was also a good correlation between DMR and other standard methods (e.g., microscopy, flow cytometry and Western blotting; Figs. 5B, C).
Through these sets of early experiments, the DMR detection platform was shown to not only require far fewer cells than standard alternative approaches, but also produce results much faster (< 15 minutes). In addition, the DMR platform is ideally suited for rapid multi-target detection, since it can make measurements on few cells in small sample volumes and in a multiplexed manner. The use of BOND strategy has further advanced DMR profiling capabilities. As a universal labeling approach, BOND simplifies the preparation of the targeted MNPs for multiplexing and amplifies nanoparticle binding to cells. As a proof-of-principle analysis, cancer cells were targeted with CLIO nanoparticles via BOND. At a low cell count (~1000 cells per sample), parallel DMR measurements could be performed rapidly [26].
Through the remarkable integration of the DMR and BOND technologies, this chip-based NMR detection platform has been recently applied in clinical trials of cancer cell profiling [25]. Single fine-needle aspirates were obtained from a cohort of 50 patients and analyzed directly with the DMR platform for real-time quantification of multiple protein markers. A surprising degree of heterogeneity in protein expression both across the different patient samples and even with the same tumor was observed (Fig. 6A).
Clinical applications of DMR in tumor diagnostics. Single fine-needle aspirates were obtained from a cohort of patients, and were tagged via the BOND strategy for DMR detection. (A) Real-time quantification of multiple protein markers with the DMR platform indicates a high degree of heterogeneity in protein expression both across the different patient samples and even with the same tumor. (B) Using a four-protein signature (HER2/neu, EGFR, EpCAM and Muc-1), the DMR platform reports a 96% accuracy for establishing a cancer diagnosis, surpassing conventional clinical analyses by immunohistochemistry. (Reproduced with permission from [25]. Copyright 2011 American Association for the Advancement of Science AAAS.)
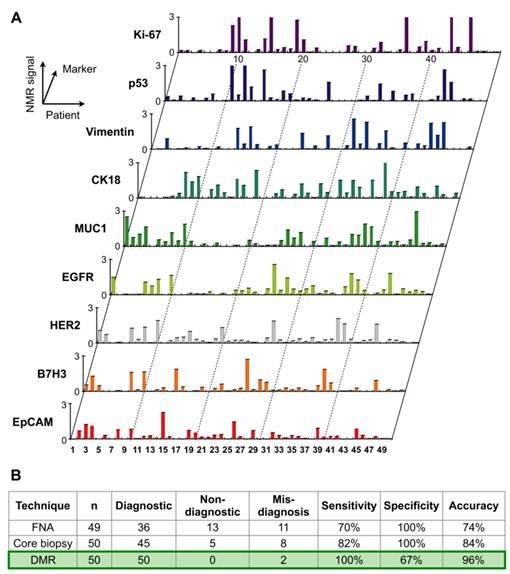
This first clinical study indicated that 1) single fine-needle aspirates contain sufficiently high numbers of cells to enable real-time quantitative analysis of many molecular markers with DMR; 2) using a four-protein signature (HER2/neu, EGFR, EpCAM and Muc-1), the DMR platform reports a 96% accuracy for establishing a cancer diagnosis, surpassing conventional clinical analyses by immunohistochemistry (Fig. 6B). Based on these results, a number of additional clinical trials are currently ongoing. The DMR platform thus represents a novel system for real-time molecular diagnostics close to the patient bedside, and a valuable tool for investigating and establishing tumor heterogeneity and therapeutic drug targeting.
CONCLUSIONS AND PERSPECTIVES
The DMR biosensor platform seamlessly integrates several cutting-edge technologies, namely nanomaterials, bioconjugation chemistry and microfabrication. As a novel technique, it offers a number of synergistic advantages such as high detection sensitivity, rapid target measurement with minimal sample processing, and the ability to profile a wide range of targets in a multiplexed manner from small sample volumes. With recent developments such as the advent of chip-based μNMR devices, optimized magnetic nanomaterials and advanced conjugation techniques, the DMR technology is a highly attractive platform to enable low-cost, and sensitive biomolecular detection in a point-of-care setting.
In addition to having significant practical use for quantitative and molecular analyses in biomedical research, the DMR platform also has far-reaching impact on clinical disease management. The capability to detect and molecularly profile cells with minimal false negatives would accelerate the advance of personalized treatment, by providing valuable information on cellular/molecular signature of individual patients. We further envision a broader application of the DMR platform in global healthcare. As DMR sensing does not require extensive sample purification as do optical methods, and the platform can be packaged as a portable device, the system is thus well-suited for rapid and point-of-care testing, especially in resource-limited primary clinics.
Acknowledgements
The authors thank N. Sergeyev for providing cross-linked dextran-coated iron oxide nanoparticles. H. Shao acknowledges financial support from the B.S.-Ph.D. National Science Scholarship awarded by the Agency for Science, Technology and Research, Singapore. This work was supported in part by NIH Grants (2RO1EB004626, U01-HL080731, HHSN268201000044C, U54-CA119349 and T32-CA79443).
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Kingsmore SF. Multiplexed protein measurement: technologies and applications of protein and antibody arrays. Nat Rev Drug Discov. 2006;5:310-20
2. Sawyers C. Targeted cancer therapy. Nature. 2004;432:294-7
3. Etzioni R, Urban N, Ramsey S. et al. The case for early detection. Nat Rev Cancer. 2003;3:243-52
4. Nagrath S, Sequist LV, Maheswaran S. et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235-9
5. Cheng MM, Cuda G, Bunimovich YL. et al. Nanotechnologies for biomolecular detection and medical diagnostics. Curr Opin Chem Biol. 2006;10:11-9
6. Tibbe AG, de Grooth BG, Greve J, Liberti PA, Dolan GJ, Terstappen LW. Optical tracking and detection of immunomagnetically selected and aligned cells. Nat Biotechnol. 1999;17:1210-3
7. Wang MM, Tu E, Raymond DE. et al. Microfluidic sorting of mammalian cells by optical force switching. Nat Biotechnol. 2005;23:83-7
8. Stern E, Klemic JF, Routenberg DA. et al. Label-free immunodetection with CMOS-compatible semiconducting nanowires. Nature. 2007;445:519-22
9. Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nat Biotechnol. 2005;23:1294-301
10. Baselt DR, Lee GU, Natesan M, Metzger SW, Sheehan PE, Colton RJ. A biosensor based on magnetoresistance technology. Biosens Bioelectron. 1998;13:731-9
11. Osterfeld SJ, Yu H, Gaster RS. et al. Multiplex protein assays based on real-time magnetic nanotag sensing. Proc Natl Acad Sci U S A. 2008;105:20637-40
12. Tamanaha CR, Mulvaney SP, Rife JC, Whitman LJ. Magnetic labeling, detection, and system integration. Biosens Bioelectron. 2008;24:1-13
13. Millen RL, Nordling J, Bullen HA, Porter MD, Tondra M, Granger MC. Giant magenetoresistive sensors. 2. Detection of biorecognition events at self-referencing and magnetically tagged arrays. Anal Chem. 2008;80:7940-6
14. Chemla YR, Grossman HL, Poon Y. et al. Ultrasensitive magnetic biosensor for homogeneous immunoassay. Proc Natl Acad Sci U S A. 2000;97:14268-72
15. Schaller V, Sanz-Velasco A, Kalabukhov A. et al. Towards an electrowetting-based digital microfluidic platform for magnetic immunoassays. Lab Chip. 2009;9:3433-6
16. Baselt DR, Lee GU, Natesan M, Metzger SW, Sheehan PE, Colton RJ. A biosensor based on magnetoresistance technology. Biosens Bioelectron. 1998;13:731-9
17. Osterfeld SJ, Yu H, Gaster RS. et al. Multiplex protein assays based on real-time magnetic nanotag sensing. Proc Natl Acad Sci U S A. 2008;105:20637-40
18. Millen RL, Kawaguchi T, Granger MC, Porter MD, Tondra M. Giant magnetoresistive sensors and superparamagnetic nanoparticles: a chip-scale detection strategy for immunosorbent assays. Anal Chem. 2005;77:6581-7
19. Gaster RS, Hall DA, Nielsen CH. et al. Matrix-insensitive protein assays push the limits of biosensors in medicine. Nat Med. 2009;15:1327-32
20. Aytur T, Foley J, Anwar M, Boser B, Harris E, Beatty PR. A novel magnetic bead bioassay platform using a microchip-based sensor for infectious disease diagnosis. J Immunol Methods. 2006;314:21-9
21. Lee H, Sun E, Ham D, Weissleder R. Chip-NMR biosensor for detection and molecular analysis of cells. Nat Med. 2008;14:869-74
22. Gueron M. Nuclear-relaxation in macromolecules by paramagnetic-ions - novel mechanism. J Magn Reson. 1975;19:58-66
23. Perez JM, Josephson L, O'Loughlin T, Hogemann D, Weissleder R. Magnetic relaxation switches capable of sensing molecular interactions. Nat Biotechnol. 2002;20:816-20
24. Lee JH, Huh YM, Jun YW. et al. Artificially engineered magnetic nanoparticles for ultra-sensitive molecular imaging. Nat Med. 2007;13:95-9
25. Haun JB, Castro CM, Wang R. et al. Micro-NMR for rapid molecular analysis of human tumor samples. Sci Transl Med. 2011;3:71ra16
26. Haun JB, Devaraj NK, Hilderbrand SA, Lee H, Weissleder R. Bioorthogonal chemistry amplifies nanoparticle binding and enhances the sensitivity of cell detection. Nat Nanotechnol. 2010;5:660-5
27. Haun JB, Devaraj NK, Marinelli BS, Lee H, Weissleder R. Probing intracellular biomarkers and mediators of cell activation using nanosensors and bioorthogonal chemistry. ACS Nano. 2011;5:3204-13
28. Lee H, Yoon TJ, Figueiredo JL, Swirski FK, Weissleder R. Rapid detection and profiling of cancer cells in fine-needle aspirates. Proc Natl Acad Sci U S A. 2009;106:12459-64
29. Liong M, Tassa C, Shaw SY, Lee H, Weissleder R. Multiplexed magnetic labeling amplification using oligonucleotide hybridization. Adv Mater. 2011;23:H254-7
30. Agasti S, Liong M, Tassa C. et al. Supramolecular host-guest interaction for labeling and detection of cellular biomarkers. Angew Chem Int Ed Engl. 2011 DOI: 10.1002/anie
31. Issadore D, Min C, Liong M, Chung J, Weissleder R, Lee H. Miniature Magnetic Resonance System for Point-of-Care Diagnostics. Lab Chip. 2011;11:2282-7
32. Bean CP, Livingston JD. Superparamagnetism. J Appl Phys. 1959;30:S120
33. Kittel C. Physical Theory of Ferromagnetic Domains. Rev Mod Phys. 1949;21:541-83
34. Brooks RA. T(2)-shortening by strongly magnetized spheres: a chemical exchange model. Magn Reson Med. 2002;47:388-91
35. Josephson L, Kircher MF, Mahmood U, Tang Y, Weissleder R. Near-infrared fluorescent nanoparticles as combined MR/optical imaging probes. Bioconjug Chem. 2002;13:554-60
36. Josephson L, Tung CH, Moore A, Weissleder R. High-efficiency intracellular magnetic labeling with novel superparamagnetic-Tat peptide conjugates. Bioconjug Chem. 1999;10:186-91
37. Sun S, Zeng H, Robinson DB. et al. Monodisperse MFe2O4 (M = Fe, Co, Mn) nanoparticles. J Am Chem Soc. 2004;126:273-9
38. Jun YW, Lee JH, Cheon J. Chemical design of nanoparticle probes for high-performance magnetic resonance imaging. Angew Chem Int Ed Engl. 2008;47:5122-35
39. Jang JT, Nah H, Lee JH, Moon SH, Kim MG, Cheon J. Critical enhancements of MRI contrast and hyperthermic effects by dopant-controlled magnetic nanoparticles. Angew Chem Int Ed Engl. 2009;48:1234-8
40. Jun YW, Huh YM, Choi JS. et al. Nanoscale size effect of magnetic nanocrystals and their utilization for cancer diagnosis via magnetic resonance imaging. J Am Chem Soc. 2005;127:5732-3
41. Huh YM, Jun YW, Song HT. et al. In vivo magnetic resonance detection of cancer by using multifunctional magnetic nanocrystals. J Am Chem Soc. 2005;127:12387-91
42. Miguel OB, Gossuin Y, Morales MP, Gillis P, Muller RN, Veintemillas-Verdaguer S. Comparative analysis of the 1H NMR relaxation enhancement produced by iron oxide and core-shell iron-iron oxide nanoparticles. Magn Reson Imaging. 2007;25:1437-41
43. Peng S, Wang C, Xie J, Sun S. Synthesis and stabilization of monodisperse Fe nanoparticles. J Am Chem Soc. 2006;128:10676-7
44. Lee H, Yoon TJ, Weissleder R. Ultrasensitive detection of bacteria using core-shell nanoparticles and an NMR-filter system. Angew Chem Int Ed Engl. 2009;48:5657-60
45. Yoon TJ, Lee H, Shao H, Weissleder R. Highly magnetic core-shell nanoparticles with a unique magnetization mechanism. Angew Chem Int Ed Engl. 2011;50:4663-6
46. Devaraj NK, Upadhyay R, Haun JB, Hilderbrand SA, Weissleder R. Fast and sensitive pretargeted labeling of cancer cells through a tetrazine/trans-cyclooctene cycloaddition. Angew Chem Int Ed Engl. 2009;48:7013-6
47. Devaraj NK, Hilderbrand S, Upadhyay R, Mazitschek R, Weissleder R. Bioorthogonal Turn-On Probes for Imaging Small Molecules inside Living Cells. Angew Chem Int Ed Engl. 2010;49:2869-72
48. Haun JB, Yoon TJ, Lee H, Weissleder R. Molecular detection of biomarkers and cells using magnetic nanoparticles and diagnostic magnetic resonance. Methods Mol Biol. 2011;726:33-49
49. Karver MR, Weissleder R, Hilderbrand SA. Synthesis and Evaluation of a Series of 1,2,4,5-Tetrazines for Bioorthogonal Conjugation. Bioconjug Chem. 2011;22:2263-70
50. Olson DL, Peck TL, Webb AG, Magin RL, Sweedler JV. High-Resolution Microcoil 1H-NMR for Mass-Limited, Nanoliter-Volume Samples. Science. 1995;270:1967-70
51. G. Webb A. Radiofrequency microcoils in magnetic resonance. Prog Nucl Magn Reson Spectrosc. 1997;31:1-42
52. Sun N, Liu Y, Lee H, Weissleder R, Ham D. CMOS RF Biosensor Utilizing Nuclear Magnetic Resonance. IEEE J Solid-State Circuits. 2009;44:1629
53. Sun N, Yoon T-J, Lee H. et al. Palm NMR and one-chip NMR. IEEE ISSCC Digest Tech Papers. 2010:488- 489
54. Grimm J, Perez JM, Josephson L, Weissleder R. Novel nanosensors for rapid analysis of telomerase activity. Cancer Res. 2004;64:639-43
55. Kim GY, Josephson L, Langer R, Cima MJ. Magnetic relaxation switch detection of human chorionic gonadotrophin. Bioconjug Chem. 2007;18:2024-8
56. Perez JM, O'Loughin T, Simeone FJ, Weissleder R, Josephson L. DNA-based magnetic nanoparticle assembly acts as a magnetic relaxation nanoswitch allowing screening of DNA-cleaving agents. J Am Chem Soc. 2002;124:2856-7
57. Zhao M, Josephson L, Tang Y, Weissleder R. Magnetic sensors for protease assays. Angew Chem Int Ed Engl. 2003;42:1375-8
58. Tsourkas A, Hofstetter O, Hofstetter H, Weissleder R, Josephson L. Magnetic relaxation switch immunosensors detect enantiomeric impurities. Angew Chem Int Ed Engl. 2004;43:2395-9
59. Sun EY, Weissleder R, Josephson L. Continuous analyte sensing with magnetic nanoswitches. Small. 2006;2:1144-7
60. Perez JM, Simeone FJ, Saeki Y, Josephson L, Weissleder R. Viral-induced self-assembly of magnetic nanoparticles allows the detection of viral particles in biological media. J Am Chem Soc. 2003;125:10192-3
61. Perez JM, Simeone FJ, Tsourkas A, Josephson L, Weissleder R. Peroxidase substrate nanosensors for MR imaging. Nano Lett. 2004;4:119-22
Author contact
![]() Corresponding author: R. Weissleder, MD, PhD or H. Lee, PhD., Center for Systems Biology, Massachusetts General Hospital, 185 Cambridge St, CPZN 5206, Boston, MA, 02114. Tel. 617-726-8226 rweisslederharvard.edu or hleeharvard.edu
Corresponding author: R. Weissleder, MD, PhD or H. Lee, PhD., Center for Systems Biology, Massachusetts General Hospital, 185 Cambridge St, CPZN 5206, Boston, MA, 02114. Tel. 617-726-8226 rweisslederharvard.edu or hleeharvard.edu
 Global reach, higher impact
Global reach, higher impact