13.3
Impact Factor
Theranostics 2013; 3(1):76-84. doi:10.7150/thno.4835 This issue Cite
Review
Molecular Imaging of Chemokine Receptor CXCR4
1. Goldyne Savad Institute of Gene Therapy, Hadassah University Hospital, Jerusalem, 91120, Israel.
2. Cyclotron/Radiochemistry Unit/Nuclear Medicine Department, Hadassah Hebrew University Hospital, Jerusalem, 91120, Israel.
Received 2012-7-7; Accepted 2012-8-28; Published 2013-1-15
Abstract
CXCR4 was found to be expressed by many different types of human cancers and its expression has been correlated with tumor aggressiveness, poor prognosis and resistance to chemotherapy. CXCR4 was also shown to contribute to metastatic seeding of organs that express its ligand CXCL12 and support the survival of these cells. These findings suggest that CXCR4 is a potentially attractive therapeutic target, and several antagonists and antibodies for this receptor were developed and are under clinical evaluation. Quantifying CXCR4 expression non-invasively might aid in prognostication as a mean for personalized therapy and post treatment monitoring. Multiple attempts were done over the recent years to develop imaging agents for CXCR4 using different technologies including PET, SPECT, fluorescent and bioluminescence, and will be reviewed in this paper.
Keywords: CXCR4, molecular imaging, diagnostics, cancer.
Introduction
CXCR4 is a 7-transmembrane G-coupled receptor which belongs to the chemokine receptors family, and is expressed by variety of cells during development and adult life. CXCR4 was initially cloned as an orphan chemokine receptor, and when it was identified as one of the co-receptors for T-tropic HIV, it drew much attention from both the research community as well as pharmacological companies [1-3]. The important role of CXCR4 in HIV entry to CD4 T cells initiated the development of different antagonists, both small molecules and peptide-based.
The sole universally accepted natural vertebrate ligand for CXCR4 is the chemokine CXCL12 (also known as stromal-derived factor, SDF-1) which was primarily isolated from a bone marrow stromal cell line and can also bind another chemokine receptor, CXCR7. Under normal condition, CXCR4/CXCL12 axis plays a role in leukocyte recruitment and is critical for homing and retention of hematopoietic cells, including stem cells, in the bone marrow (BM) [1, 4, 5]. Blocking of CXCR4 with antagonists such as AMD3100, BKT140 (T140) or others ligands, induces mobilization of progenitor cells from the BM into the blood, and enables collection of these cells for clinical procedures such as BM transplantation [6].
In addition to its extensive physiological roles, CXCR4 has also been found to be expressed by various human cancers including breast, prostate, lung, colon and multiple myeloma [7-14], and has been suggested to be involved in the process of cancer cell metastasis [15, 16]. The first report regarding CXCR4 role in metastasis was published by Muller et al. who showed that the metastasis of breast cancer cell line is dependent on CXCR4/CXCL12 axis [9]. Since then, multiple publications have shown that CXCR4 can provide survival and proliferation signals to cancer cells, direct these cells to specific metastatic sites and provide some resistance to chemotherapy [4, 6, 13]. CXCR4 expression, whether shown by mRNA or by immunohistochemistry, was suggested to correlate to poor prognosis in several cancers [17, 18], making it a potential target for anti-CXCR4 targeted therapy. However, currently there is no ability to evaluate CXCR4 expression non-invasively and in whole tumors, and tests are limited to biopsies, which are restricted to a specific site and might give false negative results.
This review will mainly focus on the research that was done to enable evaluation of CXCR4 expression in whole tumors non-invasively, by development of imaging agents using bioluminescence, fluorescent molecules or radiolableled molecules, some of which are summarized in Table 1.
Imaging agents targeting CXCR4.
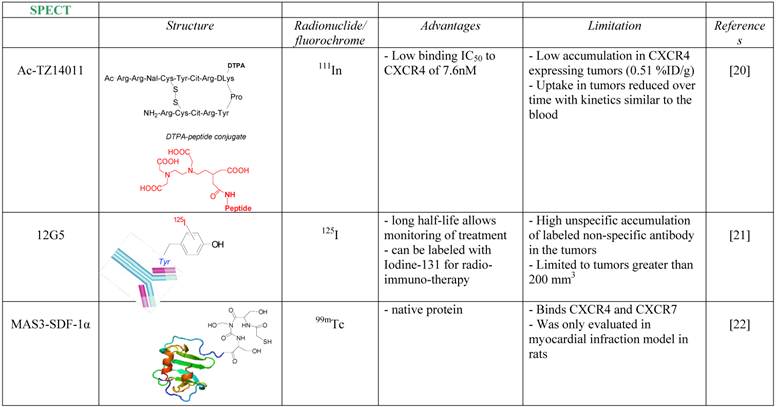
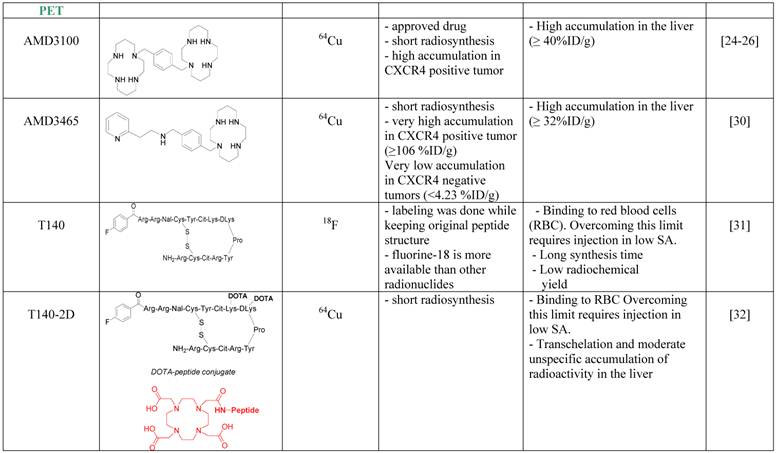
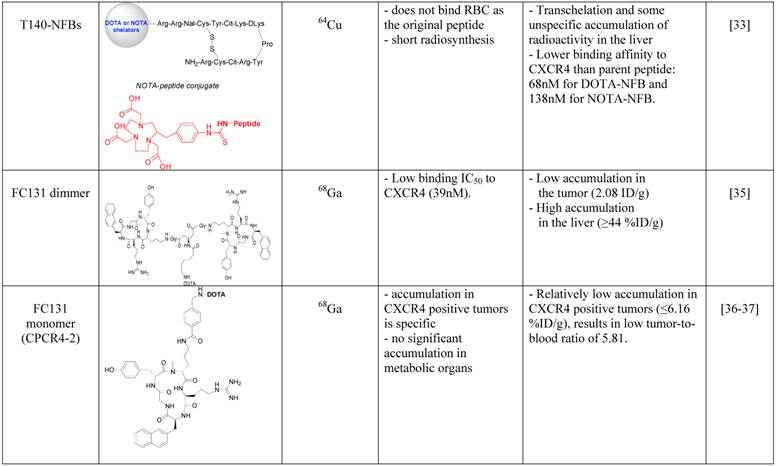
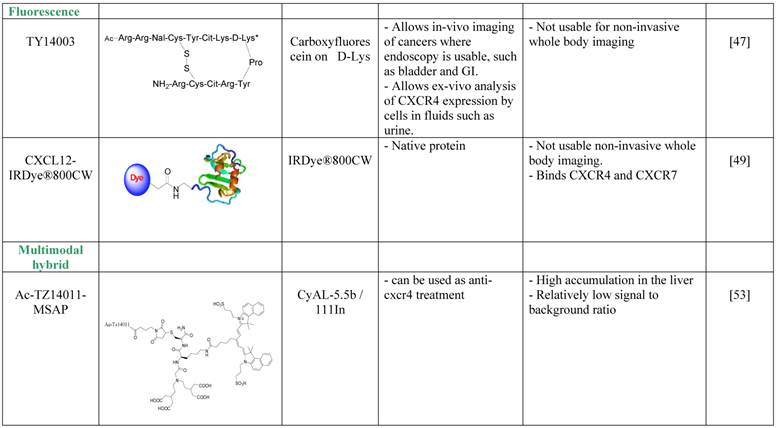
SPECT tracers targeting CXCR4
Single photon emission computed tomography, SPECT, is a technology that utilizes radioactive isotopes which emit a photon upon decay. Detection of single photon requires physical collimators, which exhibit low geometric efficiencies, in order to reject photons that are not within a small angular range (otherwise the angle of incidence will not be known) [19]. This method requires injection of radioactive tracer, and the distribution of the tracer is thereafter monitored using a SPECT machine. The SPECT is usually combined with a computed tomography (CT) for anatomical information.
One of the first attempts to detect CXCR4 in tumors using a radiolabeled antagonist was done by Hanaoka et al., who labeled the peptide antagonist Ac-TZ14011 with Inidium-111 [20]. The researchers conjugated the antagonist to Diethylenetriaminepentaacetic acid (DTPA) which enabled chelation of 111In, to give 111In-DTPA-Ac-TZ14011. The tracer was able to inhibit CXCL12 binding to CXCR4 in similar magnitude to its parent molecule, with IC50 of 7.6 nM. The accumulation in CXCR4 expressing tumor, however, was limited (0.51% Injected Dose (ID)/g), reduced over time with kinetics similar to the blood, and no in-vivo specificity was proven.
A more successful attempt to develop CXCR4 targeted tracer was done using CXCR4-specific antibody, 12G5 (Figure 1A). The antibody was labeled with Iodine-125, and injected to mice bearing glioblastoma tumors U87 and U87-transfected with CXCR4. The labeled antibody nicely accumulated in CXCR4 positive tumors, however the researchers reported on several limitations of the tracer, including relatively high unspecific accumulation of labeled non-specific antibody in the tumors, and inability to see different accumulation between the unspecific antibody and 12G5 in tumors smaller than 200 mm3 [21].
Another noteworthy research to image CXCR4 was done in rats undergoing myocardial infraction (MI), using 99mTc labeled CXCL12. CXCR4 was shown previously to be elevated after MI, and indeed Misra et al. were able to show significant accumulation of the tracer in the heart of rats post MI [22]. Another point that was not addressed is whether CXCR7, which is expressed in the heart valves and can bind CXCL12, had any contribution to the accumulation of labeled CXCL12 in the heart.
PET tracers targeting CXCR4
Positron emission tomography (PET) is a nuclear medicine technology that, similarly to SPECT, uses injected radiolabeled tracers for imaging their accumulation in target organs. The radionuclides which can be used for PET are different in that they emit a positron when undergoing decay. During the annihilation process between the positron and an electron in the tissue, two photons are released simultaneously in opposite direction [23]. The detection of two photons gives 2-3 orders of magnitude more sensitive than SPECT ensuing superior resolution, however production of the radioisotopes is usually more expensive and the radionuclides typically have shorter half-lives.
The first CXCR4 antagonist to be labeled with PET radionuclide was AMD3100, which was labeled with copper-64. AMD3100 is a bicyclam, that can chelate metal ions and therefore the synthesis of 64Cu-AMD3100 is quick and relatively simple resulting in high radiochemical yield. 64Cu-AMD3100 was first evaluated by us in normal mice [24], and showed rapid clearance from the blood and accumulation in CXCR4 expressing organs such as the BM and spleen. The tracer was later reported by us and others to specifically accumulate in CXCR4 expressing tumors (Figure 1B) [25, 26]. The main drawback of the tracer was high accumulation (>40% ID/g) in the liver, which was specific to the parent molecule, and masked some of the adjacent organs. This phenomenon is not CXCR4-specific binding in the liver because (a) high CXCR4 expressing organs such as the spleen do not display such high uptake of the tracer; (b) only limited liver cells express CXCR4 [27-29]; and (c) other tracers such as labeled T140 discussed below do not show high accumulation in the liver. Another similar small molecule, AMD3465, which structure contains one cyclam that can chelate copper-64, was also evaluated as a PET tracer for imaging CXCR4 and showed very high accumulation in CXCR4 expressing tumor (~100% ID/g), and lower but significant (40% ID/g) accumulation in the liver (Figure 1D) [30].
A peptide based CXCR4 antagonist, similar to Ac-TZ14011 previously labeled with 111In, but had a flouro-benzyl group at the N-terminus, was also labeled by us using the nuclide fluorine-18 [31]. The peptide, named T140, was labeled without changing it chemical structure to give 18F-T140, and evaluated in-vivo. Surprisingly, we found that the molecule had CXCR4-independent binding to red blood cells, which we were able to overcome by injecting the tracer in low specific activity (Figure 1C). Another drawback of this labeled tracer was its long synthesis time and relatively low radiochemical yield. To shorten the synthesis, we have introduced the chelator 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA), on the lysine groups of the peptide and labeled it with copper-64 to give 64Cu-T140-2D. This peptide tracer showed similar binding to red blood cells as the fluorine-18 labeled T140 [32] [33], and in low specific activity form could be used for imaging CXCR4 positive tumors. Similar method was used by us and others to label T140 with gallium-68 without significant differences [32, 34]. It is important to note that the accumulation of radioactivity in the liver was significantly higher when T140 was labeled with chelated radiometals than when it was labeled with 18F. This suggests that some of the radioactivity in the liver is due to transchelation of the radiometal and the chelating moiety should be improved to prevent this phenomenon.
SPECT and PET imaging of subcutaneous tumors using CXCR4 specific tracers. (A) SPECT imaging of tumors of U87 cells transfected with human CXCR4 using the anti-CXCR4 antibody 12G5 (upper) or isotype antibody (lower) labeled with Iodine-125, 24, 48 and 73h post injection [21]. (B) PET imaging of lung metastasis of CHO cells transfected with CXCR4 using CXCR4 antagonist AMD3100 labeled with copper-64, 1h post injection [25]. (C) PET imaging of subcutaneous tumors of CHO cells transfected with CXCR4 using CXCR4 peptide antagonist T140 labeled with fluorine-18. The tracer was injected in low specific activity (SA) after adding 10μg of unlabeled peptide, images were taken 2h post injection [31]. (D) PET imaging of subcutaneous tumors U87 cells transfected with CXCR4 using CXCR4 antagonist AMD3465 labeled with copper-64, 90min post injection [30]. (E) PET imaging of subcutaneous tumors of OH1 cells using cyclic CXCR4-binding pentapeptide CPCR4-2 labeled with Galium-68, at 60, 90 and 110min post injection [37]. (F,G) PET imaging of subcutaneous tumors of CHO cells transfected with CXCR4 using CXCR4 peptide antagonist T140 after substitution of the 4F-benzyl group at the N-terminus of the peptide with DOTA (F) or NOTA (G) labeled with copper-64, up to 24h post injection. The substitution with either chelators allowed using high SA peptide unlike the original peptide shown in (C) [55]. Reprinted by permission of the Society of Nuclear Medicine.
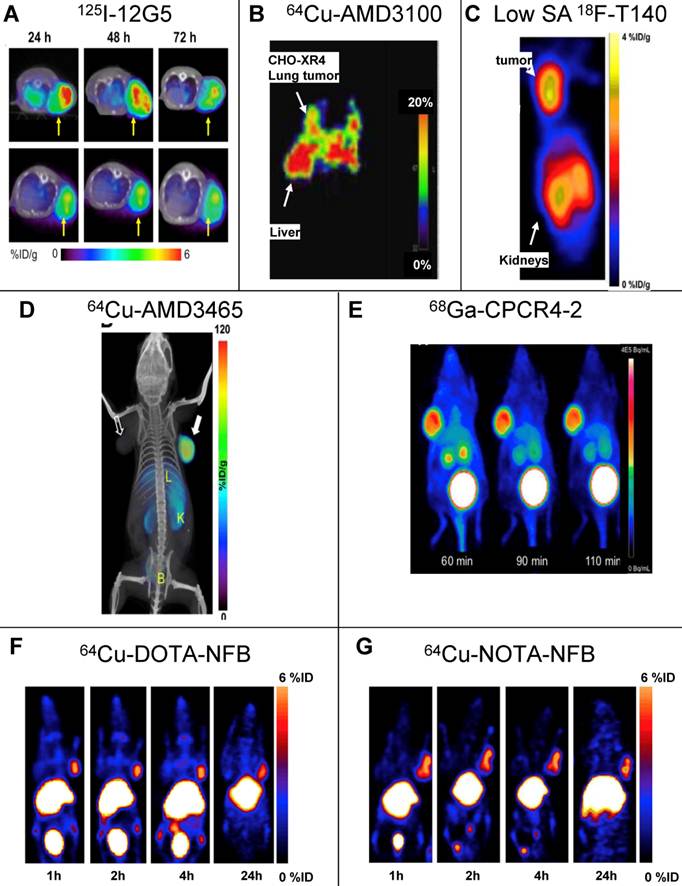
In an attempt to limit undesired binding of labeled T140 to red blood cells, we synthesized the peptide with DOTA or 1,4,7-triazacyclononane-N,N',N''-triacetic acid (NOTA) chelators at the N-terminus, replacing the fluoro-benzoyl group and thereafter labeled the tracers with copper-64. Replacing the fluoro-benzoyl group with either DOTA or NOTA diminished the binding of labeled T140 to red blood cells, while only slightly reducing the binding to CXCR4 (Figure 1F and 1G, respectively) [33]. These two optimized tracers, used in high specific activity, showed high accumulation in CXCR4 positive tumors but not in CXCR4-negative tumors, and high tumor-to-background ratio, while having short radiosynthesis time, high radiochemical yield and no non-specific binding to red blood cells.
Another Tracer that was developed for PET imaging of CXCR4 was based on a dimer of cyclic penta-peptide FC131, which was conjugated with DOTA, and thereafter labeled with Galium-68. The dimer tracer gave a limited accumulation in the tumor, while displaying very high accumulation in the liver (~44%ID/g) [35]. The researchers suggested that the dimmer has suboptimal biodistribution due to its high lipophilicity, and evaluated the FC131 cyclic penta-peptide as a monomeric form (CPCR4-2) [36] [37]. Although the monomeric tracer hypothetically have lower polyvalency, it showed better biodistribution in vivo, with significantly lower accumulation in the liver, higher accumulation in the tumor and tumor to muscle ratio of about 18 (Figure 1E) [36] [37].
An interesting small molecule that was described by Liang Z et al. called MSX-122 and displayed CXCR4 antagonist properties, was labeled with fluorine-18, to give 18F-MSX-122F [38]. The labeled derivative showed binding to CXCR4 in vitro however the authors did not report in vivo experiments thus far.
Bioluminescence tracers targeting CXCR4
Bioluminescence is based on detection of light produced by the catalytic activity of luciferase using small substrate molecules, luciferins. The absence of noise in the reporter system when used in animals gives extremely high sensitive due to the extraordinary signal-to-noise ratios [39]. The major limit of this method is that the luciferase gene is not of mammalian origin and has to be introduced into the cells, making unusable for human patients, can only be used for research..
The usage of bioluminescence as a reporter gene for evaluating the role of CXCR4 in different settings such as tumor development was reported by various laboratories, and had a significant impact on understanding the role of CXCR4 in various biological systems [40-42]. However, those indirect methods where luciferase was used only as a mean to evaluate differences between CXCR4 positive and negative cells and was not used to image CXCR4 itself will not be discussed herein.
A very interesting method for direct imaging of CXCR4 and its interaction with its ligand CXCL12 using bioluminescence was recently developed by Luker et al. [43]. These researchers made CXCL12 fused to Gaussia luciferase, and were able to detect binding of the fused chemokine to its receptors CXCR4 and CXCR7 in vitro. The fusion protein was transduced into human mammary fibroblasts that were co-injected with breast cancer cells into mice, and this bioluminescense system was elegantly used to show that CXCR7-positive breast cancer cells enhanced proliferation of CXCR4-positive breast cancer cells in orthotopic tumors and spontaneous metastases[44]. The same researchers also published an improved method for imaging CXCL12/CXCR4 and CXCL12/CXCR7 interaction in vivo. In order to achieve this, Luker et al. used a method of Gaussia luciferase (GLuc) complementation, where CXCL12 is fused to the C-terminus fragment of GLuc and either CXCR4 or CXCR7 are fused to the N-terminus fragment [45]. This is a fully reversible system that allowed the researchers to compare in a semi-quantitative manner the interaction of CXCL12 with either CXCR4 or CXCR7, which was reported to favor towards CXCR7 in MDA-MB-231 breast cancer cells. This system also allowed the researchers to evaluate CXCL12/CXCR4 inhibition in vivo by using CXCR4 antagonist AMD3100. On the other hand, inhibition of CXCL12/CXCR7 interactions, was not inhibited using CXCR7 inhibitor CCX771, which, as the authors suggest, might be due to short exposure to the chemical and the low amounts that were used [45].
Luker et al. have established another method using similar technique - firefly luciferase complementation - to test CXCR4 activation both in vivo and in vitro, by fusing CXCR4 to the N-terminus of luciferase and fusing the C-terminus to β-arrestin [46]. The researchers were able to show inhibition of CXCR4 activation in vitro using both CXCR4 antagonists AMD3100 and TF14013, and inhibition of CXCR4 activation in vivo using AMD3100.
Both luciferase complementation methods established by Luker et al. [45, 46] are very elegant and allow assessment of CXCL12/CXCR4 signaling in vitro and in vivo, however as mentioned above they can only be used in animal models. Each of the technique also have its own downside when compared to the other - CXCR4/β-arrestin complementation offer usage of one transfected cell line, however baseline bioluminescence seems to be relatively high. CXCL12/CXCR4 complementation, on the other hand, requires the use of two transfected cell lines but has low background and can also allow testing the inhibition of CXCL12/CXCR7.
Fluorescent tracers targeting CXCR4
Fluorescent labeling and staining, when combined with an appropriate imaging instrument, is a sensitive and quantitative method. Fluorescence results from a process that occurs when certain molecules (generally polyaromatic hydrocarbons or heterocycles) called fluorophores, fluorochromes, or fluorescent dyes absorb light. The absorption of light by a population of these molecules raises their energy level to a brief excited state. As they decay from this excited state, they emit fluorescent light. The major downside of fluorescence imaging is the limited depth that the fluorescent light can penetrate before it is absorbed by the tissue; hence the imaging is limited to superficial organs or tumors [47].
Nishizawa et al. avoided the depth limitation by aiming at imaging of bladder cancer and of urothelial cells found in the urine of bladder cancer patients [48]. The researchers used a derivative of the T140 CXCR4 antagonist, TY14003, which was labeled with carboxyfluorescein on the D-Lysine group [49]. Fluorescent TY14003 was shown to specifically bind to urothelial cancer cells in the urine sample but also to white blood cells; however the cell types were clearly distinct from one another by shape and size. The fluorescent tracer showed great potential under the microscope but was less convincing when the authors tried to image bladder cancer in mice using fluorescent endoscopic observation. Nishizawa et al. did not evaluate fluorescent TY14003 in low-grade bladder cancers due to low CXCR4 expression by these tumors and suggested that the tracer might prove to be very helpful tool in cytoscopy of urine samples. It might also be interesting to test the tracer as a reagent for flow cytometry analysis of urine samples, and compare it to commercially available fluorescent labeled anti CXCR4 antibodies.
A different method to overcome the depth limitation of fluorescent imaging was done by using near-infrared (NIR) probe, which theoretically allows detection of tracers up to few cm into the tissue. Meincke et al. conjugated CXCL12 to NIR dye, and evaluated whether this setting allows imaging of tumors expressing CXCR4 and CXCR7 [50]. Although the NIR dye allows dipper imaging, the researchers were limited by tissue penetration, and used subcutaneous tumor models. CXCL12 conjugated to the NIR dye was shown to be very sensitive, and allowed visualization of the tumor for up to 4 days, while some of the background and unspecific accumulation of the tracer reduced after 2 days. This method for imaging CXCR4 or CXCR7 positive tumors is not usable for whole body imaging, however it might be useful for live detection of tumor cells and metastasis to tumor draining lymph nodes under surgery, and allow the surgeon to remove all CXCR4 positive malignant cells.
Several other reagents were developed by different groups, including T140 derivative TZ14011 coupled to iridium complex [51], anti-CXCR4 antibody labeled with fluorescent metal nanoshell [52], and a quantum dot conjugated to proteinA-anti CXCR4 antibody [53]. These reports, although very interesting, did not show in vivo imaging and limited the experimental design to cell binding and cell imaging hence will not be reviewed in this paper.
CXCR4 multimodal hybrid imaging
The development of multimodal probes is attaining a significant interest in the field of molecular imaging. Such probes can be used in more than one imaging modality. Each modality has its own strengths and weaknesses, and therefore, a combination of modalities can be used in the clinic for optimal detection. Kuil et al. developed a multimodality probe based on T140 derivative, Ac-Tz14011, by conjugating the D-lys of the peptide to multifunctional single attachment point reagent that contains a DTPA chelate and a CyAL-5.5b fluorochrome [54]. The researchers chelated Inidium-111 into the DTPA and performed SPECT/CT and fluorescent imaging of tumor bearing mice. The reagent showed a tumor-to-muscle ratio of 4.55 ±0.68 in mice bearing CXCR4-positive tumors and 1.2 ±0.12 ratio in mice bearing CXCR4-negative tumors, a difference that was significant, however seem to be less impressive than similar ratios reported for molecules described above. Another setback was the relatively high accumulation in the liver, of about 20%ID/g. Due to the bimodality of the tracer, the researchers were able to show the accumulation of the tracer in the tumor by fluorescent imaging after surgically revealing the tumor [54]. To try and improve the multimodal tracer, the same researchers tried to combine the multifunctional single attachment point reagent with dimmer and tetramer of Ac-Tz14011, with the rational that multimerization will enhance binding to CXCR4 while reducing the influence of the labeling dye on tracer biodistribution and non-specific binding [54]. Unfortunately, the multimerization of Ac-Tz14011 showed inconclusive results in vivo, with lower %ID/g in the tumor compared to the monomer and slightly higher tumor-to-muscle ratio for the dimer.
Conclusions
Multiple attempts to develop imaging agents targeting CXCR4 were done in the recent years. A limit that is evident for all the tracers is the normal expression of CXCR4 in several tissues such as the BM, spleen and lymph nodes. Some of the agents are not usable for whole body human imaging because of the technology being use, such as bioluminescent and fluorescence; In the future, fluorescent probes can prove useful in clinical procedures employing endoscopic examination of GI and bladder and might also be applied as guidance for tumor and metastatic cells resection. Currently, only PET and SPECT nuclear medicine imaging agents can be utilized clinically. Of the different nuclear medicine tracer reviewed above, the PET tracers are more promising agents for CXCR4 non-invasive imaging in the clinic. Imaging CXCR4 in patients might give guidance to physicians as to the nature of the tumor in regard to aggressiveness and potential to metastasis and help in the decision of suitable treatment. In addition, anti-CXCR4 therapy is currently being evaluated in clinical trials, and application of CXCR4 imaging tracers described herein might benefit these trials as well as future treatment of patients with CXCR4-positive tumors, and avoiding unnecessary treatment of patients with CXCR4-negative tumors. It is our hope that the use of molecular imaging modalities with specific tracers in the diagnosis and management of patients with cancer may lead to more effective interventions tailored to an individual's needs.
Acknowledgements
Reprinted by permission of the Society of Nuclear Medicine from:
Nimmagadda S, Pullambhatla M, and Pomper MG. Immunoimaging of CXCR4 Expression in Brain Tumor Xenografts Using SPECT/CT. J Nucl Med. 2009; 50(7): 1124-1130. Figure 3
De Silva RA, Peyre K, Pullambhatla M, Fox JJ, Pomper MG, and Nimmagadda S. Imaging CXCR4 Expression in Human Cancer Xenografts. J Nucl Med. 2011; 52(6): 986-993. Figure 3
Gourni E, Demmer O, Schottelius M, et al. PET of CXCR4 Expression by a 68Ga-Labeled Highly Specific Targeted Contrast Agent. J Nucl Med. 2011; 52(11): 1803-1810. Figure 3
Competing Interests
The authors have declared that no competing interest exists.
References
1. Murdoch C. CXCR4: chemokine receptor extraordinaire. Immunol Rev. 2000;177:175-84
2. Horuk R. Chemokine receptors. Cytokine Growth Factor Rev. 2001;12:313-35
3. Phillips RJ, Burdick MD, Lutz M, Belperio JA, Keane MP, Strieter RM. The stromal derived factor-1/CXCL12-CXC chemokine receptor 4 biological axis in non-small cell lung cancer metastases. Am J Respir Crit Care Med. 2003;167:1676-86
4. Balkwill F. The significance of cancer cell expression of the chemokine receptor CXCR4. Semin Cancer Biol. 2004;14:171-9
5. Peled A, Petit I, Kollet O, Magid M, Ponomaryov T, Byk T. et al. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 1999;283:845-8
6. Peled A, Wald O, Burger J. Development of novel CXCR4-based therapeutics. Expert Opin Investig Drugs. 2012;21:341-53
7. Furusato B, Mohamed A, Uhlen M, Rhim JS. CXCR4 and cancer. Pathol Int. 2010;60:497-505
8. Tanaka T, Bai Z, Srinoulprasert Y, Yang BG, Hayasaka H, Miyasaka M. Chemokines in tumor progression and metastasis. Cancer Sci. 2005;96:317-22
9. Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME. et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50-6
10. Redjal N, Chan JA, Segal RA, Kung AL. CXCR4 inhibition synergizes with cytotoxic chemotherapy in gliomas. Clin Cancer Res. 2006;12:6765-71
11. Taichman RS, Cooper C, Keller ET, Pienta KJ, Taichman NS, McCauley LK. Use of the stromal cell-derived factor-1/CXCR4 pathway in prostate cancer metastasis to bone. Cancer Res. 2002;62:1832-7
12. Vicari AP, Caux C. Chemokines in cancer. Cytokine Growth Factor Rev. 2002;13:143-54
13. Azab AK, Runnels JM, Pitsillides C, Moreau AS, Azab F, Leleu X. et al. CXCR4 inhibitor AMD3100 disrupts the interaction of multiple myeloma cells with the bone marrow microenvironment and enhances their sensitivity to therapy. Blood. 2009;113:4341-51
14. Kurtova AV, Tamayo AT, Ford RJ, Burger JA. Mantle cell lymphoma cells express high levels of CXCR4, CXCR5, and VLA-4 (CD49d): importance for interactions with the stromal microenvironment and specific targeting. Blood. 2009;113:4604-13
15. Yoon Y, Liang Z, Zhang X, Choe M, Zhu A, Cho HT. et al. CXC chemokine receptor-4 antagonist blocks both growth of primary tumor and metastasis of head and neck cancer in xenograft mouse models. Cancer Res. 2007;67:7518-24
16. Li JK, Yu L, Shen Y, Zhou LS, Wang YC, Zhang JH. Inhibition of CXCR4 activity with AMD3100 decreases invasion of human colorectal cancer cells in vitro. World J Gastroenterol. 2008;14:2308-13
17. Lee HJ, Kim SW, Kim HY, Li S, Yun HJ, Song KS. et al. Chemokine receptor CXCR4 expression, function, and clinical implications in gastric cancer. Int J Oncol. 2009;34:473-80
18. Oda Y, Tateishi N, Matono H, Matsuura S, Yamamaoto H, Tamiya S. et al. Chemokine receptor CXCR4 expression is correlated with VEGF expression and poor survival in soft-tissue sarcoma. Int J Cancer. 2009;124:1852-9
19. Rahmim A, Zaidi H. PET versus SPECT: strengths, limitations and challenges. Nucl Med Commun. 2008;29:193-207
20. Hanaoka H, Mukai T, Tamamura H, Mori T, Ishino S, Ogawa K. et al. Development of a 111In-labeled peptide derivative targeting a chemokine receptor, CXCR4, for imaging tumors. Nucl Med Biol. 2006;33:489-94
21. Nimmagadda S, Pullambhatla M, Pomper MG. Immunoimaging of CXCR4 expression in brain tumor xenografts using SPECT/CT. J Nucl Med. 2009;50:1124-30
22. Misra P, Lebeche D, Ly H, Schwarzkopf M, Diaz G, Hajjar RJ. et al. Quantitation of CXCR4 expression in myocardial infarction using 99mTc-labeled SDF-1alpha. J Nucl Med. 2008;49:963-9
23. Saleem A, Charnley N, Price P. Clinical molecular imaging with positron emission tomography. Eur J Cancer. 2006;42:1720-7
24. Jacobson O, Weiss ID, Szajek L, Farber JM, Kiesewetter DO. 64Cu-AMD3100--a novel imaging agent for targeting chemokine receptor CXCR4. Bioorg Med Chem. 2009;17:1486-93
25. Weiss ID, Jacobson O, Kiesewetter DO, Jacobus JP, Szajek LP, Chen X. et al. Positron emission tomography imaging of tumors expressing the human chemokine receptor CXCR4 in mice with the use of (6)(4)Cu-AMD3100. Mol Imaging Biol. 2011;14:106-14
26. Nimmagadda S, Pullambhatla M, Stone K, Green G, Bhujwalla ZM, Pomper MG. Molecular imaging of CXCR4 receptor expression in human cancer xenografts with [64Cu]AMD3100 positron emission tomography. Cancer Res. 2010;70:3935-44
27. Li W, Gomez E, Zhang Z. Immunohistochemical expression of stromal cell-derived factor-1 (SDF-1) and CXCR4 ligand receptor system in hepatocellular carcinoma. J Exp Clin Cancer Res. 2007;26:527-33
28. Mavier P, Martin N, Couchie D, Preaux AM, Laperche Y, Zafrani ES. Expression of stromal cell-derived factor-1 and of its receptor CXCR4 in liver regeneration from oval cells in rat. Am J Pathol. 2004;165:1969-77
29. Hatch HM, Zheng D, Jorgensen ML, Petersen BE. SDF-1alpha/CXCR4: a mechanism for hepatic oval cell activation and bone marrow stem cell recruitment to the injured liver of rats. Cloning Stem Cells. 2002;4:339-51
30. De Silva RA, Peyre K, Pullambhatla M, Fox JJ, Pomper MG, Nimmagadda S. Imaging CXCR4 expression in human cancer xenografts: evaluation of monocyclam 64Cu-AMD3465. J Nucl Med. 2011;52:986-93
31. Jacobson O, Weiss ID, Kiesewetter DO, Farber JM, Chen X. PET of tumor CXCR4 expression with 4-18F-T140. J Nucl Med. 2010;51:1796-804
32. Jacobson O, Weiss ID, Szajek LP, Niu G, Ma Y, Kiesewetter DO. et al. PET imaging of CXCR4 using copper-64 labeled peptide antagonist. Theranostics. 2011;1:251-62
33. Jacobson O, Weiss ID, Szajek LP, Niu G, Ma Y, Kiesewetter DO. et al. Improvement of CXCR4 tracer specificity for PET imaging. J Control Release. 2012;157:216-23
34. Hennrich U, Seyler L, Schafer M, Bauder-Wust U, Eisenhut M, Semmler W. et al. Synthesis and in vitro evaluation of 68Ga-DOTA-4-FBn-TN14003, a novel tracer for the imaging of CXCR4 expression. Bioorg Med Chem. 2012;20:1502-10
35. Demmer O, Dijkgraaf I, Schumacher U, Marinelli L, Cosconati S, Gourni E. et al. Design, synthesis, and functionalization of dimeric peptides targeting chemokine receptor CXCR4. J Med Chem. 2011;54:7648-62
36. Demmer O, Gourni E, Schumacher U, Kessler H, Wester HJ. PET imaging of CXCR4 receptors in cancer by a new optimized ligand. ChemMedChem. 2011;6:1789-91
37. Gourni E, Demmer O, Schottelius M, D'Alessandria C, Schulz S, Dijkgraaf I. et al. PET of CXCR4 expression by a (68)Ga-labeled highly specific targeted contrast agent. J Nucl Med. 2011;52:1803-10
38. Liang Z, Zhan W, Zhu A, Yoon Y, Lin S, Sasaki M. et al. Development of a unique small molecule modulator of CXCR4. PLoS One. 2012;7:e34038
39. Keyaerts M, Caveliers V, Lahoutte T. Bioluminescence imaging: looking beyond the light. Trends Mol Med. 2012;18:164-72
40. Darash-Yahana M, Pikarsky E, Abramovitch R, Zeira E, Pal B, Karplus R. et al. Role of high expression levels of CXCR4 in tumor growth, vascularization, and metastasis. FASEB J. 2004;18:1240-2
41. Hong YL, Wu LH, Cui M, McMaster G, Hunt SW 3rd, Chung FZ. New reporter cell lines to study macrophage-tropic HIV envelope protein-mediated cell-cell fusion. AIDS Res Hum Retroviruses. 1999;15:1667-72
42. Gonzalez N, Perez-Olmeda M, Mateos E, Cascajero A, Alvarez A, Spijkers S. et al. A sensitive phenotypic assay for the determination of human immunodeficiency virus type 1 tropism. J Antimicrob Chemother. 2010;65:2493-501
43. Luker K, Gupta M, Luker G. Bioluminescent CXCL12 fusion protein for cellular studies of CXCR4 and CXCR7. Biotechniques. 2009;47:625-32
44. Luker KE, Lewin SA, Mihalko LA, Schmidt BT, Winkler JS, Coggins NL. et al. Scavenging of CXCL12 by CXCR7 promotes tumor growth and metastasis of CXCR4-positive breast cancer cells. Oncogene. 2012
45. Luker KE, Mihalko LA, Schmidt BT, Lewin SA, Ray P, Shcherbo D. et al. In vivo imaging of ligand receptor binding with Gaussia luciferase complementation. Nat Med. 2011;18:172-7
46. Luker KE, Gupta M, Luker GD. Imaging CXCR4 signaling with firefly luciferase complementation. Anal Chem. 2008;80:5565-73
47. Leblond F, Davis SC, Valdes PA, Pogue BW. Pre-clinical whole-body fluorescence imaging: Review of instruments, methods and applications. J Photochem Photobiol B. 2010;98:77-94
48. Nishizawa K, Nishiyama H, Oishi S, Tanahara N, Kotani H, Mikami Y. et al. Fluorescent imaging of high-grade bladder cancer using a specific antagonist for chemokine receptor CXCR4. Int J Cancer. 2010;127:1180-7
49. Oishi S, Masuda R, Evans B, Ueda S, Goto Y, Ohno H. et al. Synthesis and application of fluorescein- and biotin-labeled molecular probes for the chemokine receptor CXCR4. Chembiochem. 2008;9:1154-8
50. Meincke M, Tiwari S, Hattermann K, Kalthoff H, Mentlein R. Near-infrared molecular imaging of tumors via chemokine receptors CXCR4 and CXCR7. Clin Exp Metastasis. 2011;28:713-20
51. Kuil J, Steunenberg P, Chin PT, Oldenburg J, Jalink K, Velders AH. et al. Peptide-functionalized luminescent iridium complexes for lifetime imaging of CXCR4 expression. Chembiochem. 2011;12:1897-903
52. Zhang J, Fu Y, Li G, Zhao RY, Lakowicz JR. Detection of CXCR4 receptors on cell surface using a fluorescent metal nanoshell. J Biomed Opt. 2011;16:016011
53. Jin T, Tiwari DK, Tanaka S, Inouye Y, Yoshizawa K, Watanabe TM. Antibody-protein A conjugated quantum dots for multiplexed imaging of surface receptors in living cells. Mol Biosyst. 2010;6:2325-31
54. Kuil J, Buckle T, Oldenburg J, Yuan H, Borowsky AD, Josephson L. et al. Hybrid peptide dendrimers for imaging of chemokine receptor 4 (CXCR4) expression. Mol Pharm. 2011;8:2444-53
55. Jacobson O, Weiss ID, Szajek LP, Niu G, Ma Y, Kiesewetter DO. et al. Improvement of CXCR4 tracer specificity for PET imaging. J Control Release. 2012;157:216-23
Author contact
![]() Corresponding author: idoweicom.
Corresponding author: idoweicom.
 Global reach, higher impact
Global reach, higher impact