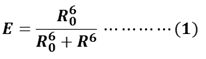13.3
Impact Factor
Theranostics 2013; 3(6):395-408. doi:10.7150/thno.5935 This issue Cite
Review
Molecular Beacons of Xeno-Nucleic Acid for Detecting Nucleic Acid
1. Key Lab for Advanced Materials and Institute of Fine Chemicals, East China University of Science and Technology, China.
2. College of Public Health, Nantong University, China.
Received 2013-1-23; Accepted 2013-4-10; Published 2013-5-5
Abstract
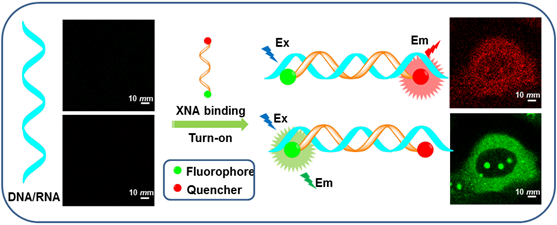
Molecular beacons (MBs) of DNA and RNA have aroused increasing interest because they allow a continuous readout, excellent spatial and temporal resolution to observe in real time. This kind of dual-labeled oligonucleotide probes can differentiate between bound and unbound DNA/RNA in homogenous hybridization with a high signal-to-background ratio in living cells. This review briefly summarizes the different unnatural sugar backbones of oligonucleotides combined with fluorophores that have been employed to sense DNA/RNA. With different probes, we epitomize the fundamental understanding of driving forces and these recognition processes. Moreover, we will introduce a few novel and attractive emerging applications and discuss their advantages and disadvantages. We also highlight several perspective probes in the application of cancer therapeutics.
Keywords: Xeno-nucleic acid, Nucleic acid, Molecular beacon, Diagnostics.
Introduction
Nuclear acids including DNA and RNA play crucial roles in biological activities. The track of nucleic acid greatly helped us study biological evolution, gene analysis, disease diagnosis, as well as drug discovery. More importantly, oligonucleotides with unnatural sugar backbones that can also be replicated in vitro often achieve excellent sensitivity and selectivity. In addition, with the innovation of oligonucleotide synthesis technology, a large number of novel oligonucleotide probes produced in the laboratory can be used to target a wide range of analysis (DNA, RNA) with high affinity and specificity. In this review, we will demonstrate several oligonucleotide probes with different sugar backbones (Xeon-nucleic acids, or XNAs) [1] that could be used in the detection of nuclear acid. Among these analogs, peptide nucleic acid (PNA) and locked nucleic acid (LNA) are the two most studied XNAs. Besides, other oligonucleotide derivatives such as 1', 5'- anhydrohexitol nucleic acids (HNA) [2], altritol nucleic acids (ANA) [3], 2'-fluoroarabinose nucleic acids (FNA) [4], threose nucleic acids (TNA) [5] and cyclohex-ene nucleic acids (CeNA) [6] also play important roles in the field of nucleic acids.
Considering the measurement of output signal upon binding the target molecules, probes for the nuclear acid to date rely on monitoring changes in the adsorbed mass, charge, or index of refraction. Microcantilevers [7,8], quartzcrystal microbalances [9], field-effect transistors [10] and surface-plasmon-resonance-based sensors [11], which have been used to probe nuclear acid achieved admirable operational convenience. However, those methods are not successfully applied in clinical settings for the high background signals [12]. In contrast, MBs perform that function by monitoring themselves stem-loop conformation changes to achieve admirable operational convenience. They allow a continuous readout, excellent spatial and temporal resolution in real time, which makes them the promising probes in lab research and clinical applications in future. Based on the change of fluorescence intensity via two complementary stem sequences binding to the target sequence, a new kind of label-free probes termed molecular beacon (MB) was proposed by Tyagi and Kramer [13,14]. Traditional MBs are designed with doubly end-labeled oligonucleotides. They are stable stem-loop structures in which the fluorescence of a reporter dye (chromophore) is quenched through transferring energy to a proximate quencher in solution [15]. Upon hybridizing with target DNA/RNA sequence, the stem-loop structure is destroyed which leads to spatial separation of fluorophores. The unique thermodynamic stability of stem-loop structure, the diversity of fluorophore-quencher pair and the highly efficient intrinsic signal switching enable the detection of nucleic acids [16,17] in real time with excellent sensitivity and selectivity [18,19,20].
In the past few decades, the findings with newly designed oligonucleotide beacons (OBs) were reported [21,22,23], which mainly focused on the investigation of XNA in the applications of proteins and enzymes, such as anti-HIV antibodies [24], bacterial lipopolysaccharide and DNA [25,26], etc.. However, XNAs as probes used to recognize and sense nucleic acid, by far, are still attractive, especially, when combined with facile detectable fluorescent reporter dye.
In this overview, we will also summarize the important features of different oligonucleotide-based MBs that were characterized thus far. Then, we will discuss their contribution to the understanding of binding and function as a nucleic acid MB, as well as provide a new horizon in the fabrication of new oligonucleotide-based MBs for applications.
Modes of nucleic acid recognition
In recent years, the detection of genetic mutations at the molecular level opens up the possibility of performing reliable diagnostics even before any symptom of a disease appears. DNA recognition might reveal the interaction of nucleic acids and proteins, furthering our insight into the regulation of gene expression.
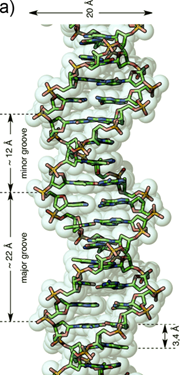
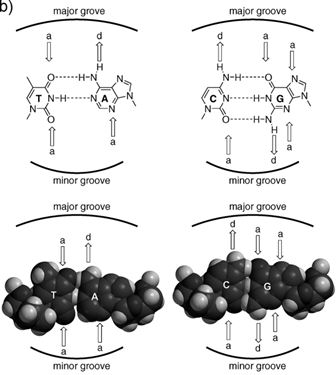
The recognition of single stranded nucleic acid could be easily understood. As compared with single stranded nucleic acid, the recognition of double stranded DNA (dsDNA) could be classified into two categories: the recognition in major groove and the recognition in minor groove. The reason is that the dsDNA structure consists in two different grooves with different geometric attributes. The wide and relatively shallow one is called major groove and the narrower and deeper one is termed minor groove (Figure 1) [27].
a) Representation of an ideal-DNA conformation with the main structural dimensions. b) DNA base-pairs indicating the pattern of hydrogen bonds. The minor groove is the side of the base pairs facing towards the sugar-phosphate backbone, and the major groove is the side facing away. (Reprinted with permission from ref. 27. Copyright 2011, Wiley-VCH Verlag GmbH & Co. KGaA.)
The available structure information of dsDNA-oligonucleotide complexes indicates that the major groove is the preferential interaction site of DNA and XNAs. In this case, DNA was recognized via a specific sequence through insertion of two stranded anti-parallel β-sheet oligonucleotide analogues into the major groove [28]. Although most oligonucleotides bind DNA in major groove, it is also well known that some molecular beacons, such as high mobility group (HMG-I) family, interact with DNA in the minor groove. The minor groove is much narrower than the major groove, which allows DNA to bind the oligonucleotide/peptide chains instead of the secondary structures.
XNA molecular beacons for nucleic acids
The synthesis of various oligonucleotide-based MBs that confers enhanced high-affinity recognition of DNA and RNA targets has been an ongoing endeavor. A variety of nucleic acid MBs have been developed which displays increased thermal stabilities and specific hybridization ability when hybridized with complementary DNA/RNA [29,30][31]. In particular, not all the XNAs are sufficiently specific to detect nucleic acid, gene mutations or real-time polymerase chain reaction (PCR) etc... Different XNAs have different specialties due to their unique chemical structures. Here we summarized the chemical structures, advantages, disadvantages of several most studied XNAs (Table 1). It should be well noted that PNA and LNA are the two most studied XNAs among these analogues, others like HNA, ANA, TNA and CeNA are also excellent candidates for nucleic acid probes, however, by now, they have rarely been reported in the application of the detection of nucleic acid.
The mechanism of oligonucleotide-based MBs in recognizing nucleic acid lies on a fine balance of interactions including hydrogen bonds between bases and surrounding probes, and base-stacking interactions between adjacent bases.
XNA based MBs usually have three recognizing modes for nucleic acids. The first light-up probe consists of a sequence-recognizing element-XNA linked to a single fluorescent dye (usually they are thiazole orange derivatives) that serves as a reporter group. When free in solution, the probe has very low fluorescence due to the intrinsic properties of the reporter dye. Binding the XNA to its complementary target allows interaction between the dye and the nucleobases, which causes the light-up probe to fluoresce brightly (Figure 2a) [71].
The second XNA based MBs with a dual-labeled oligonucleotide is capable of forming a stem-loop structure in the absence of target. Specifically, a target-binding domain is flanked by two complementary stem sequences that are usually (but not necessarily) unrelated to the target sequence. One end of the oligonucleotide is labeled with a fluorescent reporter dye and the other one is labeled with a fluorescent quencher molecule. When a MB is in its stem-loop conformation the reporter fluorophore is effectively quenched. Upon hybridization with target, the stem-loop hairpin structure of XNA opens, which separates the reporter dye and the quencher and results in increased fluorescence intensity (Figure 2b) [32,33].
FRET-based MBs are the third kind of MBs which used to detect nucleic acid. FRET is a nonradioactive process in which an excited dye donor transfers energy to a dye acceptor in the ground state through long-range dipole-dipole interactions. The donor emission has efficient spectral overlap with the acceptor absorption in FRET systems. FRET efficiency (E) is given by equation (1), where R0 is the Forster distance at which the transfer efficiency E = 50%; R is the distance between the energy donor and acceptor.
To construct FRET MBs, it needs to develop a FRET platform with favorable photophysical properties. The design criteria of a FRET platform include (1) well-resolved absorption spectra of the donor and acceptor, (2) well-separated emission spectra of the donor and acceptor, (3) donors and acceptors with comparable brightness, (4) rigid linkers, and (5) near-perfect efficiency in energy transfer. By modulating the donor-acceptor distance or spectral overlap integral in an analyze-dependent fashion, researchers could develop FRET based MBs. Normally, a FRET based MB is composed of a donor, an acceptor and a complementary sequence XNA toward the target nucleic acid. In the single-stranded state, the donor and acceptor tend to be separated and the probe shows week fluorescence upon exciting at the donor absorption. However, when the probe hybridizes with the sequence of target nucleic acid, a conformational reorganization of MBs shortens the distance between the donor and the acceptor (Figure 2c). In this situation, the probe could emit a fluorescent emission [34].
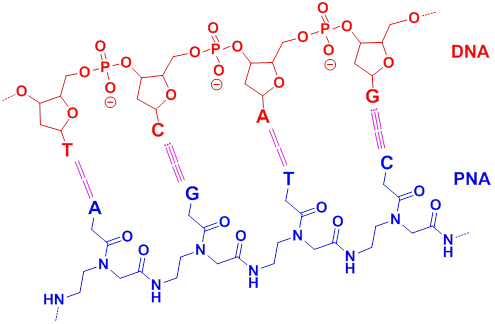
Peptide nucleic acids (PNA). Oligonucleotides-based MBs have shown great advantages in imaging of nucleic acids, however, it is still a challenge for chemists to construct a similar structure which can present the same function in monitoring the biochemistry process. Through efforts, Peter Nielsen and Ole Buchardt discovered peptide nucleic acid (PNA), a nucleobase oligomer, in the 1990s [35]. The structure of PNA is similar to oligonucleotides-based MBs, whose backbone is made from repeating N-(2-aminoethyl) glycine units linked by peptide bonds instead of a negatively charged sugar-phosphate backbone [36,37]. Different bases (purines and pyrimidines) are joined to the backbone with methylene carbonyl linkages (Figure 3) [38]. PNAs have specific structure and similar nucleobases with natural DNA molecules. This makes them have a strong hybridizing ability with target DNA or RNA.
Summary of chemical structures (aB: adenine, guanine, cytosine, or thymine), advantages and disadvantages of XNA-based biosensors.
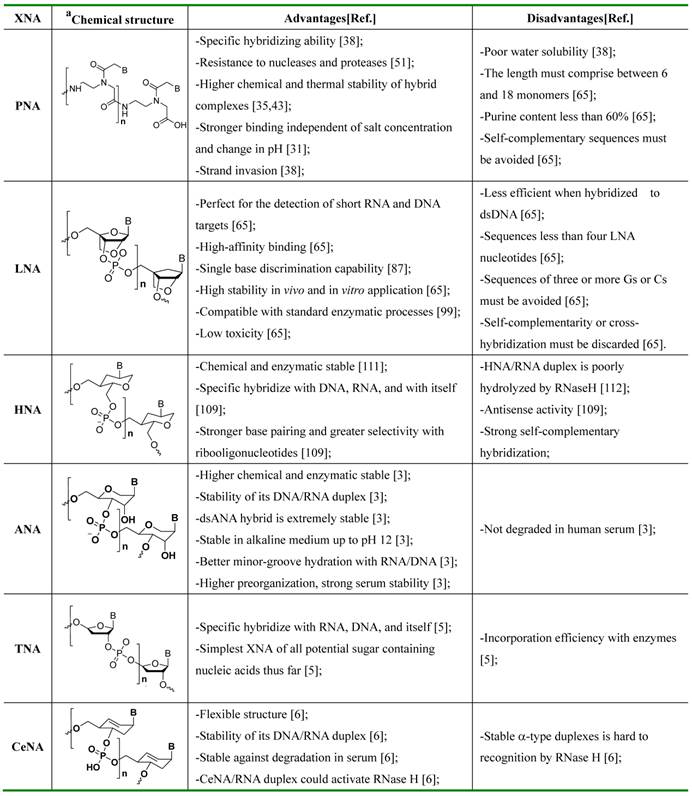
The mechanism of MBs in sensing a DNA/RNA sequence (probes refer to immobilized sequences; DNA/RNA targets refer to sequences in the sample being captured). a) The intercalated dye of probe can fold back and intercalate between the formed Watson-Crick base pairs or serves as a base surrogate that is forced to intercalate adjacent to the expected mutation site. b) Upon hybridization with complementary sequence, the stem-loop hairpin structure of MB opens, which separates the reporter dye and the quencher and results in increased fluorescence intensity; c) In the single-stranded state the donor and the acceptor are separated from each other. When the probe encounters a target RNA, the MB undergoes a spontaneous conformational reorganization that forces the stem together, leading to a fluorescence resonance energy transfer (FRET) signal change.
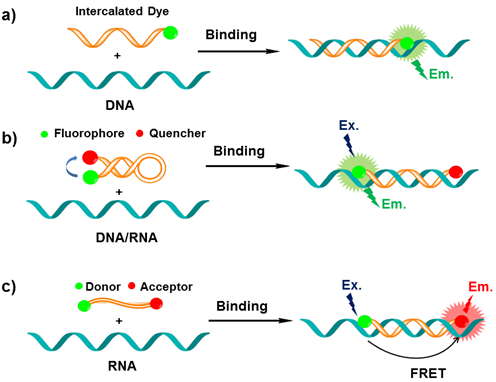
Schematic chemical model of PNA (blue) hybridized with DNA (red), with the hydrogen bonding between complementary nucleobases depicted by dotted lines (A= adenine, G= guanine, C= cytosine and T= thymine). (Reprinted with permission from ref. 38. Copyright 2006, Wiley-VCH Verlag GmbH & Co. KGaA.)
PNA have been widely used in the fields of chemistry, molecular biology, antisense therapy and gene-based diagnostics [38]. As a mature method, PNAs show great advantages over traditional approaches in the imaging and expression of DNA and RNA [39,40,41,42]. They have excellent thermal and chemical stability and their hybrid complexes for the lack of charge repulsion between the neutral PNA and the DNA/RNA [43,44]; specific hybridization ability [45] and resistance to nucleases and proteases [46]. Therefore, the lifetime of PNA/DNA or PNA/RNA is much longer both in vivo and in vitro.
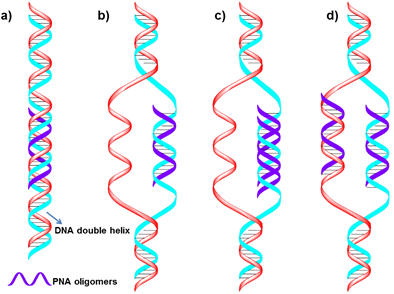
Targeting of dsDNA with PNA usually occurs via four different binding modes with the helical backbone in the major groove (Figure 4). Three of these modes (triplex formation, duplex invasion and triplex invasion) require homopurine/homopyrimidine DNA targets, whereas double duplex invasion requires targets of at least 50% A (adenine)-T (thymine) content [47].
Schematic representation of four different complexes formed by PNA binding to duplex DNA. a) Conventional triplex. b) Triplex invasion complexes are formed at homopurine DNA targets with complementary homopyrimidine PNAs. c) Duplex invasion complexes are formed with some homopurine PNAs. d) Double duplex invasion complexes. (Reprinted with permission from ref. 47. Copyright 2001, Elsevier B. V.)
Considering the chemical and biological stability, PNAs could be used to design gene therapeutic drugs [48,49,50] and other molecular biology and functional genomics [51,52]. Besides, PNA probes are extremely useful in situ hybridization and provide very good chromosome images [53,54,55,56,57,58]. A few early reports have examined the properties of PNAs as both specific [59,60,61] as well as general [62] nucleic acid capture probes. Recently, PNA-based MBs, optoelectronic [63], microarray or electrochemical probes [64] have been developed for different biochemical and biotechnological applications [65]. Fang et al. developed an electrochemical nucleic acids probe made of PNA which exhibits high sensitivity and specificity when challenged with heterogeneous samples of RNA. They further used the probes to detect a newly identified cancer biomarker-a gene fusion associated with prostate cancer. The system could detect specific mRNAs in unamplified patient samples in as little as 10 ng [66]. PNA was also successfully used to label-free DNA/PNA hybridization detection when combined with silicon-based platform [67].
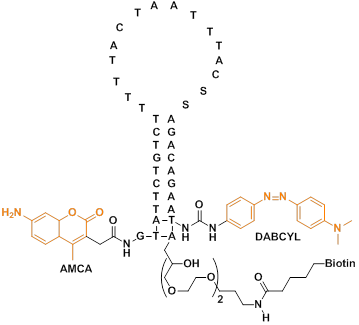
Due to the good stability and specific hybridization ability of PNA, MBs constructed from PNA have been used to improve hybridization and disfavor stem reformation [68,69]. Ortiz et al synthesized a PNA-DNA molecular beacon using 7-amino-4-methylcoumarin-3-acetic acid (AMCA) as a fluorescent moiety and a quencher moiety p-(dimethylaminophenylazo) benzoic acid (DABCYL) on the other side (Figure 5). The probe undergoes a fluorescent change in the presence of a complementary DNA target. They further used the molecular beacon for rapid detection of PCR amplicons by adding a PCR reaction to a microtitre-well containing the probe and reading the generated fluorescence.
The base sequence and chemical structure of the PNA-DNA adduct. The DNA part of the adduct has three functional moieties: (1) a 3'-terminal biotin which serves to immobilize the entire structure by binding to streptavidin-coated mitrotitre wells; (2) a quencher moiety (DABCYL) at the 3'-terminus of the DNA stem structure; (3) a sequence which pairs with 10 bases in the PNA part of the adduct, forming the 10-base pair stem structure. The PNA part of the adduct, which is joined to the DNA part via a disulfide bond, also comprises three functional moieties: (1) a 15-base probe sequence in the unstructured loop domain; (2) 10 bases capable of pairing with the DNA part, where one base in the PNA probe performs double duty, also being part of the 10-base stem structure; (3) a fluorescent moiety (AMCA), coupled to the free terminus of the PNA. (Reprinted with permission from ref. 68. Copyright 1998, Elsevier B. V.)
Recently researchers found some new fluorescent dyes very suitable for the PNA MBs. For example, thiazole orange (TO) is one of the most promising fluorophore used in MBs. Socher et al designed FIT probes in which one base is replaced by a thiazole orange (TO) dye molecule, when a DNA molecule that is complementary to the FIT-PNA molecule hybridizes to the probe, the TO dye exhibits intense fluorescence because each other stacking in the duplexes enforces a coplanar arrangement even in the excited state. They use the probes in real-time DNA quantification and single nucleotide polymorphism discovery (Figure 6) [70].
FIT probes with TO bind to Aeg and D-Orn backbones that are employed in PNA. In both molecules, the chromophores are aligned in a comparable six-atom distance from adjacent base pairs. B= adenine, thymine, guanine, cytosine; R, Ac, Ac-Lys-Lys; R', Gly-NH2. (Reprinted with permission from ref. 70. Copyright 2008, Elsevier B. V.)
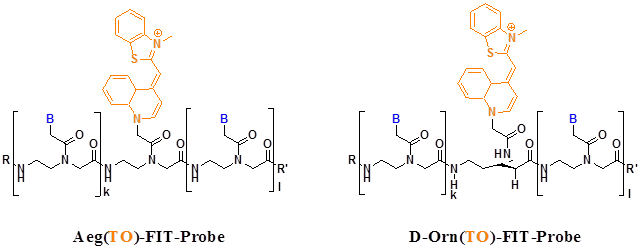
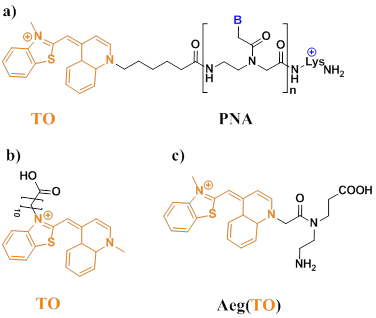
Svanvik et al found TO dye often displays enhanced fluorescence quantum yields upon binding dsDNA (Figure 7a, b) [71,72]. TO in solution binds calf thymus DNA (ctDNA) with an enhancement of quantum yield from 2 × 10-4 to 0.11 (a fluorescent increase of ∼550-fold) [73]. Upon hybridization from PNA, the quantum yield was increased from 2.2 to 6.0-fold [74]. Furthermore, Socher et al developed dual labeled peptide nucleic acid probes, which can make the presence of complementary DNA/RNA by up to 450-fold enhancements of fluorescence intensity. This enabled the detection of a DNA target at less than 0.1% of the beacon concentration. In contrast to existing DNA-based molecular beacons, this PNA-based method does not require a stem sequence to enforce dye-dye communication. The method relies on the energy transfer between a “smart” thiazole orange (TO) nucleotide that requires formation of the probe-target complex in order to become fluorescent (Figure 7c) [75].
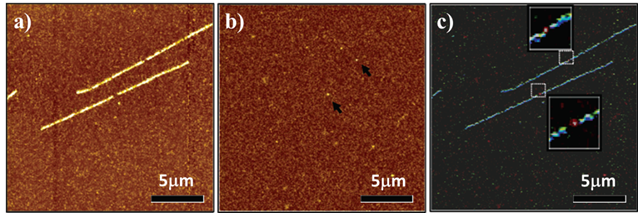
Besides the traditional fluorescent dyes, some new technologies, such as scanning near-field optical (SNOM) and atomic force microscopy (AFM), have also been used to design the PNA probes. Kim et al reported an Alexa 532 pigment labeled PNA probe to hybridize with the top part of the ea47 gene within the DNA molecule. The topographic images were concomitantly obtained by the near-field fluorescence image (Figure 8) [76].
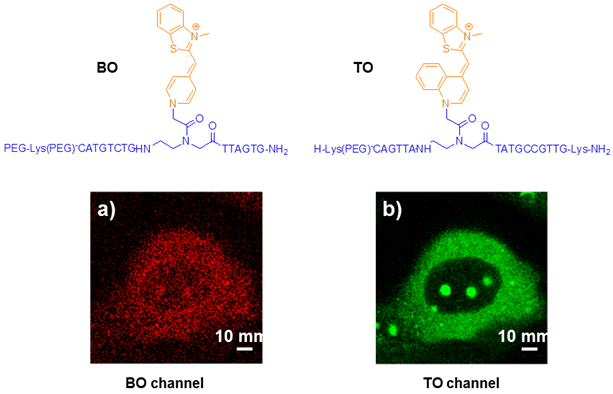
PNA MBs as fluorogenic hybridization probes could show the synthesis and transport of particular RNA molecules in living cells. Kummer et al explored the PNA-based FIT-probes in the simultaneous imaging of two different viral mRNA molecules expressed during the replication cycle of the H1N1 influenza virus (Figure 9) [77].Confocal laser scanning microscopy (CLSM) images showed that the TO fluorescence signal corresponding to NA mRNA was mainly concentrated in the nucleoli at the beginning and moved to the cytosol at later stages. Control experiments suggest that the TO signals in Figure 9 reflect the localization of NA mRNA rather than a localization bias of the probe. The BO signals revealed a different expression pattern for M1 mRNA. This mRNA localized to the nucleoli as well as cytosolic parts as soon as the BO fluorescence was detectable. These experiments provide evidence for the usefulness of PNA FIT-probes in investigations on the temporal and spatial progression of mRNA synthesis in living cells.
a) Structure of the PNA probe (B denotes nucleobases). The asymmetric cyanine dye thiazole orange (TO) is conjugated to a PNA with the sequence lys+-CCTTTTTCTT. b) Chemical structure of the thiazole orange derivative (TO-N'-10-COOH). c) Chemical structure of Aeg(TO) employed in PNA. (Reprinted with permission from ref. 71, 72. Copyright 2000, 2000. Elsevier B. V., Elsevier B. V.)
Near-field fluorescence images for the single target DNA fluorescence in situ hybridization samples by fluorescence resolution of 50 nm. a) The fluorescence images of DNA molecule (stained with molecular probe); b) The fluorescence images of Alexa 532 pigment, and c) The overlapped images of a and b. In the image of c, the YOYO-1 and A532 fluorescence signals are artificially colored in blue and red, respectively. (Reprinted with permission from ref. 76. Copyright 2004, American Chemical Society.)
Chemical structures of PNA intercalator dye (BO, TO) and CLSM images of influenza A infected MDCK cells simultaneously stained with the matrix protein 1 (M1) specific BO-probe (a) and the neuraminidase (NA) specific TO probe (b) at indicated time points post infection. White bars correspond to 10 μm. (Reprinted with permission from ref. 77. Copyright 2012, American Chemical Society.)
PNA probes could also be targeted to quadruplex DNA. Datta et al synthesized a short PNA probe which shows high thermal and thermodynamic stabilities for the PNA and the quadruplex structure of thrombin binding aptamer (TBA) hybrid. The ability of PNA to invade a structured DNA target expands its potential utility as an antigene agent or hybridization probe [78].
Short PNA molecular beacons were used for real-time PCR allelic discrimination of single nucleotide polymorphisms. Petersen et al. reported a real-time PCR assay for the genotyping of single nucleotide polymorphisms using short PNA molecular beacons. The length of PNA MBs is significantly shorter than the probe, making probe design and genotype discrimination easier [79].
More recently, Kam et al used a PNA molecular beacon to detect endogenous K-ras mRNA in living cells. The fast hybridization kinetics and the single mismatch discrimination of PNA MBs make such MBs promising for in vivo real-time imaging of mRNA with single nucleotide polymorphism (SNP) resolution [80].
Locked Nucleic Acids (LNA). The future challenges in diagnostics and treatment of diseases by DNA/RNA call for novel technologies in vitro and in vivo targeting nucleic acid molecules. Obika et al [81]. (1997) and Koshkin et al [82]. (1998) reported a minimal alteration of the pentose sugar of ribo- and deoxyribonucleotides that constrained, or “locked,” the sugar in the N-type conformation seen in A-form DNA. This alteration then served as the foundation for synthesizing locked nucleotide phosphoramidite monomers (table 1). Oligonucleotides containing one or more of monomers were given the name “lock nucleic acid” (LNA). LNA nucleotides can be mixed with DNA or RNA residues in the oligonucleotide whenever desired. Since then, LNA draw more and more attention in molecular biology techniques especially in field of nucleic acid.
As shown in table 1, the restricted 3'-endo conformation of ribose ring drastically reduced the conformational flexibility of LNA, thus, LNA shows remarkably thermodynamic stability. According to Watson-Crick rules [83,84], LNA has high affinity for complementary DNA and RNA. The Tm increase for LNA-DNA hybridization in solution ranges from 2.0 to 6.0 °C per LNA monomer [85]. LNA shows the good discrimination of mismatch base pairs for the native nucleic acids, especially for less than 10 bases [86]. Besides, LNAs have other excellent properties for nucleic acid biosensor, as below:
1. Short probes with high Tm: Perfect for detection of short RNA.
2. Increased discriminatory power: Single base discrimination capability [87].
3. Resistant to exo- and endo-nucleases: High stability both in vivo and in vitro.
4. Increased target specificity: Fast binding to targets.
5. Strand invasion: Detect “hard to access” samples.
LNAs have been widely used in microarrays to detect DNA [88], miRNAs [89] and PCR amplicons [90]. For its specific detection and good stability, LNA was also used in electrochemical biosensors [91,92] and carbon nanotube biosensors [93].
For the application in MBs, LNA has been used to overcome traditional limitations of molecular beacons (MB). Wang et al. designed a novel MB of LNA which exhibits very high melting temperature, enhanced single base mismatch discrimination capability, stability against digestion by nuclease and no binding with single-stranded DNA binding proteins [94].
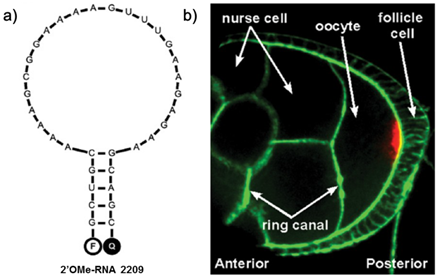
In order to obtain image dynamic processes involving RNAs in living cells, Irina et al synthesized another MB of LNA from 2'-O-methylribonucleotides which established a reliable approach for visualizing native mRNAs in real time. They found that the incorporation of just a few LNA nucleotides enabled these shorter probes to stably anneal to more structured regions of the RNA than conventional molecular beacons (Table 2) [95]. Spinning disk confocal microscopy images showed the tiny LNA that specific for oskar mRNA could access target regions and hybridize to target mRNA efficiently in living Drosophila melanogaster oocytes (Figure 11).
Schematic chemical model of LNA (blue) hybridized with DNA (red) in antiparallel orientation, with the hydrogen bonding between complementary nucleobases depicted by dotted lines.
a) Predicted structure for 2'OMe-RNA 2209 that specifically targets oskar mRNA. b) Oskar mRNA localization (red) in a fixed Drosophila melanogaster egg chamber at mid-oogenesis, as detected with the 2'OMe-RNA 2209 molecular beacon. Oskar mRNA is transported from the nurse cells to the oocyte through ring canals via a microtubule-dependent process. Action was stained with Phalloidin-FITC (green). (Reprinted with permission from ref. 95. Copyright 2012, American Chemical Society.)
Molecular beacon sequences and their labelsa
| Name | Sequence (5'-3') |
|---|---|
| 2'OMe-RNA 1214 | TMR - ggucg UUGUAGGUUCCACUGGUU cgacc - BHQ-2 |
| 2'OMe-RNA 2209 | Cy5 - gcugc AAAAGCGGAAAAGUUUGAAGAGAA gcagc - BHQ-2 |
| 2'OMe-RNA 2213 | TMR - cggc AAGUUUGAAGA gccg - dabcyl |
| LNA/2'OMe-RNA 1227 | TMR - gccg AAUCGUUGUAG cggc - dabcyl |
| LNA/2'OMe-RNA 2210-1 | TMR - gcc GAAGAGAA ggc - dabcyl |
| LNA/2'OMe-RNA 2210-2 | TMR - gcc GAAGAGAA ggc - dabcyl |
| LNA/2'OMe-RNA 2213-1 | TMR - cggc AAGUUUGAAGA gccg - dabcyl |
| LNA/2'OMe-RNA 2213-2 | TMR - cggc AAGUUUGAAGA gccg - dabcyl |
aunderlined nucleotides = LNA; italic nucleotides = stem sequence; UPPER CASE nucleotides = probe sequence; TMR = tetramethylrhodamine; BHQ-2 = Black Hole Quencher 2. (Reprinted with permission from ref. 95. Copyright 2012, American Chemical Society.)
Traditional MBs for DNA exhibit poor stability and low signal enhancement once immobilized onto a solid surface. The reason is that the signals of MBs were usually blocked when the hairpin structure immobilized on to a solid surface. Martinez et al [96] reported a new LNA molecular beacon that overcomes limitations of MBs for surface immobilization. They achieved a 25-fold enhancement with detection limits reaching the nanomolar range. The LNA-based biosensor was shown to possess better stability, reproducibility, selectivity, and robustness when compared with the traditional MBs. Since then, LNA was also applied to gold (111) surface with a stronger DNA recognition signal (4-4.5 times) than its counterpart, and could be differentiated between a fully complementary DNA target and a single base mismatch, where the mismatch discrimination ratio is almost two times comparing with the ratio relevant in case of DNA-based detection [97].
In order to check the hybridization ability of LNA nucleotides in the unpaired region of the MBs, Han et al. designed a DNA hairpin containing a 19-mer loop and a six base-pair stem [98]. They evaluated the single-base mismatch recognition power of surface immobilized. By AFM nanolithography technique, they can directly measure the variations of overall film thickness or miniaturized nano-spot height before and after the interfacial molecules hybridization.
LNA nucleotides have also been introduced as biosensors in DNA/RNA aptamers. The traditional nucleic acid aptamers are not only susceptible to nucleases but also subjective to degradation by proteases. To date, chemical modifications which were employed to overcome those problems showed insufficient efficacy and specificity in vivo. LNA's rigid conformation could overcome the shortcomings that were mentioned above. Darfeuille et al [99] studied LNA nucleotides as nuclease resistant aptamers into the RNA targeted against the HIV-1 trans-activation response (TAR) RNA. Although most of their efforts resulted in non-functional chimeric nucleic acids, one of LNA derivatives behaves as the parent RNA aptamer and retains similar key structural determinants for binding to TAR. This example indicates that LNA modifications alternated with DNA could increase their stability and nuclease resistance without necessarily reducing their affinity for the target molecule.
Moreover, Astakhova et al. found that insertion of the LNA nucleotides into the probes resulted in high binding affinity to cDNA and improved fluorescence quantum yields. Mixer LNA/DNA fluorescent probes containing the 1-(phenylethynyl)pyrene fluorophore were synthesized. The conjugates displayed significantly higher hybridization affinity to target DNA; increased fluorescence quantum yields of single-stranded oligonucleotides and their duplexes; and improved ability to form an interstrand excimer compared with analogous non-LNA probes [100].
In addition, LNA also displayed potential applications as aptamers when it has been introduced into a sequence [101,102] or incorporated with exciton-controlled hybridization-sensitive fluorescent oligonucleotide (ECHO) probes [103]. This makes LNA nucleotides as one of the future directions of chemical conjugations to aptamers.
The applications of LNA in real-time PCR were investigated by several groups. Morandi et al used the LNA based real time PCR in particular situations where there are difficulties in primer design due to sequence complexity. They studied a new real time RT-PCR assay using LNA modified primers and LNA MB probes to monitor hepatitis C virus (HCV) viral load in plasma and serum samples. The LNA based RT-PCR assay was successful to measure quantitatively the level of HCV RNA with high sensitivity (50 IU/ml), the wide range of genotype detection and increased specificity [104]. On that basis, they further reported a novel assay called Allele Specific LNA quantitative PCR (ASLNAqPCR) containing LNA-modified allele specific primers and LNA-modified beacon probes. The probes show increase sensitivity, specificity and accurately quantify mutations in diagnose and quantify mutations. Moreover, ASLNAqPCR can be performed in any laboratory with real-time PCR equipment and can easily be adapted to detect hot spot mutations in other oncogenes [105]. Besides, Hillyer et al also reported a quantitative real-time PCR assay that incorporates the use of molecular beacon and LNA probes to overcome the impediment of 12 human interferon-alpha (IFN-α) and 3 IFN-λ subtypes high homology [106].
1', 5'- anhydrohexitol nucleic acid (HNA). 1', 5'-anhydrohexitol nucleic acids (HNA) are oligonucleotides that were built up from natural nucleobases and a phosphorylated 1, 5-anhydro-D-arabino-2, 3-dideoxyhexitol building blocks with a base moiety positioned in the 2-position (Table 1). HNA is able to hybridize sequence selectively as well with DNA, RNA, and with itself [107]. HNA could form highly selective and exceptionally stable duplexes with RNA. This is due to the positioning of the base moiety in the 2'-position, instead of the anomeric position, allowing the oligomer to fold in a helix-like structure with the same geometry as found in the α-form of dsRNA [108]. Besides, they are stable toward nuclease degradation.
The conformational preference of the hexitol monomers is driven by the fact that the ring oxygen atom only contains unshared pairs and that steric strains should be avoided in selecting the energetically most favorable conformation of a molecule. The base moiety of the hexitol nucleosides is axially oriented. When these monomers are polymerized, oligomers are obtained that form helical duplex structures as well with DNA and RNA and with itself (HNA), with a geometry resembling those of the Watson-Crick pairing natural nucleic acids [109].
The HNA/RNA duplex is more stable than all other associations of natural nucleic acids (dsDNA, dsRNA, DNA/RNA). The order of duplex stability is given by HNA/HNA > HNA/RNA > HNA/DNA [110]. This makes HNAs excellent probes for nucleic acid especially for RNA. Abramov et al used the Cy5 fluorescent dye labeled HNA high-affinity arrays for detections of DNA and RNA single-base mismatches and found the relative intensity of the signal and match/mismatch discrimination increased up to 5-fold for DNA targets and up to 3-3.5-fold for RNA targets applying HNA arrays [111]. However, in contrast to DNA/RNA duplexes, the corresponding HNA/RNA hybrids are poorly hydrolyzed by RNAseH [112].
Threose nucleic acid (TNA). (3'-2')-a-L-Threose nucleic acid (TNA) is an unnatural nucleic acid that was identified during an extensive evaluation of alternative sugar-phosphate backbones aiming at explaining the structure of the biological nucleic acids (Table 1) [113,5]. Because threose is one of the two four-carbon monosaccharides, TNA is the simplest of all potential sugar containing nucleic acids. TNA possesses the ability to specifically base-pair with RNA, DNA, and itself. This capability with the chemical simplicity of threose relative to ribose suggests that TNA could have acted as an evolutionary competitor of RNA or even have preceded RNA as the genetic molecule of life[114,115]. Moreover, TNA hybridizes efficiently with nucleic acid in a sequence-specific manner. Thus, TNA could be a good candidate for the development of biosensors in the near future.
Altritol (ANA) and cyclohexene (CeNA). A further development of MBs is that chemical modification of the oligonucleotides is relatively easy and straightforward, which could provide an ideal framework and the flexible synthetic methods to create the novel probes, such as altritol (ANA) and cyclohexene (CeNA) that are also becoming increasingly attractive for various applications.
In order to increase minor-groove hybridization as well as in an effort to influence hybridization in a beneficial way, D-altritol nucleic acid (ANA) was designed [3]. ANA consists of a phosphorylated D-altritol backbone with nucleobases inserted in the 2'-position of the carbohydrate moiety (Table 1). They differ structurally from HNA by the presence of a supplementary hydroxyl group in the 3'-α-position. Inversion of configuration, giving the 3'-(R)-form, D-mannitol nucleic acids (MNA) lack hybridization capabilities with natural nucleic acid [116].
The introduction of an additional hydroxyl group in the 2'-position of natural furanose nucleosides or in the 3'-position of 1,5-anhydrohexitol nucleosides influence hybridization owing to the following effects: a) influence on the conformation of the nucleoside itself, which might become locked in one of the extreme furanose conformations; b) influence on the polarity of the solvent-accessible surface and c) stabilization of a particular oligonucleotide conformation due to the formation of hydrogen bonds and steric effects. Recently, the application of ANA as biosensor has employed high-affinity arrays to detect single-base mismatches of DNA and RNA [72].
Cyclohexene nucleic acid (CeNA, Table 1) as a new nucleic acid structure is the replacement of the furanose moiety of DNA by a cyclohexene ring. It can be obtained by the classical phosphoramidite chemisty starting from protected cyclohexenyl nucleoside building blocks [6]. Incorporation of cylcohexenyl nucleosides in a DNA chain increases the stability of a DNA/RNA hybrid. The complex formed between cyclohexenyl oligoadenylate and its DNA or RNA complement is of similar stability. Replacement of natural DNA and RNA with a six-membered ring increases the conformational rigidity of the oligomers. The introduction of a double bond into a cyclohexane ring gives a cyclohexene ring with a half-chair form as a global minimum. The cyclohexene system is more flexible than the cyclohexane system, and approaches more the flexibility of a furanose ring.
ANA and CeNA have already established themselves as attractive recognition candidates in nucleic acid biosensor technology. The unique structural and hybridization features of ANA and CeNA make them superior to DNA for use as a sequence-specific hybridization probe, and open up exciting opportunities for DNA diagnostics.
Conclusion and Outlook
XNA molecular beacon as a new sensor opened up new opportunities to monitor DNA/RNA in vitro and in vivo. Despite oligonucleotide-based fluorescent probes have been greatly improved, by far, there are only a few probes that could be applied in living cells. Thus, possible future directions include the improvement of the sensitivity and specificity of probes, as well as their photo physical properties (including the development of near infrared dyes and increasing their quantum yield). Radiometric and FRET probes with different excitation and emission parameters would be pretty desirable, as they could allow the dynamics of several XNA probes to bind DNA/RNA, and help the researchers to clarify more details about the interaction between XNA and DNA/RNA in the level of living cells. These active directions are great pivotal fields between chemistry and biology, which will credibly create new chemical tools that further enhance our understanding of the complex role of XNA-DNA or XNA-RNA in the cancer therapeutics. However, XNA biosensors are still not ready for large-scale decentralized testing applications. Such applications would require improvements in the sensitivity of XNA biosensors, and in their ability to recognize mutations in large DNA fragments. The applications of XNAs will still have a long way to go in the future.
Acknowledgements
We thank the National Basic Research 973 Program (2013CB733700) and the Fundamental Research Funds for the Central Universities (WJ1213007), the talent program of ECUST (YJ0142149), the Program of Shanghai Pujiang (K100-2-1275) for financial support and the National Natural Science Foundation of China (21341001).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Steele FR, Gold L. The sweet allure of XNA. Nat Biotechnol. 2012;30:624-625
2. Van Aerschot A, Verheggen I, Hendrix C, Herdewijn P. 1, 5-Anhydrohexitol nucleic acids, a new promising antisense construct. Angew Chem. 1995;34:1338-1339
3. Allart B, Khan K, Rosemeyer H, Schepers G, Hendrix C, Rothenbacher K, Seela F, Van Aerschot A, Herdewijn P. D-altritol nucleic acids (ANA): hybridisation properties, stability, and initial structural analysis. Chem Eur J. 1999;5(8):2424-2431
4. James Wilds C, Jose Damha M. Duplex recognition by oligonucleotides containing 2'-deoxy-2'-fluoro-d-arabinose and 2'-deoxy-2'-fluoro-d-ribose. Intermolecular 2'-OH-phosphate contacts versus sugar puckering in the stabilization of triple-helical complexes. Bioconjugate Chem. 1999;10(2):299-305
5. Schoning KU, Scholz P, Guntha S, Wu X, Krishnamurthy R, Eschenmoser A. Chemical etiology of nucleic acid structure: the α-threofuranosyl-(3'→2') oligonucleotide system. Science. 2000;290:1347-1351
6. Wang J, Verbeure B, Luyten I, Lescrinier E, Froeyen M, Hendrix C, Rosemeyer H, Seela F, van Aerschot A, Herdewijn P. Cyclohexene nucleic acids (CeNA): serum stable oligonucleotides that activate RNase H and increase duplex stability with complementary RNA. J Am Chem Soc. 2000;122(36):8595-8602
7. Viani MB, Pietrasanta LI, Thompson JB, Chand A, Gebeshuber IC, Kindt JH, Richter M, Hansma HG, Hansma PK. Probing protein-protein interactions in real time. Nat Struct Mol Biol. 2000;7:644-647
8. Ziegler C. Cantilever-based biosensors. Anal Bioanal Chem. 2004;379:946-959
9. Cooper MA. Label-free screening of bio-molecular interactions. Anal Bioanal Chem. 2003;377:834-842
10. Katz E, Willner I. Probing biomolecular interactions at conductive and semiconductive surfaces by impedance spectroscopy: routes to impedimetric immunosensors, DNA-Sensors, and enzyme biosensors. Electroanalysis. 2003;15:913-947
11. Homola J. Present and future of surface plasmon resonance biosensors. Anal Bioanal Chem. 2003;377:528-539
12. Asanov AN, Wilson WW, Oldham PB. Regenerable Biosensor Platform: A Total Internal Reflection Fluorescence Cell with Electrochemical Control. Anal Chem. 1998;70:1156-1163
13. Tyagi S, Kramer FR. Molecular Beacons: Probes that Fluoresce upon Hybridization. Nat Biotechnol. 1996;14:303-308
14. Tyagi S, Bratu D, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat Biotechnol. 1998;16:49-53
15. Hwang GT, Seo YJ, Kim BH. A highly discriminating quencher-free molecular beacon for probing DNA. J Am Chem Soc. 2004;126:6528-6529
16. Sandhya S, Chen W, Mulchandani A. Molecular beacons: A real-time polymerase chain reaction assay for detecting Escherichia coli from fresh produce and water. Anal Chim Acta. 2008;614:208-212
17. Takács T, Jeney C, Kovács L, Mózes J, Benczik M, Sebe A. Molecular beacon-based real-time PCR method for detection of 15 high-risk and 5 low-risk HPV types. J Virol Methods. 2008;149:153-162
18. Fang XH, Li JWJ, Perlette J, Tan WH, Wang KM. Peer reviewed: molecular beacons: novel fluorescent probes. Anal Chem. 2000;72:747A-753A
19. Tan WH, Wang KM, Drake TJ. Molecular beacons. Curr Opin Chem Biol. 2004;8:547-553
20. Wang KM, Tang ZW, Yang CJ, Kim YM, Fang XH, Li W, Wu YR, Medley D C, Cao ZH, Li J, Colon P, Lin H, Tan WH. Molecular engineering of DNA: molecular beacons. Angew Chem. 2009;48:856-870
21. Oh KJ, Cash KJ, Hugenberg V, Plaxco KW. Peptide beacons: a new design for polypeptide-based optical biosensors. Bioconjugate Chem. 2007;18:607-609
22. Cheng W, Chen YL, Yan F, Ding L, Ding SJ, Ju HX, Yin YB. Ultrasensitive scanometric strategy for detection of matrix metalloproteinases using a histidine tagged peptide-Au nanoparticle probe. Chem Commun. 2011;47:2877-2879
23. Zhu L, Xie J, Swierczewska M, Zhang F, Lin X, Fang X, Niu G, Lee S, Chen X. Dual-functional, receptor-targeted fluorogenic probe for in vivo imaging of extracellular protease expressions. Bioconjugate Chem. 2011;22:1001-1005
24. Oh KJ, Cash KJ, Plaxco KW. Excimer-based peptide beacons: a convenient experimental approach for monitoring polypeptide-protein and polypeptide-oligonucleotide interactions. J Am Chem Soc. 2006;128:14018-14019
25. Wu JC, Zawistowski A, Ehrmann M, Yi T, Schmuck C. Peptide functionalized polydiacetylene liposomes act as a fluorescent turn-on sensor for bacterial lipopolysaccharide. J Am Chem Soc. 2011;133:9720-9723
26. Wu JC, Zou Y, Li C, Sicking W, Piantanida I, Yi T, Schmuck C. A molecular peptide beacon for the ratiometric sensing of nucleic acids. J Am Chem Soc. 2012;134:1958-1961
27. Pazos E, Mosquera J, Vazquez ME, Mascarenas JL. DNA recognition by synthetic constructs. ChemBioChem. 2011;12:1958-197
28. Somers WS, Phillips SEV. Crystal structure of the met represser-operator complex at 2.8 Å resolution reveals DNA recognition by β-strands. Nature. 1992;359:387-393
29. Yang RH, Jin JY, Long LP, Wang YX, Wang H, Tan WH. Reversible molecular switching of molecular beacon: controlling DNA hybridization kinetics and thermodynamics using mercury (II) ions. Chem Commun. 2009;3:322-324
30. Lin YW, Ho HT, Huang CC, Chang HT. Fluorescence detection of single nucleotide polymorphisms using a universal molecular beacon. Nucleic Acids Res. 2008;36:e123
31. Nielsen PE. Applications of peptide nucleic acids. Curr Opin Biotechnol. 1999;10(1):71-75
32. Tsuji A, Koshimoto H, Sato Y, Hirano M, Sei-Iida Y, Kondo S, Ishibashi K. Direct observation of specific messenger RNA in a single living cell under auorescence microscope. Biophys J. 2000;78:3260-3274
33. Tsuji A, Sato Y, Hirano M, Suga T, Koshimoto H, Taguchi T, Ohsuka S. Development of a time-resolved-uorometric method for observing hybridization in living cells using-uorescence resonance energy transfer. Biophys J. 2001;81:501-515
34. Endoh T, Funabashi H, Mie M, Kobatake E. Method for detection of specific nucleic acids by recombinant protein with fluorescent resonance energy transfer. Anal Chem. 2005;77:4308-4314
35. Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497-1500
36. Antsypovitch SI. Peptide nucleic acids: structure, properties, applications, strategies and practice of chemical synthesis. Russian Chem Rev. 2002;71(1):71-83
37. Nielsen PE. Peptide nucleic acid: a versatile tool in genetic diagnostics and molecular biology. Anal biotechnol. 2001;12:16-20
38. Shakeel S, Karim S, Ali A. Peptide nucleic acid (PNA)-a review. J Chem Technol Biotechnol. 2006;81:892-899
39. Ali M, Neumann R, Ensinger W. Sequence-specific recognition of DNA oligomer using peptide nucleic acid (PNA)-modified synthetic ion channels: PNA/DNA hybridization in nanoconfined environment. ACS Nano. 2010;4(12):7267-7274
40. Hüsken N, Gębala M, Schuhmann W, Metzler-Nolte N. A single-electrode, dual-potential ferrocene-PNA biosensor for the detection of DNA. ChemBioChem. 2010;11:1754-1761
41. Kim SK, Cho H, Jeong J, Kwon JN, Jung Y, Chung BH. Label-free and naked eye detection of PNA/DNA hybridization using enhancement of gold nanoparticles. Chem Commun. 2010;46:3315-3317
42. Lao AIK, Su X, Aung KMM. SPR study of DNA hybridization with DNA and PNA probes under stringent conditions. Biosens Bioelectrons. 2009;24:1717-1722
43. Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. PNA hybridizes to complementary oligonucleotides obeying the watson-crick hydrogen-bonding rules. Nature. 1993;365(6446):566-568
44. Nielsen PE. Structural and biological properties of peptide nucleic acid (PNA). Pure Appl Chem. 1998; 70: 105-110. d) Hyrup B and Nielsen PE, Peptide nucleic acids (PNA): synthesis, properties and potential applications. Bioorg Med Chem. 1996;4:5-23
45. Demidov V, Frank-Kamenetskii MD. Two sides of the coin: affinity and specificity of nucleic acid interactions. Trends Biochem Sci. 2004;29:62-71
46. Demidov V, Potaman VN, Frank-Kamenetskii MD, Egholm M, Buchardt O, Sonnichsen SH. et al. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem Pharmacol. 1994;48:1310-1313
47. Nielsen PE. Peptide nucleic acid: a versatile tool in genetic diagnostics and molecular biology. Curr Opin Biotechnol. 2001;12:16-20
48. Demidov V, PNA comes of age. from infancy to maturity. Drug Discov Today. 2002;7:153-155
49. Nielsen PE. Peptide nucleic acids as therapeutic agents. Curr Opin Struct Biol. 1999;9:353-357
50. Hanvey JC, Peffer NJ, Bisi JE. Antisense and antigene properties of peptide nucleic acids. Science. 1992;258:1481-1485
51. Demidov V, Frank-Kamenetskii MD, Egholm M, Buchardt O, Nielsen PE. Sequence specific double strand DNA cleavage by peptide nucleic acid (PNA) targeting using nuclease S1. Nucleic Acids Res. 1993;21:2103-2107
52. Veselkov AG, Demidov V, Nielsen PE, Frank-Kamenetskii MD. A new class of genome rare cutters. Nucleic Acids Res. 1996;24:2483-2487
53. Ulhmann V, Prasad M, Silva I, Luettich K, Grande L, Alonso L, Thisted M, Pluzek KJ, Gorst J, Ring M. et al. Improved in situ detection method for telomeric tandem repeats in metaphase spreads and interphase nuclei. Mol Pathol. 2000;53:48-50
54. Kajstura J, Pertoldi B, Leri A, Beltrami CA, Deptala A, Darzynkiewicz Z, Anversa P. Telomere shortening is an in vivo marker of myocyte replication and aging. Am J Pathol. 2000;156:813-819
55. Deng W, Lucas JN. Combined fish with pan-telomeric PNA and whole chromosome-specific DNA probes to detect complete and incomplete chromosomal exchanges in human lymphocytes. Int J Radiat Biol. 1999;75:1107-1112
56. Fomina J, Darroudi F, Boei JJWA, Natarajan AT. Discrimination between complete and incomplete chromosome exchanges in X-irradiated human lymphocytes using FISH with pan-centromeric and chromosome specific DNA probes in combination with telomeric PNA probe. Int J Radiat Biol. 2000;76:807-813
57. Boei JJWA, Vermeulen S, Natarajan AT. Analysis of radiationinduced chromosomal aberrations using telomeric and centromeric PNA probes. Int J Radiat Biol. 2000;76:163-167
58. Wang M, Holmes-Davis R, Rafinski Z, Jedrzejewska B, Choi KY, Zwick M, Bupp C, Izmailov A, Paczkowski J, Warner B, Koshinsky H. Accelerated photobleaching of a cyanine dye in the presence of a ternary target DNA, PNA probe, dye catalytic complex: a molecular diagnostic. Anal Chem. 2009;81(6):2043-2052
59. Orum H, Nielsen PE, Jorgensen M, Larsson C, Stanley C, Koch T. Sequence-specific purification of nucleic acids by PNA-controlled hybrid selection. BioTechniques. 1995;19:472-480
60. Chandler DP, Stults JR, Anderson KK, Cebula S, Schuck BL, Brockman FJ. Affinity capture and recovery of DNA at femtomolar concentrations with peptide nucleic acid probes. Anal Biochem. 2000;283:241-249
61. Chandler DP, Stults JR, Cebula S, Schuck BL, Weaver DW, Anderson KK, Egholm M, Brockman FJ. Affinity purification of DNA and RNA from environmental samples with peptide nucleic acid clamps. Appl Environ Microbiol. 2000;66:3438-3445
62. Seeger C, Batz H-G, Orum H. PNA-mediated purification of PCR amplifiable human genomic DNA from whole blood. BioTechniques. 1997;23:512-516
63. Ferreira GNM, Silva DAC, Tome B. Acoustic wave biosensors: physical models and biological applications of quartz crystal microbalance. Trends Biotechnol. 2009;27:689-697
64. Rogero C, Chaffey BT, Mateo-Marti E, Sobrado JM, Horrocks BR, Houlton A, Lakey JH, Briones C, Martin-Gago JA. Silicon surface nanostructuring for covalent immobilization of biomolecules. J Phys Chem C. 2008;112:9308-9314
65. Briones C, Moreno M. Applications of peptide nucleic acids (PNAs) and locked nucleic acids (LNAs) in biosensor development. Anal Bioanal Chem. 2012;402:3071-3089
66. Fang ZC, Kelley SO. Direct electrocatalytic mRNA detection using PNA-nanowire sensors. Anal Chem. 2009;81(2):612-617
67. Cattani-Scholz A, Pedone D, Dubey M, Neppl S, Nickel B, Feulner P, Schwartz J, Abstreiter G, Tornow M. Organophosphonate-based PNA-functionalization of silicon nanowires for label-free DNA detection. ACS Nano. 2008;2(8):1653-1660
68. Ortiz E, Estrada G, Lizardi PM. PNA molecular beacons for rapid detection of PCR amplicons. Mol Cell Probes. 1998;12:219-226
69. Tsourkas A, Behlke MA, Bao G. Structure-function relationships of shared-stem and conventional molecular beacons. Nucleic Acids Res. 2002;30:4208
70. Socher E, Jarikote DV, Knoll A, Roglin L, Burmeister J, Seitz O. FIT probes: Peptide nucleic acid probes with a fluorescent base surrogate enable real-time DNA quantification and single nucleotide polymorphism discovery. Anal Biochem. 2008;375:318-330
71. Isacsson J, Cao H, Ohlsson L, Nordgren S, Svanvik N, Westman G, Kubista M, Sjoback R, Sehlstedt U. Rapid and specific detection of PCR products using light-up probes. Mol Cell Probes. 2000;14:321-328
72. Svanvik N, Stahlberg A, Sehlstedt U, Sjoback R, Kubista M. Detection of PCR products in real time using light-up probes. Anal Biochem. 2000;287:179
73. Nygren J, Svanvik N, Kubista M. The interactions between the fluorescent dye thiazole orange and DNA. Biopolymers. 1998;46:39-51
74. Svanvik N, Westman G, Wang DY, Kubista M. Light-up probes: thiazole orange-conjugated peptide nucleic acid for detection of target nucleic acid in homogeneous solution. Anal Biochem. 2000;281:26-35
75. Socher E, Knoll A, Seitz O. Dual fluorophore PNA FIT-probes-extremely responsive and bright hybridization probes for the sensitive detection of DNA and RNA. Org Biomol Chem. 2012;10:7363-7371
76. Kim JM, Hirose T, Sugiyama S, Ohtani T, Muramatsu H. Visualizing a hybridized PNA probe on a DNA molecule with near-field optical microscopy. Nano Letters. 2004;4(11):2091-2097
77. Kummer S, Knoll A, Socher E, Bethge L, Herrmann A, Seitz O. PNA FIT-probes for the dual color imaging of two viral mRNA targets in influenza H1N1 infected live cells. Bioconjugate Chem. 2012;23(10):2051-2060
78. Datta B, Armitage BA. Hybridization of PNA to structured DNA targets: quadruplex invasion and the overhang effect. J Am Chem Soc. 2001;123(39):9612-9619
79. Petersen K, Vogel U, Rockenbauer E, Nielsen KV, Kolvraa S, Bolund L, Nexo B. Short PNA molecular beacons for real-time PCR allelic discrimination of single nucleotide polymorphisms. Mol Cell Probes. 2004;18(2):117-22
80. Kam Y, Rubinstein A, Nissan A, Halle D, Yavin E. Detection of endogenous K-ras mRNA in living cells at a single base resolution by a PNA molecular beacon. Mol Pharm. 2012;9(3):685-93
81. Obika S, Nanbu D, Hari Y, Morio K-i, In Y, Ishida T, Imanishi T. Synthesis of 2'-O,4'-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3, -endo sugar puckering. Tetrahedron Lett. 1997;38(50):8735-8738
82. Koshkin AA, Rajwanshi VK, Wengel J. Novel convenient syntheses of LNA bicyclo nucleosides. Tetrahedron Lett. 1998;39(24):4381-4384
83. Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5- methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54(14):3607-3630
84. Obika S, Nanbu D, Hari Y, Andoh J, Morio K, Doi T, Imanishi T. Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation, 2'-O,4'-C-methyleneribonucleosides. Tetrahedron Lett. 1998;39(30):5401-5404
85. Natsume T, Ishikawa Y, Dedachi K, Tsukamoto T, Kurita N. Effect of base mismatch on the electronic properties of DNA-DNA and LNA-DNA double strands: density-functional theoretical calculations. Chem Phys Lett. 2007;446(1-3):151-158
86. Petersen M, Wengel J. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 2003;21(2):74-81
87. Ratilainen T, Holmen A, Tuite E, Nielsen PE, Norden B. Thermodynamics of sequence-specific binding of PNA to DNA. Biochemistry. 2000;39(26):7781-7791
88. Diercks S, Gescher C, Metfies K, Medlin LK. Evaluation of locked nucleic acids for signal enhancement of oligonucleotide probes for microalgae immobilised on solid surfaces. J Appl Phycol. 2009;21(6):657-668
89. Fang S, Lee HJ, Wark AW, Corn RM. Attomole microarray detection of MicroRNAs by nanoparticle-amplified SPR imaging measurements of surface polyadenylation reactions. J Am Chem Soc. 2006;128(43):14044-14046
90. Orum H, Jakobsen MH, Koch T, Vuust J, Borre MB. Detection of the factor V Leiden mutation by direct allelespecific hybridization of PCR amplicons to photoimmobilized locked nucleic acids. Clin Chem. 1999;45(11):1898-1905
91. Chen J, Zhang J, Wang K, Lin X, Huang L, Chen G. Electrochemical biosensor for detection of BCR/ABL fusion gene using locked nucleic acids on 4-aminobenzenesulfonic acidmodified glassy carbon electrode. Anal Chem. 2008;80(21):8028-8034
92. Lin LQ, Lin XH, Chen JH, Chen W, He M, Chen YZ. Electrochemical biosensor for detection of BCR/ABL fusion gene based on hairpin locked nucleic acids probe. Electrochem Commun. 2009;11(8):1650-1653
93. Berti F, Eisenkolbl C, Minocci D, Nieri P, Rossi AM, Mascini M, Marrazza G. Cannabinoid receptor gene detection by electrochemical genosensor. J Electroanal Chem. 2011;656(1-2):55-60
94. Wang L, Yang CJ, Medley CD, Benner SA, Tan W. Locked nucleic acid molecular beacons. J Am Chem Soc. 2005;127(45):15664-15665
95. Catrina IE, Marras SAE, Bratu DP. Tiny molecular beacons: LNA/2'-o-methyl RNA chimeric probes for imaging dynamic mRNA processes in living cells. ACS Chem. Biol. 2012;7:1586-1595
96. Martinez K, Estevez MC, Wu YR, Phillips JA, Medley CD, Tan WH. Locked nucleic acid based beacons for surface interaction studies and biosensor development. Anal Chem. 2009;81(9):3448-3454
97. Mishra S, Ghosh S, Mukhopadhyay R. Ordered self-assembled locked nucleic acid (LNA) structures on Gold(111) surface with enhanced single base mismatch recognition capability. Langmuir. 2012;28(9):4325-4333
98. Han WH, Liao JM, Chen KL, Wu SM, Chiang YW, Lo ST, Chen CL, Chiang CM. Enhanced recognition of single-base mismatch using locked nucleic acid-integrated hairpin DNA probes revealed by atomic force microscopy nanolithography. Anal Chem. 2010;82(6):2395-2400
99. Darfeuille F, Hansen JB, Orum H, Primo1 CD, Toulmea JJs. LNA/DNA chimeric oligomers mimic RNA aptamers targeted to the TAR RNA element of HIV-1. Nucleic Acids Res. 2004;32:3101-3107
100. Astakhova IV, Ustinov AV, Korshun VA, Wengel J. LNA for optimization of fluorescent oligonucleotide probes: improved spectral properties and target binding. Bioconjugate Chem. 2011;22:533-539
101. Virno A, Randazzo A, Giancola C, Bucci M, Cirino G, Mayol L. A novel thrombin binding aptamer containing a G-LNA residue. Bioorg Med Chem. 2007;15(17):5710-5718
102. Hernandez FJ, Kalra N, Wengel J, Vester B. Aptamers as a model for functional evaluation of LNA and 2′-amino LNA. Bioorg Med Chem Lett. 2009;19:6585-6587
103. Sugizaki K, Okamoto A. ECHO-LNA conjugates: hybridization-sensitive fluorescence and its application to fluorescent detection of various RNA strands. Bioconjugate Chem. 2010;21(12):2276-2281
104. Morandi L, Ferrari D, Lombardo C, Pession A, Tallini G. Monitoring HCV RNA viral load by locked nucleic acid molecular beacons real time PCR. J Virol Methods. 2007Mar;140(1-2):148-54
105. Morandi L, de Biase D, Visani M, Cesari V, De Maglio G, Pizzolitto S, Pession A, Tallini G. Allele specific locked nucleic acid quantitative PCR (ASLNAqPCR): an accurate and cost-effective assay to diagnose and quantify KRAS and BRAF mutation. PLoS One. 2012;7(4):e36084
106. Hillyer P, Mane VP, Schramm LM, Puig M, Verthelyi D, Chen A, Zhao Z, Navarro MB, Kirschman KD, Bykadi S, Jubin RG, Rabin RL. Expression profiles of human interferon-alpha and interferon-lambda subtypes are ligand-and cell-dependent. Immunol Cell Biol. 2012Sep;90(8):774-83
107. Hendrix C, Verheggen I, Rosemeyer H, Seela F, Van Aerschot A, Herdewijn P. 1', 5'-anhydrohexitol oligonucleotides: synthesis, base pairing and recognition by regular oligodeoxyribonucleotides and oligoribonucleotides. Chem Eur J. 1997;3:110-120
108. Herdewijn P. Targeting RNA with conformationally restricted oligonucleotides. Liebigs Ann. 1996;9:1337-1348
109. De Winter H, Lescrinier E, Aerschot AVan, Herdewijn P. Molecular Dynamics Simulation To Investigate Differences in Minor Groove Hydration of HNA/RNA Hybrids As Compared to HNA/DNA Complexes. J Am Chem Soc. 1998;120:5381-5394
110. Hendrix C, Rosemeyer H, De Bouvere B, Van Aerschot A, Seela F, Herdewijn P. 1',5'-anhydrohexitol oligonucleotides: hybridisation and strand displacement with oligoribonucleotides, interaction with RNase H and HIV reverse transcriptase. Chem Eur J. 1997;3:1513-1520
111. Abramov M, Schepers G, Van Aerschot A, Hummelenb PV, Herdewijn P. HNA and ANA high-affinity arrays for detections of DNA and RNA single-base mismatches. Biosens and Bioelectron. 2008;23:1728-1732
112. Hendrix C, Rosemeyer H, De Bouvere B, Van Aerschot A, Seela A, Herdewijn P. 1',5'-Anhydrohexitol oligonucleotides: hybridisation and strand displacement with oligoribonucleotides, interaction with RNase H and HIV reverse transcriptase. Chem Eur J. 1997;3:1513-1520
113. Eschenmoser A. Chemical etiology of nucleic acid Structure. Science. 1999;284:2118-2124
114. Carothers JM, Oestreich SC, Davis JH, Szostak JW. Informational complexity and functional activity of RNA structures. J Am Chem Soc. 2004;126:5130-5137
115. Chaput JC, Ichida JK, Szostak JW. DNA polymerase-mediated DNA synthesis on a TNA template. J Am Chem Soc. 2003;125:856-857
116. Hossain N, Wroblowski B, Van Aerschot A, Rozenski J, DeBruyn A, Herdewijn P. Oligonucleotides composed of 2'-deoxy-1',5-anhydro-d-mannitol nucleosides with a purine base moiety. J Org Chem. 1998;63:1574-1582
Author contact
![]() Corresponding author: Junchen Wu, Meilong 130, Shanghai 200237, China. Telephone: (+86) 21-6425-3674, fax: (+86) 21-6425-2258, E-mail: jcwuedu.cn.
Corresponding author: Junchen Wu, Meilong 130, Shanghai 200237, China. Telephone: (+86) 21-6425-3674, fax: (+86) 21-6425-2258, E-mail: jcwuedu.cn.
 Global reach, higher impact
Global reach, higher impact