13.3
Impact Factor
Theranostics 2017; 7(6):1511-1523. doi:10.7150/thno.18401 This issue Cite
Research Paper
Novel Linear Peptides with High Affinity to αvβ3 Integrin for Precise Tumor Identification
State Key Laboratory of Natural Medicines, Department of Biomedical Engineering, School of Engineering, China Pharmaceutical University, Nanjing, 24 Tongjia Road, 210009 (China).
* These authors contributed equally to this work
Received 2016-12-18; Accepted 2017-1-12; Published 2017-4-6
Abstract
Development of alternative linear peptides for targeting αvβ3 integrin has attracted much attention, as the traditional peptide ligand, cyclic RGD, is limited by inferior water-solubility and complex synthesis. Using pharmacophore-based virtual screening and high-throughput molecular docking, we identified two novel linear small peptides RWr and RWrNM with high affinity and specificity to αvβ3 integrin. The competitive binding with cyclic RGD (c(RGDyK)) and cellular uptake related to the integrin expression levels verified their affinity to αvβ3 integrin. The intermolecular interaction measurement and dynamics simulation demonstrated the high binding affinity and stability, especially for RWrNM. In vivo peptide-guided tumor imaging and targeted therapy further confirmed their specificity. Results indicated that the newly identified small linear peptide RWrNM, with high affinity and specificity to αvβ3 integrin, better water-solubility, and simplified synthetic process, could overcome limitations of the current cyclic RGD peptides, paving the way for diverse use in diagnosis and therapy.
Keywords: αvβ3 integrin, linear peptide, binding affinity.
Introduction
αvβ3 integrin, a receptor for vitronectin, is one of the integrin family members consisting of dimeric alpha v and beta 3 subunits [1, 2]. The αvβ3 integrin mediates the intravasation and extravasation of tumor cells and has been implicated in the malignant spread of some tumor cell types such as breast cancer and glioblastoma [3, 4]. Therefore, the dimeric αvβ3 integrin has become an essential molecular target for early cancer diagnosis and intervention [5, 6].
Selective tumor targeting in vivo can be achieved by monoclonal antibodies [7, 8], which specifically bind to target receptors. However, most antibodies are immunogenic and have long plasma half-life rendering them suboptimal for molecular imaging [9, 10]. Small peptides and biomolecules are therefore preferred for biological imaging because of their low immunogenicity, reduced barriers to topical delivery, high affinity and selectivity for receptors, and desirable pharmacokinetic properties.
Cyclic RGD peptides are small molecules that bind αvβ3 integrin with high affinity. For this reason, a variety of RGD containing peptides has been developed for targeting tumor-induced angiogenic blood vessels or tumor-associated integrin. Conjugation of these peptides to imaging agents or drugs affords bioactive molecules for cancer imaging [11, 12] and targeted therapy [13], respectively. However, the cyclic RGD structure requires complicated peptide synthesis leading to increase in production cost and difficulty in quality control. Also, recent studies have demonstrated the strong binding affinity of RGD-containing peptides not only to αvβ3 integrin receptor but also to αvβ5 and α5β1 integrins [14, 15]. Therefore, efforts to develop alternative small linear peptides with similar or even higher affinity and specificity to αvβ3 integrin than cyclic RGD motif peptide have attracted much attention.
Computer-assisted virtual screening [16, 17, 18] is an effective method for drug discovery of small molecules with binding affinity to target receptors [19, 20, 21]. Structure-based pharmacophore strategy has been successfully used to screen small molecule leading compounds in drug development [22, 23]. Molecular docking and dynamic simulation are also considered practical methods to analyze the intermolecular interaction and explain the binding affinity and stability [24, 25]. Therefore, the combination of pharmacophore models with molecular docking will render more efficient hits. Although the compounds obtained from virtual screening have the potential specificity for the targets, it is necessary to confirm the feasibility of this approach by in vitro and in vivo experiments.
In this study, we have integrated structure-based pharmacophore method with molecular docking to screen the linear bioactive peptides for identifying αvβ3 integrin. Two novel small linear peptides (RWr, RWrNM) were selected with strong molecular interactions with αvβ3 integrin. To evaluate the affinity of these two peptides to αvβ3, cell lines with different expression levels of αvβ3 were cultured with fluorescence dye-labeled RWr and RWrNM. Confocal imaging and flow cytometry were used to identify their affinity and specificity to αvβ3. Microscale thermophoresis (MST) was performed to quantify affinity of both peptides to αvβ3 integrin. Furthermore, the effects of RWrNM and RWr on cell migration, angiogenesis, and downstream signaling pathways of αvβ3 were investigated. The tumor targeting ability and the therapeutic efficacy of peptide conjugates were further studied.
Results
Molecular dynamics of docking conformation and binding affinity
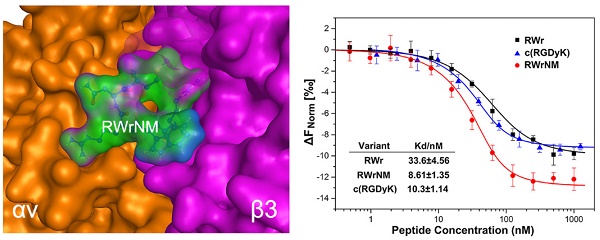
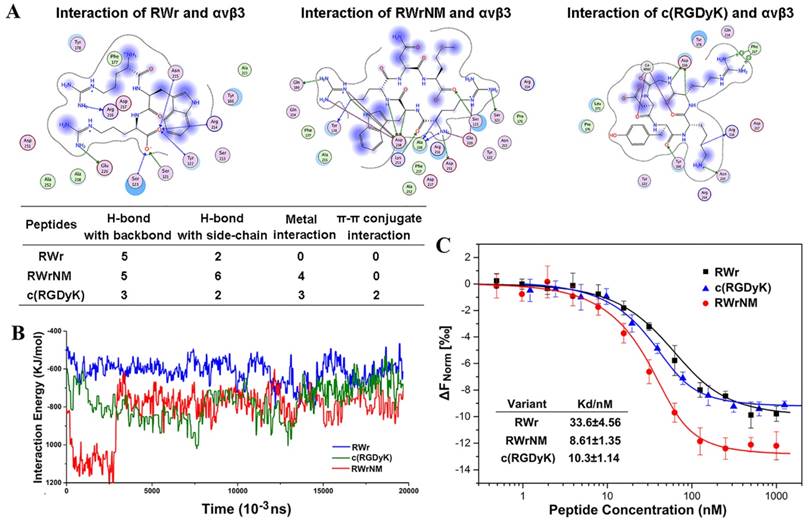
We identified two novel linear peptides, RWr and RWrNM, by using structure-based pharmacophore method integrated with molecular docking that had the highest docking score and potentially high binding affinity with αvβ3 integrin. The integrin-peptide binding modes were visualized through the docking interaction and compared with the well-established αvβ3-targeting cyclic peptide, c(RGDyK) (Figure 1A). The interaction diagrams indicated that the amino acids of αvβ3 protein interacted with the peptides and different ligands formed different interaction bonds. The bonding interactions between the peptides and integrin were in the following order: RWrNM (15) > c(RGDyK) (10) > RWr (7) implying potentially higher affinity of RWrNM than that of c(RGDyK). We also analyzed the molecular stability of the three peptides with integrin. As displayed in Figure 1B, molecular interactions between the integrin receptor and peptides were unstable in the initial 15 ns. Subsequently, the interactions were smooth and constant. The interaction energy between RWrNM and αvβ3 integrin was slightly lower than that of c(RGDyK), implying more stable binding of RWrNM to αvβ3 than to RWr and c(RGDyK). The stable interaction conformations of these three peptides to αvβ3 integrin at 20ns were also output (Figure S1). Interestingly, like c(RGDyK), RWrNM peptide exhibited a similar cyclic structure in the stable conformation. These findings encouraged us to further investigate the binding affinity of the new peptides, especially RWrNM.
Although the peptides obtained from the virtual screening have the potential targeting capability for αvβ3 integrin, it was necessary to obtain experimental confirmation. The screened peptides were synthesized and purified as proved by their mass spectra and high-performance liquid chromatography (Figure S2). MicroScale Thermophoresis (MST) was applied to determine the affinity of αvβ3 integrin to RWr, RWrNM and c(RGDyK) (Figure 1C). The sigmoidal binding curves were fitted to yield the Kd values of RWr, RWrNM and c(RGDyK) as 33.6±4.56 nM, 8.61±1.35 nM, and 10.3±1.14 nM, respectively, which confirmed the strong affinity of RWrNM for integrin and was in agreement with the molecular docking results.
Uptake of the peptides by different tumor cells
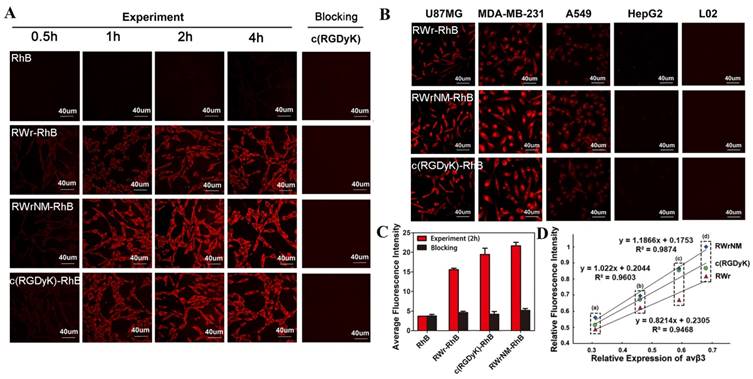
Since the conformation of integrin, when it is integrated in the cell surface membrane, may be different from that of the free molecule, we evaluated the binding ability of the peptides with αvβ3 integrin at the cellular level. As shown in Figure 2A, the fluorescence RhB-labeled peptides were taken up byU87MG tumor cells, which highly express αvβ3, with the bright fluorescence displayed in the cytoplasm after 2 hours of incubation; no fluorescence was observed when free RhB was incubated with U87MG cells. Longer incubation (4 hours) did not increase the cellular uptake of these peptides which made 2 hours the appropriate time point for cell imaging. Specifically, RWrNM-RhB resulted in the most abundant cellular accumulation, followed by c(RGDyK)-RhB and RWr-RhB. To confirm the binding of the peptides to αvβ3 receptors on tumor cell membranes, blocking experiment was carried out with U87MG cells pre-incubated with αvβ3 standard ligand c(RGDyK). No fluorescence signal was observed indicating that c(RGDyK) occupied the receptors mediating the uptake of RWr and RWrNM. This suggested that both linear peptides bind to the same cell membrane receptor αvβ3 as the well-established peptide c(RGDyK). The semi-quantitative analysis of the cellular uptake imaging gave a more direct comparison of the fluorescence intensities (Figure 2C) with the order of RWrNM > c(RGDyK) > RWr. Similar results were observed in the MDA-MB-231 cell line with high expression of αvβ3 (Figure S3). When a control RhB-labeled peptide (NWMrR) with scrambled sequence was evaluated, only weak fluorescence was observed in U87MG cells during the 4h incubation (Figure S4). Also, the blocking experiment using c(RGDyK) had no influence on the cellular uptake of the control peptide. These data further proved the high binding affinity and specificity of the screened peptides to αvβ3.
To confirm that the cell affinity of the new peptides correlated with the expression of αvβ3 integrin, five cell lines with different levels of αvβ3 integrin expression were used. The αvβ3 integrin expression in U87MG, MDA-MB-231, A549, HepG2 and L02 cells were measured by Western blotting (Figure S5); the expression levels were in the order of U87MG > MDA-MB-231 > A549 > HepG2 > L02 cells. After 2h incubation, the three peptides groups showed a gradient decrease in fluorescence from U87MG to L02 cells (Figure 2B). Only weak fluorescence could be detected in HepG2 cells and no signal was found in normal cells, L02. Figure 2D shows a good linear fitting (R2>0.9) between the fluorescence intensity of RWrNM-RhB, c(RGDyK)-RhB and RWr-RhB and relative αvβ3 expressions in the tumor cell lines. These findings revealed that the new peptides, RWrNM and RWr could be specifically internalized by tumor cells mediated through αvβ3 integrin and a higher integrin level resulted in greater cellular accumulation. More importantly, RWrNM demonstrated a slightly stronger affinity to tumor cells than that of c(RGDyK) which motivated us to further investigate its properties.
Binding affinity of the peptides to αvβ3 integrin at molecular level. (A) The binding interaction mode between c(RGDyK), RWr and RWrNM peptide with αvβ3 integrin receptor after molecular docking. Residues are annotated with the 3-letter amino acid code. The active site residues are represented as follows: polar residues in light purple, hydrophobic residues in green, acidic residues with a red contour ring, basic residues with a blue contour ring. Green and blue arrows indicate hydrogen bonding to side chain and backbone atoms, respectively. The cation-π stacking interactions are represented in green dotted lines. Metal ions interaction network is represented in cyan dotted lines. Light-blue halos around residues indicate the degree of interaction with ligand atoms (larger and darker halos means more interaction). (B) Interaction energies between different peptides (RWr, c(RGDyK) and RWrNM) and αvβ3 integrin protein under 20ns. (C) The affinity of αvβ3 integrin protein to RWr, RWrNM and c(RGDyK) measured by MicroScale Thermophoresis and the Kd values of RWr, RWrNM and c(RGDyK) were calculated respectively.
Uptake of the peptides by different tumor cells. (A) Time course of the cellular uptake of different peptide-RhB conjugates in U87MG cells with free RhB as control, and cellular uptake of these conjugates after c(RGDyK) blocking. Red: fluorescence of RhB. Scale bar: 40 μm (B) Cellular uptake of RWr-RhB, RWrNM-RhB and c(RGDyK)-RhB at 2h in U87MG, MDA-MB-231, A549, HepG2 and L02 cells. Red: fluorescence of RhB. Scale bar: 40μm. (C) Average fluorescent intensities of U87MG cells treated with free RhB, RWr-RhB, c(RGDyK)-RhB and RWrNM-RhB before and after c(RGDyK) blocking. (D) linear fitting between the relative fluorescent intensity of peptide (RWr, RWrNM and c(RGDyK)) and relative αvβ3 expressions in four tumor cell lines (a: HepG2, b: A549, c: MDA-MB-231, d: U87MG).
Binding affinities of the peptides to tumor cells
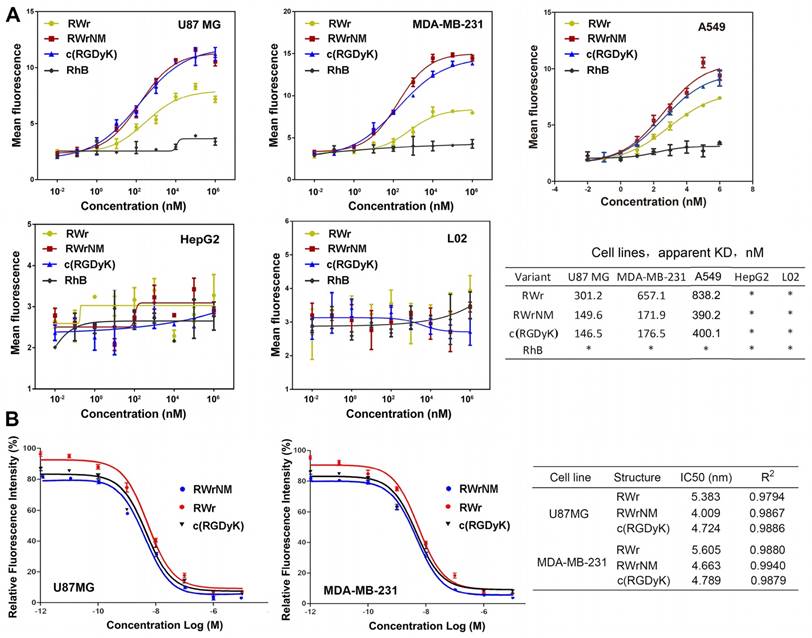
Quantitative analysis of the binding affinity of the peptides to U87MG, MDA-MB-231, A549, HepG2 and L02 cell lines was performed by flow cytometry following incubation with different peptide concentrations (Figure 3A). Based on the mean fluorescence curves, the KD values were calculated. The KD values of RWr binding to U87MG cells, MDA-MB-231 and A549 cells were much higher than that of RWrNM and c(RGDyK), indicating a higher binding level of RWrNM and c(RGDyK). On the contrary, no binding was observed with HepG2 and L02 cells that barely express αvβ3 integrin confirming the ability of RWr and RWrNM to selectively bind to cells with high αvβ3 expression.
The affinity of RWr and RWrNM to αvβ3 integrin receptor was further quantified by competitive binding analysis. The RhB-labeled c(RGDyK) was incubated with the cells followed by the addition of different concentrations of free RWr, RWrNM and c(RGDyK) peptides (Figure 3B). As the concentration of the peptides increased, the relative fluorescence intensity of U87MG cells or MDA-MB-231 cells treated with RhB-c(RGDyK) decreased in a biphasic mode. The IC50 values were determined by nonlinear regression fitting using GraphPad Prism software. In both U87MG and MDA-MB-231 cells, RWrNM demonstrated the lowest IC50 values compared to c(RGDyK) and RWr indicating the strong affinity of RWrNM for integrin.
To exclude the possibility that αvβ5 and α5β1 receptors may be involved in the interaction with RWrNM, monoclonal antibody (mAbs) against αvβ5 and α5β1 integrins were used to prove the specificity of RWrNM to αvβ3 integrin. As shown in Figure S6, compared with c(RGDyK), the pre-incubation of mAbs against αvβ5 and α5β1 integrins did not cause a significant reduction in the cellular uptake of RWrNM in U87MG cells verifying the specificity of RWrNM to αvβ3 integrin.
Effects of peptides on tumor cell migration and angiogenesis
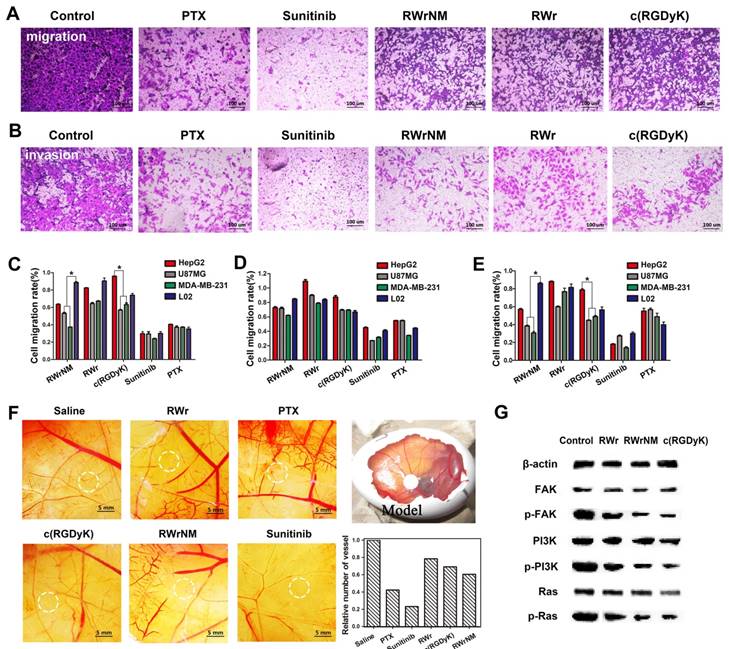
Having verified the strong affinity of RWrNM to αvβ3 integrin, we investigated whether RWrNM could exert similar biological effects as c(RGDyK) on various cellular functions, including migration, invasion, and adhesion, which are closely associated with integrin [26, 27, 28]. The presence of RWrNM, RWr, and c(RGDyK) caused an obvious decrease in cell migration compared with the controls (Figure 4A, Figure S7). The quantification of the cells crossing the transwell membrane was plotted as shown in Figure 4C. No significant difference existed between the three peptides, although RWrNM displayed a slightly stronger inhibitory effect on cell migration than that of c(RGDyK) and RWr. The migration rates of U87MG and MDA-MB-231 cells displayed an obvious decrease after incubation with RWrNM while the migration of HepG2 and L02 cells only marginally decreased, possibly as a result of low expression of αvβ3 integrin that mediates cell binding of the peptide. In contrast, the anticancer drug PTX and Sunitinib demonstrated their strong inhibition on cell migration in all experimental cell lines with no selectivity for tumor cell lines. We then evaluated the ability of the peptides to block tumor cell invasiveness, an important factor for angiogenesis. RWrNM showed a similar anti-invasiveness effect as RWr and c(RGDyK) (Figure 4B, Figure S8). The quantification of cell invasion rate was also plotted (Figure 4D) with the invasion rates above 60% for all peptides indicating the weak effect of the peptides on cell invasion. As cell adhesion is closely related to the αvβ3 integrin, the effects of peptides on the cell adhesion were also investigated and displayed in Figure 4E. Similar to the results of cell migration, the peptides demonstrated inhibitory effects on cell adhesion with apparent selectivity. The effects of these peptides on angiogenesis were also studied by using chick chorioallantoic membrane (CAM) model. Compared to the saline control, all three peptides could inhibit angiogenesis to some extent (Figure 4F). The quantification of vessels within the experimental region showed that RWrNM had a slightly higher inhibitory effect than RWr and c(RGDyK) peptides.
Given the results from above experiments, we hypothesize that RWrNM shared the same underlying molecular mechanisms with c(RGDyK) after binding with αvβ3 integrin. It is well-known that αvβ3 integrin directly acts on the FAK protein and impacts the downstream PI3K and Ras signaling pathways [29, 30, 31]. Accordingly, Western blot analysis showed that both RWrNM and c(RGDyK) decreased the levels of p-FAK, p-PI3K, and p-Ras proteins (Figure 4G). These results confirmed that RWrNM, by binding to αvβ3 integrin, was functionally equivalent or stronger than c(RGDyK) and acted on same signaling pathways.
Binding affinities and specificity of the peptides to tumor cells. (A) Quantification of the binding affinity of the peptides to U87MG, MDA-MB-231, A549, HepG2 and L02 cells by flow cytometry with different concentrations of the peptides. Data shown are the average of triplicate experiments and error bars represent standard deviations. Cell binding data were fitted to sigmoidal curves to calculate apparent dissociation constants. *: Not measured. (B) Competitive binding curves of RWr, RWrNM and c(RGDyK) in U87MG and MDA-MB-231 cells, which were obtained by incubation of cells with RhB labelled c(RGDyK) and different concentrations of RWr, RWrNM and c(RGDyK).
Inhibition effects of the peptides on tumor cell migration and angiogenesis. (A) Microscopic images of the migration of U87MG cells treated with PTX, Sunitinib RWrNM, RWr and c(RGDyK). Scale bar: 100 μm (B) Microscopic images of the invasion of U87MG cells treated with PTX, Sunitinib, RWrNM, RWr and c(RGDyK). Scale bar: 100 μm (C) Quantitative analysis of the different cells crossing the transwell membrane after the treatments with peptides and anticancer drugs. *P < 0.05 (D) Quantification of the invasion rates of different cell lines after treatments with peptides and anticancer drugs. (E) Quantification of cell adhesion rates in different cell lines treated with PTX, Sunitinib, RWrNM, RWr and c(RGDyK). *P < 0.05 (F) Effects of these peptides on angiogenesis in Chick chorioallantoic membrane model. Relative vessel numbers of different treatment groups were plotted. Scale bar: 5 mm (G) Western blot analysis of the levels of FAK, p-FAK, PI3K, p-PI3K, Ras and p-Ras proteins in U87MG cells after incubated with RWrNM, RWr and c(RGDyK).
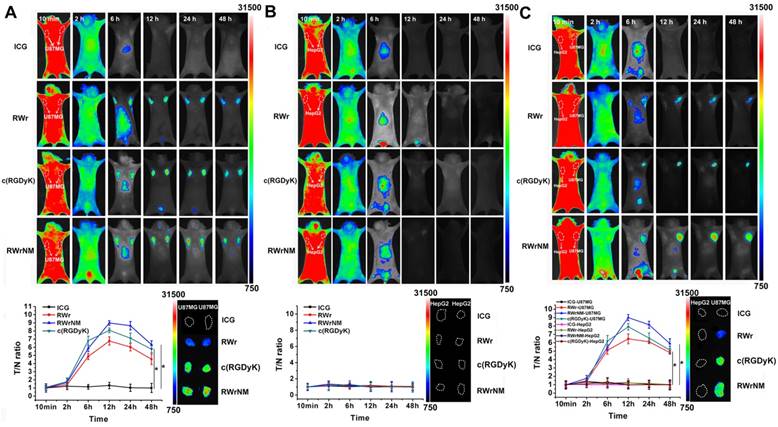
In vivo tumor targeting ability of peptides
The exceptionally superior affinity of RWrNM to avβ3 motivated us to explore whether this peptide can be used for in vivo tumor targeting as the widely applied c(RGDyK). To this end, RWrNM together with RWr and c(RGDyK) were conjugated with the near infrared fluorescence dye ICG derivative to form tumor targeting probes. In mice bearing two U87MG tumors in the bilateral symmetry axilla, the fluorescence was distributed throughout the body at 10min post injection of free ICG and peptide-ICG conjugates (Figure 5A). Free ICG treatment didn't cause any tumor retention and the fluorescence diminished after 6 hours and undetectable after 12 hours. In contrast, RWr, c(RGDyK) and RWrNM groups showed obvious signal in the symmetry tumors at 6h when the fluorescence in other parts of the body had gradually decreased. In particular, the most prominent tumor fluorescence was found in mice injected with c(RGDyK)-ICG and RWrNM-ICG. For the in vivo imaging, the tumor/normal tissue ratio (T/N ratio) was plotted against each time point. The most intense fluorescence was observed in the RWrNM group at 12h post injection, which was still present in the two bilateral tumors at 48h. In contrast, none of these peptides showed targeting ability to HepG2 tumors with low αvβ3 integrin expression implanted in the bilateral symmetry axilla of mice as displayed in Figure 5B. To further verify the selective targeting ability of peptides to tumors overexpressing αvβ3, HepG2, and U87MG were implanted respectively in the bilateral symmetry axilla of the subject mice. As displayed in Figure 5C, only U87MG tumors at the right site showed bright fluorescence while the HepG2 tumors at the left site showed nearly no fluorescence after 6h post-injection. Also, RWrNM resulted in the strongest peak signal followed by c(RGDyK) and RWr. This in vivo imaging was consistent with the cellular uptake data indicating the selective targeting ability of RWr and RWrNM to tumors with high expression of αvβ3 integrin. The results were further confirmed by the fluorescence imaging of the tumors harvested from the mice (inset). It is of note that RWrNM led to a more distinct tumor fluorescence than c(RGDyK) in U87MG tumors. The superior tumor targeting ability of RWrNM was most probably caused by the stronger affinity to αvβ3 integrin as evidenced by the MST assay above. Also, liquid chromatography showed satisfying serum stability of the three peptides in 24 hours (Figure S9). Collectively, these lines of evidence strongly suggested that RWrNM can be used as an in vivo tumor targeting probe with strong tumor affinity.
Improved therapeutic efficacy of Dox by RWrNM conjugation
Based on the successful validation of the in vivo targeting efficacy, we next investigated whether conjugation of RWrNM to the chemotherapeutic drug doxorubicin (Dox) could enhance therapeutic efficacy due to improved tumor targeting ability. The peptide-Dox conjugate was synthesized and purified as shown by its mass spectrum and high-performance liquid chromatography (Figure S10). Various conjugates were administrated into mice bearing U87MG tumors and tumor volume, animal weight and survival rate monitored during the 15-day treatment. Most notably, the RWrNM-Dox conjugate led to the longest delay in tumor growth (Figure 6A, 6B). Obvious tumor shrinkage was also observed in mice treated with c(RGDyK)-Dox, but to a lesser extent than RWrNM-Dox. Treatment with RWr-Dox resulted in a moderate improvement of tumor inhibition than free Dox, most likely due to the insufficient affinity to αvβ3 integrin. Although no significant changes in body weights were observed during treatment in these groups, the free Dox group showed a continual decrease in body weight (Figure 6C). The toxicity and the lowest body weight led to the lowest survival rate in the free Dox group (Figure 6D). The survival rate of mice in different groups is displayed in Figure 6D. Compared to the control group, which showed a survival rate of 50% on day 15, RWrNM-Dox treatment resulted in the highest survival rate of 90%. Histopathology analysis was carried out to further examine the toxicity in tumors and other organs (Figure 6E). Free Dox treatment induced a certain degree of cell apoptosis in tumor tissues, but obvious cardiotoxicity and hepatotoxicity were also observed. However, such toxicity was significantly alleviated when Dox was conjugated with RWrNM or c(RGDyK).
In vivo tumor targeting ability of peptides. (A) In vivo fluorescent imaging of mice bearing two U87MG tumors at bilateral symmetry axilla treated with free ICG, RWr-ICG, c(RGDyK)-ICG and RWrNM-ICG. *P < 0.05 (B) In vivo fluorescent imaging of mice bearing two HepG2 tumors at bilateral symmetry axilla treated with free ICG, RWr-ICG, c(RGDyK)-ICG and RWrNM-ICG. (C) In vivo fluorescent imaging of mice bearing U87MG and HepG2 tumors respectively at bilateral symmetry axilla treated with free ICG, RWr-ICG, c(RGDyK)-ICG and RWrNM-ICG. *P < 0.05 (Tumor/normal tissue (T/N) ratios at different time points after intravenous injection of different peptides conjugated with ICG, and fluorescence imaging of the tumors harvested from the mice (inset)).
Antitumor efficacy of RWr, c(RGDyK) and RWrNM conjugated with Dox on U87MG-tumor-bearing mice. (A) Images of the tumors in different treatment groups obtained after the 15-days therapy. (B) Tumor growth curves of the U87MG-tumor-bearing mice after intravenous injection with saline, Dox, RWr-Dox, c(RGDyK)-Dox and RWrNM-Dox. *P < 0.05 (C) Body weight of mice during different peptide-Dox treatments in 15 days. (D) Survival rate of mice in different peptide-Dox treatment groups. *P < 0.05 (E) Images of tumor, heart and spleen after H&E staining examined by histological analysis. Scale bar: 50μm (F) Laser confocal images of tumor, heart and liver tissue samples excised from the mice in different treatment groups. Scale bar: 200μm
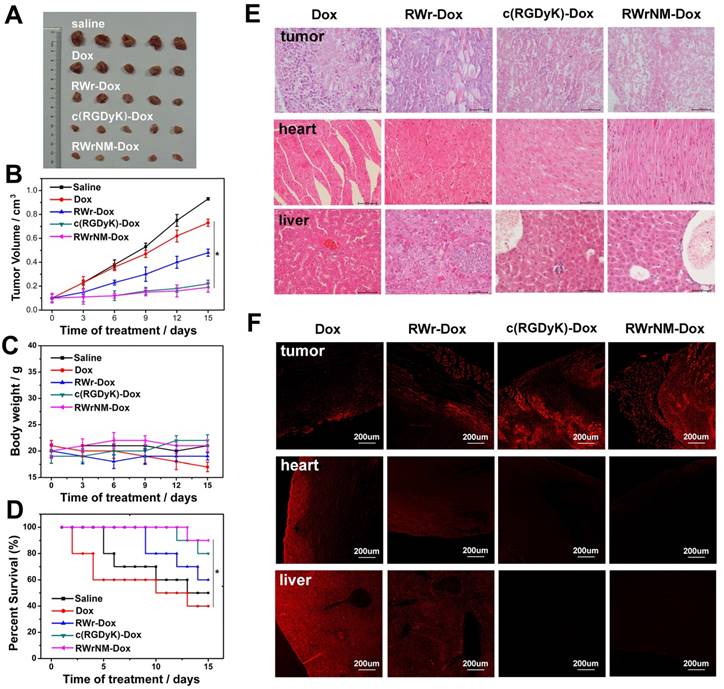
To verify that the inhibition of tumor growth and toxicity to normal tissues were indeed due to the cytotoxicity of Dox accumulation, the tissue samples were excised and examined by laser confocal microscopy (Figure 6F). The free Dox treatment resulted in a widespread distribution of Dox fluorescence in the tumor, heart and liver tissues explaining high systematic toxicity and reduced tumor inhibition. As anticipated, c(RGDyK)- or RWrNM-conjugated Dox showed nearly no Dox signal in normal organs while a prominent fluorescence signal was detected in the tumors.
We further evaluated the treatment response to different Dox conjugates in mice bearing low αvβ3 integrin-expressing HepG2-tumors (Figure S11). Compared with free Dox, the Dox conjugates resulted in no significant improvement in tumor inhibition and survival of the mice. The cell viability assay of peptide-Dox conjugates was also evaluated by using U87MG and HepG2 cell lines (Figure S12). The U87MG cells with high αvβ3 expression were more sensitive to peptide-Dox conjugates than the free Dox at the same drug concentration.
Taken together, the improved therapeutic outcome in tumors with high αvβ3 integrin expression demonstrates the superior tumor targeting ability and selectivity of RWrNM, which can potentially serve as a new targeting moiety to enhance the efficacy of current anti-tumor drugs.
Discussion
The αvβ3 integrin is considered to be an established therapeutic target for the treatment of a variety of tumors [32, 33, 34]. Cyclic RGD peptide is the well-known ligand to αvβ3 and has been extensively exploited for tumor diagnosis and treatment [35, 36, 37]. However, the complexity and high-cost of synthesizing cyclic peptides stipulate the development of alternative ligands for targeting αvβ3 integrin.
In this study, we have identified two novel linear small peptides, RWr and RWrNM. Their binding affinities were validated in different tumor cell lines exhibiting a positive correlation with αvβ3 integrin expression levels. The blocking experiments and competitive binding by c(RGDyK) confirmed the same targeting receptor for RWr and RWrNM as that of c(RGDyK). Importantly, RWrNM demonstrated a relatively higher binding ability to tumor cells than that of c(RGDyK) with lower IC50 to αvβ3 integrin over-expressing tumor cells (U87MG and MDA-MB-231). Quantitative measurement by MST also indicated that linear RWrNM had a slightly higher affinity to αvβ3 integrin than c(RGDyK). The in silico configuration analysis revealed that the high affinity of RWrNM to αvβ3 integrin was due to increased number of bonding interactions. Interestingly, the stable interaction conformation revealed that RWrNM formed a cyclic structure while interacting with αvβ3 integrin explaining its high affinity and stability. Compared with cyclic RGD peptide with strong binding affinity to αvβ3 as well as αvβ5 and α5β1 integrins, RWrNM exhibited high specificity to αvβ3 integrin. This was evident by the fact that pre-incubation of mAbs against αvβ5 and α5β1 integrins did not cause a significant reduction in the cellular uptake of RWrNM in U87MG cells.
Another significant feature of RWrNM is that it has more hydrophilic groups than c(RGDyK). The hydrophobic constants (CLogP value: RWr of -6.4483, RWrNM of -5.01828 and c(RGDyK) of -4.31882) generated by structural analysis proved the superior water solubility of RWrNM compared to c(RGDyK). Tumor targeting ligands that possess excellent water solubility are quite appealing for unlocking the full therapeutic potential of hydrophobic small molecule cancer drugs and overcoming the recurring problems of poor solubility, sub-optimal pharmacokinetics (PK), and low bioavailability. Therefore, RWrNM is a promising ligand for improving druggability, especially for the hydrophobic anticancer drugs.
These superior features of RWrNM motivated us to first investigate its in vivo tumor targeting ability. The in vivo fluorescent imaging demonstrated stronger tumor targeting ability of RWrNM, suggesting that it may serve as a versatile tumor targeting ligand with better efficacy and easier preparation. Finally, the conjugate of RWrNM to Dox was found to enhance the tumor inhibition efficiency and reduce the cardiac toxicity and hepatic toxicity of Dox.
In summary, RWrNM demonstrated strong interaction with αvβ3 integrin and improved tumor targeting ability and may serve as a new candidate for cancer identification and targeted therapy. Furthermore, the linear small peptide RWrNM could be synthesized by a relatively simple process facilitating the large scale production and decreasing its production cost and difficulty for quality control. Therefore, RWrNM could potentially substitute cyclic RGD for tumor identification and targeted therapy.
Materials and Methods
Computer-assisted virtual screening of high-affinity peptide
We used a Receptor-Ligand Pharmacophore Generation protocol (default parameters, DS software) to generate structure-based pharmacophore models derived from the crystal complexes of both integrin ανβ3 and the RGD peptide [38]. The protocol for generating the pharmacophore models is as follows: First, a set of features from the binding ligand was identified. A series of pharmacophore models were rendered based on the active sites in the αvβ3 integrin-RGD complex. The pharmacophore scores of all models were ranked and the model with the highest score was selected for further ligand screening. To this end, the pharmacophore models were enumerated and selectivity was estimated based on a Genetic Function Approximation (GFA) model, which was built from a training set of 1544 pharmacophore models. This set was then used to search the Drug-like Diverse database in Discovery Studio (DS), where the logarithmic values of the database search hits were used as our targets. Finally, the top model was screened against a 90,000,000-peptide database. The retrieved hits were further sorted through the use of filters (such as the maximum fit value of the pharmacophore model from the structure-based model) and then subjected to a molecular docking process.
The crystal structure of the extracellular segment of αvβ3 integrin when complexed with the RGD ligand (PDB ID: 1L5G) was downloaded from the Protein Data Bank (PDB). The ligand binds at the major interface between the αv and β3 subunits, making extensive contact with both. A LibDock docking algorithm and scoring function (LigScorel, LigScore2, PLP1, PLP2, Jain, PMF, PMF04) of the DS software were then used to perform the docking of αvβ3 integrin. The preprocessing of αvβ3 integrin receptor as well as all structures from earlier retrieved hits were performed according to the docking program's recommended protocols. The LibDock methodology effectively executed the docking in a high-throughput manner while maintaining the structural rigidity of αvβ3 integrin. Finally, we calculated the binding energies of the ligand-receptor interactions as well as the Libscore values for various ligand-receptor interactions.
Molecular docking and interaction analysis
Docking was performed to study the affinity of peptide and protein. We obtained ligand structure from the chemBioDraw software and protein structure (PDB ID: 1L5G) from Protein Data Bank (http://www.rcsb.org/). The peptide structures were generated and minimized using In Situ Ligand Minimization protocol of Discovery Studio v3.0 (Accelrys, San Diego,USA) [39]. The active site was defined with a 10 Å radius around the bound ligand in the crystal structure of αvβ3 integrin. The minimized structures were docked using the Dock Ligands Protocol of Discovery Studio v3.0 with CDOCKER algorithms to get the best docking conformations. Molecular docking was calculated by using 2000 Maximum Bad Orientations and 600 Orientation vdW Energy Threshold. The best poses were prioritized by cdocker_interaction_energy scoring.
Molecular dynamics simulation and output analysis
The best conformation and binding energy of the protein-ligand complex were used for the molecular dynamics (MD) simulation analysis [40]. MD simulation was performed with the GROMACS-5.0.7 package at GROMOS 96 43a1 force field (Van Gunsteren et al. 1996) in dodecahedron box of a simple point charge. Ligand topology was generated by PRODRG2 Server. Three Cl- ions were added in solvent molecules to attain system neutrality. Energy minimization was carried out with the steepest descent of 50,000 steps. To extend the equilibrium, the system was subjected to position-restrained dynamic simulation (NVT and NPT) with 50,000 steps at 300 K temperature. Finally, MD was performed at 300 K temperature for 20 ns.
The GROMACS output can be achieved in the form of trajecory files. These trajectory files were further investigated using root mean square deviation (gmx rms), root mean square fluctuation (gmx rmsf), and hydrogen bond (gmx hbond). Graphs were plotted using xmgrace. The electrostatic interaction energy and van der Waals interaction energy were generated by calculations with MD restart (mdrun -rerun).
Microscale thermophoresis assay
The peptides were dissolved in sterile water and diluted into 14 gradients of concentration (2000, 1000, 500, 250, 125, 62.5, 31.25, 15.625, 7.812, 3.906, 1.953, 0.976, 0.488 and 0.244 nM). The αvβ3 integrin was labeled with Protein Labeling Kit BLUE according to the protocol. The fluorescence-labelled avb3 integrin was mixed with peptides solution in 14 concentration gradients at 1:1 ratio, followed by 30 minutes incubation. The capillaries (Cat#K002, NanoTemper) were used to draw the mixed solutions from the 14 tubes, which were then loaded with the order of declining peptides concentration. Pre-scanning was performed to ensure the fluorescent intensities in the 14 tubes were equal. Subsequently, MST binding curves were measured for 3 times and the Kd values were calculated according to the following formulation: f(c)=unbound + (bound - unbound)/2 * (FluoConc + c + Kd - Sqrt ((FluoConc + c + Kd)^2 - 4 * FluoConc *c) [41].
Cell culture
Human tumor cell lines including breast cancer (MDA-MB-231), malignant glioma (U87MG), hepatocellular carcinoma (HepG2), and lung cancer (A549) as well as Human normal liver cells (L02) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). U87MG, MDA-MB-231, A549 and HepG2 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco) supplemented with 10% fetal calf serum (FBS, Gibco), penicillin (100 µg/mL), and streptomycin (100 µg/mL). L02 cells were maintained in RPMI1640 medium (Invitrogen) supplemented with 10% fetal calf serum (FBS, Gibco), penicillin (100 µg/mL), and streptomycin (100 µg/mL) [42].
Dye-labeling of peptide candidates
The screened peptides were synthesized and labeled with visible dye Rhodamine B by Karebay Biochem Ltd Ningbo, China. Free peptides were conjugated with near infrared fluorescence dye ICG derivative (ICG-Der-02, prepared in our laboratory) to investigate in vivo tumor targeting. Briefly, 9.97mg of ICG-Der-02, 2.88mg of EDC•HCl (1-(3-Dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride), and 1.73 mg of NHS (N-Hydroxysulfosuccinimide sodium salt) were dissolved in 0.5mL deionized water by stirring at room temperature followed by reaction in the dark for 4 h. The solution was then stirred with RWr, RGD, c(RGDyK) and RWrNM (0.01 mmol) dissolved in PBS. The reaction mixture was stirred at room temperature overnight. The crude mixture was filtered and the solution was concentrated by passing through Sephadex G-25 gel filtration [43].
Cell integrin receptor binding assay
Confocal microscopy was used to visualize the targeting ability of RWr, RWrNM, RGD and c(RGDyK) probes to tumor cells. MDA-MB-231, U87MG, HepG2, A549 and L02 cell lines were used due to their different levels of αvβ3 integrin expression. Cells were seeded on confocal Petri dishes at a density of 3×105 cells per well and incubated for 24 hours allowing for cell attachment. To observe the cellular uptake at different time points, peptide-RhB conjugates were added in the culture medium and incubated for different incubation times (0.5, 1, 2 and 4 h). After incubation, cells were washed three times with PBS (pH 7.4) and imaged using laser confocal microscopy. Cellular uptake (2h) of RWr, RWrNM, RGD and c(RGDyK) probes were quantified [44].
Binding assay of peptide candidates to αvβ3 integrin receptor
Glioblastoma cells U87MG and breast cancer cells MDA-MB-231 with high αvβ3 integrin expression were used for binding experiments. Cells were seeded in 6-well plates at a density about 106 cells per well with 2 mL of culture medium at 37 °C for 24 h. After 24 hours of cultivation, the medium was replaced by 1 mL fresh medium (without FCS) containing RhB-peptides (10μM) and incubated at 37°C. After 2 h, the medium was removed to stop the incubation and the cells were washed twice with PBS (pH 7.4) to remove the unbound peptides. Then, the cells were scrapped from the wells using a manual scrapper, transferred to centrifuge tubes (1.5 ml), and centrifuged at 1000 rpm for 5 min. The cells were re-suspended in PBS buffer (pH 7.4) for flow cytometry analysis [45].
Competitive binding curves analysis
RhB-labeled c(RGDyK) were used to determine the binding affinities of RWr, RWrNM, and c(RGDyK) peptides to surface αvβ3 integrin in U87MG and MDA-MB-231 cell lines. Briefly, U87MG and MDA-MB-231 cells were harvested, washed three times with PBS, and re-suspended in PBS (pH 7.4, 5×106 cells/mL). Six-well plates were incubated with RhB-labeled c(RGDyK) (1000 nM/well) in the presence of increasing concentrations of different free RWr, RWrNM, and c(RGDyK) peptide. Cells were then incubated for 2 h at room temperature before being washed three times with PBS (pH 7.4). Cells were transferred to centrifuge tubes and centrifuged at 1000 rpm for 5 min. The resulting pellet was re-suspended in PBS (pH 7.4) and then transferred to flow cytometry tubes for analysis. All experiments were carried out in triplicate. The best-fit IC50 values for the U87MG and MDA-MB-231 cells were calculated by fitting the data with a nonlinear regression using GraphPad Prism (GraphPad Software, Inc., San Diego, CA) [46].
Cell migration assay
Different cell lines (U87MG, MDA-MB-231, HepG2, and L02) were cultured in the 6-well plates with a cell density of about 60-70% and incubated with peptides (40μM) for 24 h. Subsequently, the cells were digested and plated in the upper chamber (100 μL; 5×105 cells), with three parallel chambers. 600μl DMEM+10% FBS was added to the lower chamber and cells were cultured for 12 hours. The unattached cells were removed from the microporous membrane with a cotton swab. The lower surface of the membrane was fixed with methanol for 30 min at room temperature, stained with 0.1% crystal violet for 30min, and then photographed. 33% glacial acetic acid was added to 96-well plates and then the OD values at 570 nm were detected. Migration rate was calculated as % migration = (experiment group OD570/ control group OD570) * 100% [47].
Chick chorioallantoic membrane model experiment
Chick embryos of 4 days old were purchased from Qian Yuan Hao biotech (Ningbo, China). The drug samples were prepared by adding 20μL of 40uM peptide solutions or 20μL positive drug solution on filter papers. After locating the areas with rich vasculature, these areas were covered with the filter papers. Fixation was performed 3 days after incubation for 15 minutes. The papers were removed gently and the chick embryos were imaged and the vessel numbers were recorded [48].
Western blot analysis
Analysis of various enzymes and their phosphorylated derivatives, FAK, PI3K, Ras, p-FAK, p-PI3K and p-Ras, was performed using Western blotting. After treatment with free RWr, RWrNM, and c(RGDyK) (64μg /mL) for 48 h, cells were lysed and equal amounts of cell lysates (50 μg of protein) were loaded and separated by SDS-PAGE on 8-12% SDS-polyacrylamide gels before being transferred to nitrocellulose membranes. Membranes were incubated with Tris-buffered saline containing 1% (w/v) nonfat milk and 0.1% (v/v) Tween-20 for 1 h to block non-specific binding, washed with Tween-20 for 30 min, and incubated with appropriate primary antibodies for 2 h. Blots were then incubated with horseradish-peroxidase-conjugated second antibody for 1 h, developed using enhanced chemiluminescence, imaged using G: BOX chemiXR5 and analyzed for gray using the Gel-Pro32 software. β-actin was used as an internal control to guarantee the uniformity of equally loaded proteins among all groups [49].
Animal experiments
Animals were treated in accordance with the Guidance Suggestions for the Care and Use of Laboratory Animals. 6-7-week old female nude mice were maintained under aseptic conditions in a small animal isolator and were housed in standard cages with free access to food and water. All animals acclimated to the animal facility for at least 7 days before experimentation. For the tumor xenograft model, female nude mice were subcutaneously inoculated with 1 × 107 U87MG and HepG2 cells in the bilateral axillary fossa under aseptic conditions. When the size of tumors reached ~100 mm3, the mice were used for therapeutic experiments and imaged by near-infrared (NIR) fluorescence imaging. A vernier caliper was used to monitor tumor size over time [50].
In vivo antitumor studies
On day 7 post-tumor cell implantation, U87MG tumor-bearing nude mice were divided into five groups, each consisting of 10 mice. Saline, Dox (Sigma-Aldrich), RWr-Dox, c(RGDyK)-Dox or RWrNM-Dox (Karebay Biochem, Ltd Ningbo, China) were intravenously injected into the tail vein every other day for a total of 7 doses. Each dose was equivalent to 5mg/kg of Dox. The body weight and tumor size of each mouse were measured. The volume was calculated as follows: V = (π/6 * longest diameter* perpendicular diameter2). After treatment, the mice were euthanized, the organs and tumors were collected and fixed in 10% neutral buffered formalin, embedded in paraffin blocks for H&E staining, and visualized using an optical microscope. To investigate drug distribution, histological sections were analyzed using CLSM [51].
Conjugation of peptide to doxorubicin
For the synthesis of RWrNM-Dox, 5 mL dry dimethylformamide (DMF) was placed into a 25 mL three-necked flask equipped with a magnetic stirrer and a 20 mL dropping funnel. This flask was immersed in an ice bath for 15 min. RWrNM (50 mg, 0.038 mmol) and triethylamine (4.5 mg, 0.045 mmol) were added to the flask under N2 atmosphere. After dissolving completely, 4-Nitrophenyl chloroformate (9.1 mg, 0.045 mmol) was added to the flask dropwise. The resulting mixture was allowed to stir at room temperature for 24 h and then filtered to remove the byproduct. The crude organic solution was concentrated, and purified by silica gel column. The product was dried under vacuum at 35 °C overnight.
The activated RWrNM (RWrNM-NC, 30 mg) was dissolved in 5 mL DMF and then reacted with hydrazine (0.3 mL) in nitrogen at RT for 24 h. The product was obtained by concentration and purified by silica gel column. The crude product was dried under vacuum at 35 °C overnight.
RWrNM-hydrazine (5 mg) and DOX·HCl (20 mg) were dissolved in 5 mL DMSO. After stirring for 5 min, a drop of trifluoroacetic acid was added into the flask. Then, the mixture was allowed to react in the dark for 48 h at room temperature. The crude product was purified by silica gel column.
The procedure for the synthesis of RWr-DOX was similar to the one for RWrNM-Dox except changing the amount of feeding.
Statistical analyses
Statistical analyses were performed with Prism software (GraphPad). P values of <0.05 (*) were considered significant. All error bars represent ± s.e.m. unless otherwise noted. No samples or animals were excluded from analyses.
Supplementary Material
Supplementary figures.
Acknowledgements
We acknowledge financial support from the National Natural Science Foundation of China (NSFC 61335007, 81220108012, 81371684, 81000666, 81171395 and 81328012); the 973 Key Project (2015CB755504); and the Priority Academic Program Development of Jiangsu Higher Education.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Desgrosellier JS, Barnes LA, Shields DJ. et al. An integrin alpha(v)beta(3)-c-Src oncogenic unit promotes anchorage-independence and tumor progression. Nat Med. 2009;15:1163-9
2. Mas-Moruno C, Beck JG, Doedens L. et al. Increasing αvβ3 selectivity of the anti-angiogenic drug cilengitide by N-methylation. Angew Chem Int Ed Engl. 2011;50:9496-500
3. Leung CS, Yeung TL, Yip KP. et al. Calcium-dependent FAK/CREB/TNNC1 signalling mediates the effect of stromal MFAP5 on ovarian cancer metastatic potential. Nat Commun. 2014;5:5092
4. Turaga RC, Yin L, Yang JJ. et al. Rational design of a protein that binds αvβ3 integrin outside the ligand binding site. Nat Commun. 2016;7:11675
5. Reynolds AR, Hart IR, Watson AR. et al. Stimulation of tumor growth and angiogenesis by low concentrations of RGD-mimetic integrin inhibitors. Nat Med. 2009;15:392-400
6. Alberici L, Roth L, Sugahara KN. et al. De novo design of a tumor-penetrating peptide. Cancer Res. 2013;73:804-12
7. Oh P, Testa JE, Borgstrom P. et al. In vivo proteomic imaging analysis of caveolae reveals pumping system to penetrate solid tumors. Nat Med. 2014;20:1062-1068
8. Tol J, Punt CJ. Monoclonal antibodies in the treatment of metastatic colorectal cancer: a review. Clin Ther. 2010;32:437-53
9. Ferris RL, Jaffee EM, Ferrone S. Tumor antigen-targeted, monoclonal antibody-based immunotherapy: clinical response, cellular immunity, and immunoescape. J Clin Oncol. 2010;28:4390-9
10. Sano K, Mitsunaga M, Nakajima T. et al. In vivo breast cancer characterization imaging using two monoclonal antibodies activatably labeled with near infrared fluorophores. Breast Cancer Res. 2012;14:R61
11. Sun X, Huang X, Yan X. et al. Chelator-free (64) Cu-integrated gold nanomaterials for positron emission tomography imaging guided photothermal cancer therapy. ACS Nano. 2014;8:8438-46
12. Pike DB, Ghandehari H. HPMA copolymer-cyclic RGD conjugates for tumor targeting. Adv Drug Deliv Rev. 2010;62(2):167-83
13. Schottelius M, Laufer B, Kessler H, Wester HJ. Ligands for mapping alphavbeta3-integrin expression in vivo. Acc Chem Res. 2009;42(7):969-80
14. Pedchenko V, Zent R, Hudson GB. αvβ3 and αvβ5 Integrins Bind Both the Proximal RGD Site and Non-RGD Motifs within Noncollagenous (NC1) Domain of the α3 Chain of Type IV Collagen. J Biol Chem. 2004;279:2772-2780
15. Sarin V, Gaffin DR, Meininger AG, Muthuchamy M. Arginine-glycine-aspartic acid (RGD)-containing peptides inhibit the force production of mouse papillary muscle bundles via α5β1 integrin. J Physiol. 2005;564:603-617
16. Park JA, Lee YJ, Lee JW. et al. Cyclic RGD Peptides Incorporating Cycloalkanes: Synthesis and Evaluation as PET Radiotracers for Tumor Imaging. ACS Med Chem Lett. 2014;5:979-82
17. Du H, Cui C, Wang L, Liu H, Cui G. Novel tetrapeptide, RGDF, mediated tumor specific liposomal doxorubicin (DOX) preparations. Mol Pharm. 2011;8:1224-32
18. Kim YH, Jeon J, Hong SH. et al. Tumor targeting and imaging using cyclic RGD-PEGylated gold nanoparticle probes with directly conjugated iodine-125. Small. 2011;7:2052-60
19. Zhu L, Guo N, Li Q. et al. Dynamic PET and Optical Imaging and Compartment Modeling using a Dual-labeled Cyclic RGD Peptide Probe. Theranostics. 2012;2:746-56
20. Danhier F, Le Breton A, Preat V. RGD-based strategies to target alpha(v) beta(3) integrin in cancer therapy and diagnosis. Mol Pharm. 2012;9:2961-73
21. King D, Clark T, Gissane C. Use of a rapid visual screening tool for the assessment of concussion in amateur rugby league: a pilot study. J Neurol Sci. 2012;320:16-21
22. Sankaranarayanan R, Nessa A, Esmy PO, Dangou JM. Visual inspection methods for cervical cancer prevention. Best Pract Res Clin Obstet Gynaecol. 2012;26:221-32
23. Sukumar S, Zhu X, Ericksen SS, Mitchell JC. DBSI Server: DNA Binding Site Identifier. Bioinformatics. 2016;32:13
24. Kitchen DB, Decornez H, Furr JR. et al. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004;3(11):935-49
25. Christ F, Voet A, Marchand A. et al. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat Chem Biol. 2010;6(6):442-448
26. Varzavand A, Hacker W, Ma D. et al. αvβ1 Integrin Suppresses Prostate Cancer Metastasis via Regulation of the Hippo Pathway. Cancer Res. 2016 Sep 28
27. Qi L, Jafari N, Li X. et al. Talin2-mediated traction force drives matrix degradation and cell invasion. J Cell Sci. 2016;129(19):3661-3674
28. Virga J, Bognár L, Hortobágyi T. et al. Prognostic Role of the Expression of Invasion-Related Molecules in Glioblastoma. J Neurol Surg A Cent Eur Neurosurg. 2017;78(1):12-19
29. Du C, Zheng Z, Li D. et al. BKCa promotes growth and metastasis of prostate cancer through facilitating the coupling between αvβ3 integrin and FAK. Oncotarget. 2016;7(26):40174-40188
30. Kishi T, Mayanagi T, Iwabuchi S, Akasaka T, Sobue K. Myocardin-related transcription factor A (MRTF-A) activity-dependent cell adhesion is correlated to focal adhesion kinase (FAK) activity. Oncotarget. 2016;7(44):72113-72130
31. Clausen TM, Pereira MA, Al Nakouzi N. et al. Oncofetal Chondroitin Sulfate Glycosaminoglycans are Key Players in Integrin Signaling and Tumor Cell Motility. Mol Cancer Res. 2016;14(12):1288-1299
32. Touihri-Barakati I, Kallech-Ziri O, Boulila A. et al. Targetting αvβ3 and α5β1 integrins with Ecballium elaterium (L.) A. Rich. seed oil. Biomed Pharmacother. 2016;84:1223-1232
33. McDonnell CJ, Garciarena CD, Watkin RL. et al. Inhibition of major integrin αV β3 reduces Staphylococcus aureus attachment to sheared human endothelial cells. J Thromb Haemost. 2016 Sep 8
34. Liu Z, Chen W, Li Y, Xu Q. Αvβ3 integrin-targeted C-dot Nanocomposites as Multifunctional Agents for Cell Targeting and Photoacoustic Imaging of Superficial Malignant Tumors. Anal Chem. 2016 Nov 2
35. Shi J, Wang F, Liu S. Radiolabeled cyclic RGD peptides as radiotracers for tumor imaging. Biophys Rep. 2016;2(1):1-20
36. Zhao ZQ, Yang Y, Fang W, Liu S. Comparison of biological properties of 99mTc-labeled cyclic RGD Peptide trimer and dimer useful as SPECT radiotracers for tumor imaging. Nucl Med Biol. 2016;43(11):661-669
37. Jin ZH, Furukawa T, Degardin M. et al. αvβ3 Integrin-Targeted Radionuclide Therapy with 64Cu-cyclam-RAFT-c(-RGDfK-)4. Mol Cancer Ther. 2016;15(9):2076-85
38. Niu M, Wang F, Li F, Dong Y, Gu Y. Establishment of a screening protocol for identification of aminopeptidase N inhibitors. J Taiwan Inst Chem E. 2014;49:19-26
39. Ai G, Tian C, Deng D. et al. A combination of 2D similarity search, pharmacophore, and molecular docking techniques for the identification of vascular endothelial growth factor receptor-2 inhibitors. Anticancer Drugs. 2015;26:399-409
40. Yu YP, Wang Q, Liu YC, Xie Y. Molecular basis for the targeted binding of RGD-containing peptide to αvβ3 integrin. Biomaterials. 2014;35:1667-75
41. Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun. 2010;1:145-60
42. Ding L, Tian C, Feng S. et al. Small Sized EGFR1 and HER2 Specific Bifunctional Antibody for Targeted Cancer Therapy. Theranostics. 2015;5:378-98
43. Azhdarinia A, Wilganowski N, Robinson H. et al. Characterization of chemical, radiochemical and optical properties of a dual-labeled MMP-9 targeting peptide. Bioorg Med Chem. 2011;19:3769-76
44. Qin Z, Wang J, Wang Y. et al. Identification of a Glypican-3-Binding Peptide for In Vivo Non-Invasive Human Hepatocellular Carcinoma Detection. Macromol Biosci. 2016 Nov 15
45. Silva VL, Debora F, Nobrega FL, Martins IM, Kluskens LD, Rodrigues LR. Selection of Novel Peptides Homing the 4T1 CELL Line: Exploring Alternative Targets for Triple Negative Breast Cancer. PLoS One. 2016;11(8):0161290
46. Sabahnoo H, Noaparast Z, Abedi SM, Hosseinimehr SJ. New small 99m Tc-labeled peptides for HER2 receptor imaging. Eur J Med Chem. 2016 Nov 9
47. Liu JJ, Huang TS, Cheng WF, Lu FJ. Baicalein and baicalin are potent inhibitors of angiogenesis: Inhibition of endothelial cell proliferation, migration and differentiation. Int J Cancer. 2003;106:559-65
48. Gibbs S, Spiekstra S, Corsini E, Mcleod J, Reinders J. Dendritic cell migration assay: a potential prediction model for identification of contact allergens. Toxicol In Vitro. 2013;27:1170-9
49. Ghitti M, Spitaleri A, Valentinis B. et al. Molecular dynamics reveal that isoDGR-containing cyclopeptides are true αvβ3 antagonists unable to promote integrin allostery and activation. Angew Chem Int Ed Engl. 2012;124:7822-5
50. Cui S, Yin D, Chen Y. et al. In Vivo Targeted Deep-Tissue Photodynamic Therapy Based on Near-Infrared Light Triggered Upconversion Nanoconstruct. ACS Nano. 2012;7:676-88
51. Cao J, Cui S, Li S. et al. Targeted cancer therapy with a 2-deoxyglucose-based adriamycin complex. Cancer Res. 2013;73:1362-73
Author contact
![]() Corresponding author: Yueqing Gu, Ph.D. Professor of State Key Laboratory of Natural Medicines, Department of Biomedical Engineering, School of Engineering, China Pharmaceutical University, Nanjing, 210009, P. R. China; e-mail: guyueqingedu.cn; phone: 86-25-83271046; fax: 86-25-83271249.
Corresponding author: Yueqing Gu, Ph.D. Professor of State Key Laboratory of Natural Medicines, Department of Biomedical Engineering, School of Engineering, China Pharmaceutical University, Nanjing, 210009, P. R. China; e-mail: guyueqingedu.cn; phone: 86-25-83271046; fax: 86-25-83271249.
 Global reach, higher impact
Global reach, higher impact