13.3
Impact Factor
Theranostics 2017; 7(7):1795-1805. doi:10.7150/thno.19217 This issue Cite
Review
Current and emerging techniques for antibiotic susceptibility tests
1. Center for Biosensors and Bioelectronics, The Biodesign Institute, Arizona State University, Tempe, Arizona 85287, USA;
2. State Key Laboratory of Analytical Chemistry for Life Science, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing 210093, China;
3. School of Electrical, Computer and Energy Engineering, Arizona State University, Tempe, Arizona 85287, USA;
4. Institute of Biomedical Science, Fudan University, Shanghai, China;
5. Department of Laboratory Medicine and Pathology, Mayo Clinic, Phoenix, Arizona 85054, USA;
6. Center for Immunotherapy, Vaccines, and Virotherapy, The Biodesign Institute, Arizona State University, Tempe, Arizona 85287, USA;
7. School of Life Sciences, Arizona State University, Tempe, Arizona 85287, USA.
Received 2017-1-16; Accepted 2017-3-3; Published 2017-4-10
Abstract
Infectious diseases caused by bacterial pathogens are a worldwide burden. Serious bacterial infection-related complications, such as sepsis, affect over a million people every year with mortality rates ranging from 30% to 50%. Crucial clinical microbiology laboratory responsibilities associated with patient management and treatment include isolating and identifying the causative bacterium and performing antibiotic susceptibility tests (ASTs), which are labor-intensive, complex, imprecise, and slow (taking days, depending on the growth rate of the pathogen). Considering the life-threatening condition of a septic patient and the increasing prevalence of antibiotic-resistant bacteria in hospitals, rapid and automated diagnostic tools are needed. This review summarizes the existing commercial AST methods and discusses some of the promising emerging AST tools that will empower humans to win the evolutionary war between microbial genes and human wits.
Keywords: AST methods, antibiotic susceptibility tests
Introduction
Antibiotic-resistant bacterial pathogens are a global health epidemic, spreading at a rapid rate. In the US alone, these pathogens cost billions of dollars in healthcare, with 2 million hospitalizations and 23,000 deaths annually [1]. This epidemic is accelerated by widespread misuse of antibiotics in clinics and agriculture over the last few decades, allowing bacteria to evolve and develop means of resistance [2,3]. Resistant bacteria are widely found in the community and can also be acquired via nosocomial infections, post-surgery complications, and contaminated food [3, 4]. Resistant bacterial infections can also cause sepsis, which has mortality rates ranging from 30% to 50% [5]. Considering the life-threatening condition of a septic patient, a key clinical task is prescribing the patient with effective antibiotics, which requires rapid diagnosis of the resistant infections and antibiotic susceptibility testing (AST) [6].
AST is widely used clinically to determine antibiotic resistance profiles of bacterial isolates, to guide antibiotic treatment decisions, and predict therapeutic outcome [5, 7]. Currently, AST is usually performed in a clinical microbiology lab, which necessitates transportation of the patient samples from the healthcare provider to the lab. Susceptibility testing requires a pure culture of the offending pathogen, a process which may take several days. This delay prolongs the time to diagnosis of resistant bacteria and decisions for appropriate and effective antibiotic therapy. Delays in timely administration of appropriate therapeutics lead to increased patient mortality, poor clinical outcomes [5], and use of broad-spectrum antibiotics, the latter of which promotes antibiotic resistance. To survive this evolutionary war against bacteria, we must pursue technologies that can rapidly perform AST to enable personalized therapies (narrow-spectrum antibiotic administration) at the earliest possible treatment stage.
After receipt of the patient sample (collected on day 0), the clinical microbiologist must isolate the potential pathogen by streaking the sample on selective culture media and incubating the inoculated media overnight (or longer) to enable growth. From a primary growth plate (day 1), isolated colonies must be obtained by subculture. Once isolated colonies from the pathogenic organism are available (day 2), the bacterial inoculum is prepared and standardized (day 2) prior to performing AST via disk diffusion [7] or broth dilution [8] methods (detailed later).
The minimum inhibitory concentration (MIC) is defined as the lowest concentration of antibiotic required preventing bacteria growth and is used to determine if the infected pathogen is susceptible or resistant to an antibiotic [7-9]. It is important to note that the MIC does not necessarily imply bacterial death, but rather lack of growth. Thus, the MIC differs from the Minimum Bactericidal Concentration (MBC), a useful value which is seldom determined in a clinical laboratory because of the additional effort required. A breakpoint is defined as the concentration of an antibiotic that enables interpretation of AST to define isolates as susceptible, intermediate, or resistant [8, 10]. If the determined MIC is less than or equal to the breakpoint, then the bacterial isolate is considered susceptible to the antibiotic. Clinical breakpoints for different antibiotics and bacteria are reviewed and updated annually by national organizations, such as the Clinical Laboratory Standards Institute (CLSI) in the USA and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [8]. In addition to characterizing bacterial isolates collected from individual patients, MIC is used in epidemiological monitoring of the evolution of antibiotic-resistant bacteria. Increasing MIC values for an antibiotic over a period of time may indicate acquired antibiotic resistance for a given bacterial species [8]. MIC values serve as an important parameter to determine phenotypic resistance in bacterial cells, to monitor the global resistance surveillance, and to determine the effectiveness of new antibiotics. MIC values obtained by the current AST techniques also serve as a gold standard to evaluate new AST methods. Another concept that is becoming a useful analytic modality is the Epidemiological Cutoff Value for resistance (ECOFFs) [11]. This relates to the MIC values of a population of isolates of a particular organism against a particular drug. It can be helpful to determine intrinsic resistance that is present in some strains of a bacterial species.
The current culture-based AST tools rely on time-consuming culturing techniques, followed by disk diffusion [7] and broth dilution susceptibility testing [8], resulting in several days before MIC values are determined and reported. Paradigm-shifting AST technologies must overcome the current bottleneck associated with the slow culturing steps. Ideally, they would be directly applicable on clinical samples without the need for selection and/or enrichment on day 1, and, preferably, be able to deliver results at the point of care (i.e., at the patient's bedside). In addition to low cost and ease of operation requirements, additional features, such as identification of bacterial strains before AST and the ability to perform AST of polymicrobial infections, will also help improve patient outcomes and reduce the selection of additional resistant organisms.
In the present mini-review, we summarize the current technologies, discuss the emerging technologies, and provide scientific opinions on future AST technologies. Given the vast number of publications in this area, we mainly focus on phenotypic AST methods. Even with this focus, we will unintentionally and inevitably exclude many exciting emerging technologies in the scientific literature due to limited page and scope. Fortunately, several reviews [7, 9, 12, 13] on related topics have been published, thus enabling readers to identify topics that are inadvertently not included here.
Current Technologies
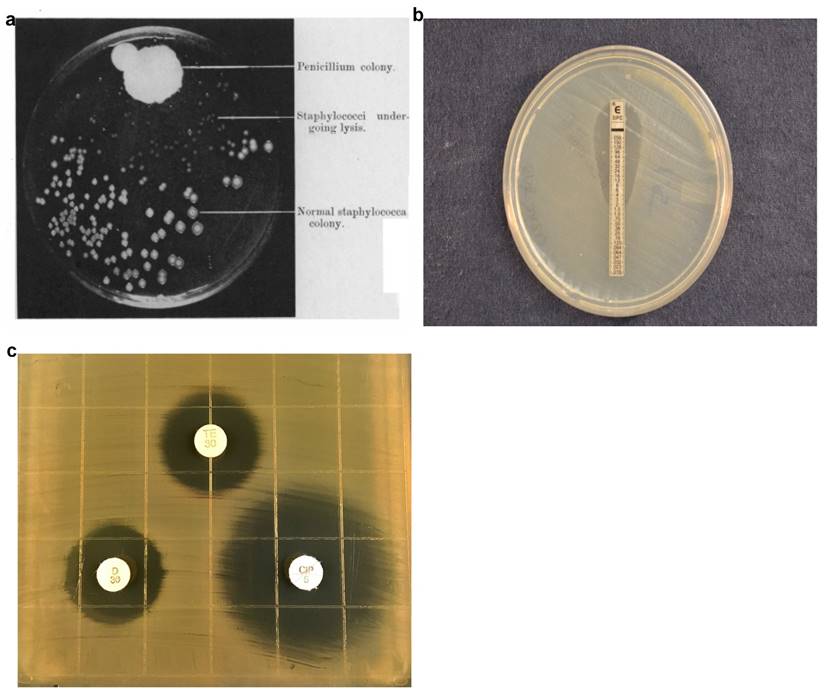
In 1928, Alexander Fleming discovered a mold that prevented the growth of staphylococci on an agar plate (Fig. 1a). The mold produced an active substance, penicillin, which become the first antibiotic and usher in the antibiotic era, a critically important milestone in modern medicine [14]. Antibiotics are commonly used to treat bacterial infections, to reduce the possibility of infections (e.g., during invasive surgeries) in hospitals, and to promote growth in food animals. The widespread use of antibiotics has accelerated the pace at which bacteria become resistant to antibiotics. While antibiotic-resistant bacteria are rapidly evolving, diagnostic technologies that can characterize the infection, guide treatment, minimize unnecessary use of antibiotics, and customize therapeutic strategies for specific patients have been slow. The mainstream technologies still rely on measuring bacterial growth in presence of antibiotics over a few days using methods such as agar dilution assays (E-test and disk diffusion), broth dilution assays, and automated systems from various manufacturers. These technologies rely on detecting bacterial growth, which is not conceptually different from how Fleming first discovered penicillin.
Agar Dilution, Disk Diffusion and Antimicrobial Gradient Assays
In the agar dilution assay, bacteria are inoculated into an agar medium containing different antibiotic concentrations. While agar dilution testing offers reproducible results, agar dilution plates are laborious to prepare and have short shelf lives. In many clinical microbiology laboratories, agar disk diffusion is routinely used for testing common, rapidly growing bacterial pathogens [7]. The disk diffusion assay involves inoculating the bacteria, enriched from clinical samples by overnight growth on selective media, onto a Mueller-Hinton agar plate, followed by placing commercially-prepared filter paper disks impregnated with predetermined concentrations of an antibiotic onto the surface of the agar medium.[12]The agar plate containing the bacteria inoculum and antibiotics disks are further incubated at 35-37°C in ambient air or 5% CO2 for 16-24 hours, depending on the suspected bacterium. During this incubation, the antibiotics diffuse into the agar with antibiotic concentration decreasing with increasing distance from the disk. Antibiotic susceptibility is determined by measuring the diameter of the zones of bacterial inhibition around the antibiotic disks and comparing the diameter with disk diffusion interpretive criteria updated annually by CLSI [12,15]. While the disk diffusion test (Fig. 1b) is technically easy, inexpensive, and flexible, it provides only categorical results (e.g., susceptible, intermediate, resistant). Since quantitative MIC results relaying the degree of susceptibility may be necessary in some cases, the gradient diffusion method offers similar flexibility and simplicity to disk diffusion and determines quantitative MICs. In the Etest, a common commercially-available gradient test, the assays are performed similarly to the disk diffusion approach except that a thin plastic strip with a continuous exponential gradient of antibiotic is used to generate diffusion of the antimicrobial agent into the agar-based medium. After overnight incubation allows bacterial growth and antibiotic diffusion, an inhibition ellipse is visible (Fig. 1c). The quantitative MIC corresponds to the point on the strip whereby the antimicrobial concentration is no longer inhibiting bacterial growth, thus revealing the inhibitory concentration. The disk diffusion and Etest methods are commonly used in clinical microbiology labs.
Broth Dilution Assay
An MIC test can also be performed using broth macrodilution, whereby broth volumes for testing each antibiotic concentration are at least 1 mL. Following incubation for 20-24 h, the MIC is the lowest concentration of antibiotic that completely inhibits bacterial growth and therefore lacks visible turbidity [8]. Due to the laborious nature of the broth macrodilution approach, the assay has been miniaturized and standardized by use of small, plastic, disposable microdilution trays which contain 96 wells to allow minimal volume (e.g.: 0.1 mL) and pre-determined antibiotic concentrations [7]. Many commercially-available systems use automatic inoculating devices, but microwells may also be inoculated with multichannel pipettors. Broth microdilution results may be determined visually or through automated instruments.
Automation of the broth microdilution assay instruments provides more precise, reliable, and quantitative AST. There are four commercially-available automated or semi-automated instruments MicroScan WalkAway, Vitek-2, BD Phoenix automated system, and Sensititre [7,10]. Each of these instruments consists of the following: 1) A single-use AST cassette, which can be a microdilution tray/test panel/card containing different antibiotics at different concentrations; 2) an AST instrument, which reads multiple cassettes over a period of time (usually overnight) to give AST results. These automated AST instruments require bacterial isolates obtained through routine culture from the patient samples.
Microscan Walkaway AST cassette, based on standard 96-well microdilution trays, is capable of handling 40-96 trays with automated sample-handling robotics, where the antibiotic susceptibility test uses a photometer to detect bacterial turbidity in the trays over 4.5-18 hours [7, 8, 16]. The Vitek-1/Vitek-2 AST instruments developed by bioMérieux, use a smaller AST cassette, called an AST card, in the 45-64 well plate format. Each Vitek-2 AST instrument is capable of handling 30-240 AST cards and detects turbidity with bacterial growth over 4-10 hours to reveal AST results. The BD Phoenix is an automated microbiology system that consists of a large AST instrument capable of reading turbidity and colorimetric changes of up to 99 AST cassettes (called panels). The BD system requires an average of 6-16 hours, starting from incubating pure bacterial cultures, to obtain MIC for the bacteria. The Sensititre system by Thermo Scientific uses the standard 96-well microdilution panels (AST cassettes), which are inoculated by the Sensititre Autoinoculator, and is capable of handling 64 panels. Bacterial growth in each panel is detected from the fluorescent intensity monitored over 18-24 hours post incubation.
Automated AST instruments, representing current state of the art technologies, are extensively used in clinical microbiology labs in the US. Compared to manual methods, these instruments provide a streamlined workflow and quantitative results, thus simplifying MIC determinations for pathogenic bacteria isolated from clinical samples [17]. However, these automated instruments still require the use of isolated bacteria grown in pure culture, and the susceptibility tests are based on measuring bacterial growth and turbidity changes. As a result, these automated technologies remain inherently slow and are severely limited by the low sensitivity of the current detection methods. Furthermore, they are limited in the number of antibiotics and concentrations tested and lack the capability of analyzing polymicrobial samples or heterogeneous response of bacterial populations to the antibiotics.
Evolution of agar dilution methods for determining antibiotic susceptibility from the discovery of antibiotics (a) to currently used disk diffusion (b) and Etest (c) assays. a) Photograph showing lack of staphylococcal colonies in the vicinity of the Penicillium mold adapted from Alexander Fleming's original research paper on the discovery of penicillin. b) E-Test uses gradient antibiotic concentrations to determine MIC of antibiotics. c) Disk diffusion assays involve placing multiple antibiotic-impregnated disks onto an agar surface inoculated with bacteria and measuring the diameter of zones of inhibition to qualitatively determine antibiotic susceptibility. Figure 1a Adapted from - Alexander Fleming. On the Antibacterial Action of Cultures of a Penicillum, with Special Reference to their use in the isolation of B. Influenze. Br J ExpPathol. 1929 Jun; 10(3): 226-236Disk diffusion assay image produced by John Popovich, Haydel Lab, ASU.E-test image produced by Rachael Liesman.
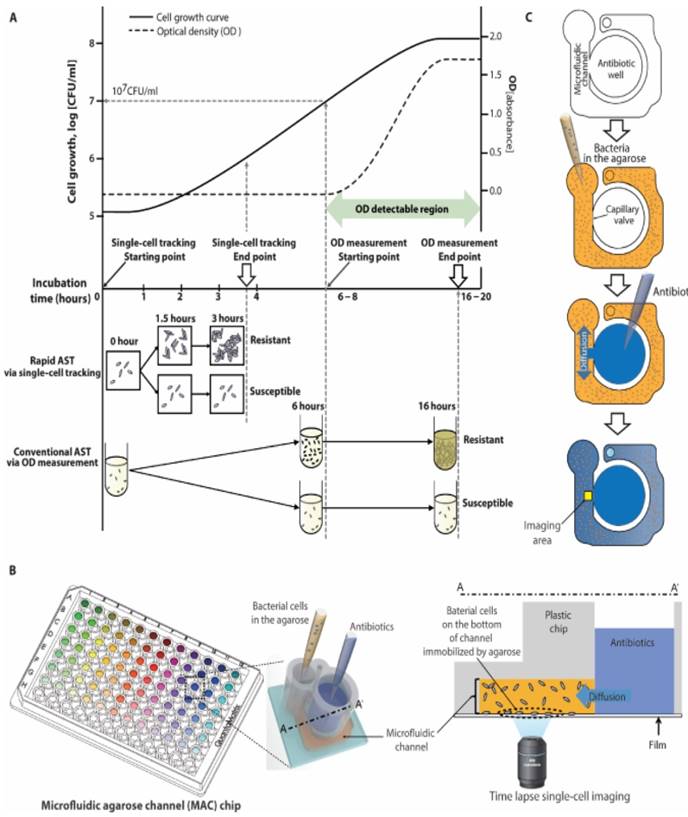
Rapid AST using an emerging imaging based tool. a) Schematic comparison of traditional AST using broth microdilution and imaging-based AST demonstrates how tracking single cell divisions can produce rapid results compared to traditional optical density (OD) tools which are limited by their sensitivity to measure only higher bacterial concentrations. b) Setup of a 96-well plate modified into a microfluidic agarose chip for concurrent addition of bacteria and antibiotics followed by microscopic imaging. c) Schematic of steps involved in adding bacteria and antibiotics and imaging a localized area to observe changes. From Choi J, Yoo J, Lee M, Kim E-G, Lee JS, Lee S, et al. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci Transl Med 2014; 6:267ra174 Reprinted with permission from AAAS.
Emerging Technologies
Newer AST techniques, which are currently and actively being pursued by commercial entities for clinical translation, are considered as emerging technologies for the purpose of this review. With the increasing clinical demand for rapid AST, various new AST techniques based on optical imaging [18-20], micro-channel resonators [21-23] and other biosensors [24, 25] have been pursued. For example, optical detection of bacterial growth via the cell lengths and numbers [18, 20, 26], forward light scattering [25], and measuring vibrational amplitude changes of magnetic beads [24, 27], have been proposed. Micro-channel resonators have also been used to detect nanoscale fluctuations associated with bacteria growth [21]. Quantitating molecular or biochemical markers, such as 16SrRNA [28], ATP [29], and luciferase [30], in bacterial cells are also being used for rapid AST. These approaches can significantly improve the current commercial AST technologies, but they still rely on culturing, which is not universally applicable for anaerobes, slow-growing bacteria, and non-cultivable microorganisms. Additionally, most of these emerging technologies still require substantial sample preparation and pre-treatment steps, such as bacterial enrichment from patient samples, and cell lysis to extract biochemical markers.
Imaging-based AST
Multiplexed automated digital microscopy (MADM) [19, 31] is an automated microscope being developed to provide rapid identification and AST of clinical samples. MADM separates bacterial cells from other substances in the clinical samples (e.g., blood or urine) using gel filters and attaches purified bacterial cells to the surface sensing surface using electro-kinetic loading [31]. After surface attachment, fluorescent in-situ hybridized (FISH) probes are used to identify bacterial cells within an hour, followed by AST [31]. To perform AST, MADM measures bacterial growth every 10 minutes as clonal aggregates multiply in Mueller-Hinton media. Since resistant cells will grow in Mueller-Hinton media with antibiotics and sensitive cells will be inhibited or killed, expansion and measurement of clonal masses over time (compared to growth controls) are used to generate growth curves and determine susceptibility. MADM also uses cell morphokinetic image analysis for differentiating bacterial species in polymicrobial infections, thus expanding clinical capability and reducing of the cost of multiple assays. While the MADM imaging approach for measuring the bacteria growth rate is faster than traditional approaches and represents a significant step forward from the current commercial tools, its universality to all antibiotics remains to be addressed [18].
Another imaging tool capable of rapid AST is single-cell morphological analysis (SCMA) [18, 32]. SCMA (Fig. 2) uses bright-field microscopy to determine antibiotic-induced morphological changes in single bacterial cells and enable rapid AST. The captured images are processed using an automated image-processing algorithm to quantify the area and number of growing bacterial cells. The classification algorithm processes several morphological characteristics to produce antimicrobial susceptibility data. Another optical imaging technique is oCelloScope [33, 34], which is based on imaging growth of a population of bacterial cells in a fluid sample with antibiotics over a period of time. The recorded images are then processed using imaging algorithms to quantify changes in the area occupied by a growing population of cells. However, unlike other high resolution imaging methods, oCelloScope does not capture the growth of individual cells, but a population of cells in liquid fluids and thus eliminates the need to attach bacterial cells to an inert surface.
Coupling of imaging-based tools with microfluidics has been reported for rapid AST. Bacterial cells are first captured in microfluidic chambers [35], micro channels [36], or droplets [37, 38] and then imaged to detect changes in the cell number [20, 39], size [18], morphology [40] and viability [37, 41] in the presence of antibiotics in order to perform AST. Novel imaging approaches, such as measuring changes in rotational frequency of magnetic beads (which is proportional to cell mass) [24, 27, 42] and electro-kinetic loading of single cells, [36] have been applied to AST using smartphone cameras and other imaging devices [41].
Although the imaging-based AST tools shorten the detection time from days to a few hours, these technologies still use replication-dependent methodologies that have a primary culture step (e.g. growth from a blood culture bottle or growth on a primary culture plate). These dependencies limit the application of imaging-based methods to slowly-growing pathogens, such as Mycobacterium tuberculosis. To perform AST on pathogens directly (i.e.: without a culture step) from clinical samples, it is necessary to separate bacteria from the patient sample matrix, and then measure a cellular attribute that is independent of replication.
Non-imaging AST
Non-imaging methods that measure the physical or biochemical signature of bacterial cells have been proposed for AST. BacterioScan detects forward laser light scattering (FLLS) [25, 43] analyzes the angular variation in the intensity of the scattered light, and determines the number and size of bacterial cells suspended in a solution. FLLS can measure bacterial concentrations as low as 103cfu/ml, which is more sensitive than other optical methods and traditional automated instruments, and may enable rapid AST (within a few hours). The FLLS technology can perform AST directly on urine samples with minimal sample preparation, thus enabling point-of-care applications. Disadvantages of FLLS include the use of a replication-dependent approach to measure AST, inability to differentiate bacteria from cell sedimentation, lack of single cell resolution, and inability to differentiate bacterial species for polymicrobial analysis.
LifeScale develops microchannel resonators for rapid AST, where the microchannel resonators are individual microcantilevers. The technology measures the mass of the bacterial cells upon passage through the microfluidics channels inside of the micro-cantilevers [23]. Microchannel resonators permit quantitation of bacterial cells and measure mass changes of the individual cells to assess antibiotic activity [21, 23, 44]. The advantages of microchannel resonator AST are the ability to perform sensitive mass and morphology measurements on single bacterial cells and the promise of AST within~3 hours. However, the applicability of the approach to clinical samples remains to be fully established.
Biochemical AST
While the AST technologies described above detect physical and morphological features of bacteria, tools that measure molecular and biochemical signatures, such as changes in 16s RNA [28, 45], DNA [46, 47] and ATP [29], of growing bacterial cells have also been studied. A biosensor-based AST (b-AST) assay being developed by Genefluidics measures bacterial growth via quantitating 16s rRNA molecules, which are specific for each bacterial species [28, 45]. After DNA probes hybridize specifically to 16S rRNA molecules, an electrochemical signal permits amplification and quantitative detection. This approach allowed for AST as short as ~4 hours using clinical urine samples from patients experiencing a urinary tract infection. Smarticles technology in development by Roche Diagnostics [30] introduces recombinant bacteriophages with DNA probes, such that a specific binding of DNA probes inside the bacterial cells leads to luciferase expression. Luciferase expression produces light, which is used to quantify the number of bacterial cells and perform rapid AST. Real-time PCR is another molecular approach, which quantifies copies of bacterial DNA and correlates this value with bacterial growth in a sample. This technique targets highly conserved regions of bacterial chromosomal DNA to ensure species specificity and has been applied to various combinations of antibiotics and bacterial species. Another approach detects bacterial genetic fingerprints that are detected upon exposure to antibiotics rather than relying on a single specific gene or DNA sequence in the other approaches described above.
While the nucleic acid-based biochemical assays, such as real-time PCR, can give faster results than the current techniques, it has several disadvantages such as relying on high bacterial concentrations to extract sufficient DNA, manual sample handling steps such as lysing bacterial cells to extract nucleic acids.[48] These manual steps make clinical adaptation of these technologies difficult, where the need of the hour is rapid automated testing. Further the extracted DNA contains DNA from both alive/dead cells leading to a higher false positive rate for these techniques [49]. Other disadvantages include the need of previously known sequences, micro-heterogeneity in the 16s RNA within a species [50], lack of correlations between genotypic and phenotypic resistance [51], and inability of performing tests on clinical samples.
Other biochemical signatures, such as ATP and NADH, have been studied as AST biomarkers with electrochemical amplification [29, 52-54]. These biochemical signatures are indicators of the metabolic activities of bacteria, thus providing critical information on bacterial viability. While some of these techniques are capable of providing rapid AST within a few hours, these techniques currently lack sensitivity to perform AST at lower antibiotic concentrations and dilution ranges. Further, the universal application of the probe molecules to multiple strains and antibiotics is also questionable. While promising, these emerging approaches require further studies and evaluations.
Future Technologies
The emerging technologies, being actively pursued by commercial entities discussed above, promise rapid AST within a few hours. Furthermore, some of the technologies can be directly applied to patient samples without any sample pretreatment. However, further shortening the test time and applying them to slowly growing organisms will require innovative approaches. We discuss future technologies that can meet these requirements below.
Microcantilevers have been recently used to perform rapid AST [22, 55, 56], whereby bacterial cells are attached to a microcantilever and deflection of the microcantilever associated with the micromotions of the bacterial cells, is detected as the signature of bacterial metabolism. This approach has led to AST within two hours for Escherichia coli and Staphylococcus aureus strains for different antibiotics [22, 55]. The correlation between the micromotions and viability (metabolism) has been studied for both prokaryotic and eukaryotic cells [57]. While sensitive, the micromotion of bacterial cells producing microcantilever deflections is affected by flowing liquids, and recent reports have also indicated inefficient transfer of antibiotics to immobilized bacterial cells under laminar flow conditions [23]. Furthermore, the sensitive cantilever deflections are caused by bacterial cells attached to the tip, so the small area of the tip limits the number of bacterial cells which are adsorbed to the surface, potentially preventing the application of the technique to lower bacterial concentrations present in clinical samples [23]. It is unclear how this approach can be applied directly to complex matrix of clinical samples and polymicrobial systems. Given that eukaryotic cells can also cause cantilever fluctuations [57], sample preparation for this technique might need extraction of bacterial cells from complex matrix along with a longer incubation of bacterial cells to attach the sensor surface, especially for low bacterial load patient samples.
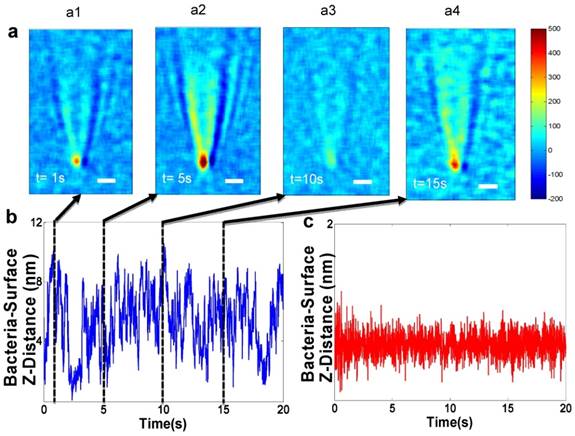
A plasmonic imaging and tracking (PIT) technique has been used to track 3D motions of single bacterial cells associated with metabolic viability, thus leading to rapid AST [58, 59]. The PIT setup is built on an inverted optical microscope, where light from a luminescence diode is directed onto the sensor chip made of gold-coated glass film with immobilized bacterial cells. Fig. 3a shows a few snapshots of the plasmonic image, which reveal large fluctuations in the image contrast of a bacterial cell. The image contrast fluctuations are due to the bacterial cell movement normal to the sensor surface (Z-direction) [60, 61], due to bacterial metabolism. Additionally, PIT has been shown to track the 3D movement of single bacterial cells and sub-cellular organelles in 3D with < 5 nm spatial resolution and <1 millisecond (ms) temporal resolution [61, 62]. This unprecedented capability enables fast tracking of the 3D movement of many bacterial cells simultaneously, providing high throughput quantification of AST with single cell detection capability. In addition, PIT could potentially be used to spatially resolve and identify bacterial cells even in a complex matrix of urine, serum, and other body fluid samples, which is critical for developing PIT into a practical solution for testing real patient samples.
Flow cytometry (FC) measures changes in morphology, cellular numbers, and viability via labeling to perform AST [63-65]. After a dye is used to stain viable cells, individual cells flow through a channel into a reader zone, where light scattering is used to measure morphology and excitation/emission spectra of cells is used to assess cell counts and viability. Multiple research studies have shown the application of this technology using various dyes [64, 65] applied to multiple bacterial species and antibiotic combinations. Although flow cytometry can produce rapid AST with 2-3 hours, it is not a widely used technique yet. Possible disadvantages are lack of use in complex patient samples, staining inefficiency of dyes, presence of autofluorescence, inability to differentiate cellular damage caused by bactericidal or bacteriostatic antibiotics, and lack of clinical databases for validation [66].
Future technologies measuring bacterial nano-motion as a measure of bacterial metabolism to perform antibiotic susceptibility. a) Snapshots of bacteria z-micro-motion. Panels a1-a4 show time differential images captured at various time points which show contrast of the bacteria versus the background. The observation of the small contrast is due to micro-motions of the live bacterial cells. b) Z-displacement vs. time - The positions of minimum contrast (a1 and a3) correspond to bacterial z-position farther away from the surface. The position of maximum contrast (a2) corresponds to z-position closest to the surface. c) z-displacement plot of a dead bacterial cell (no motion) showing a standard deviation of 0.15 nm. Reprinted with permission from Syal K, Iriya R, Yang Y, Yu H, Wang S, Haydel SE, et al. Antimicrobial Susceptibility Test with Plasmonic Imaging and Tracking of Single Bacterial Motions on Nanometer Scale. ACS Nano 2015; 10:845-852 Copyright 2017 American Chemical Society.
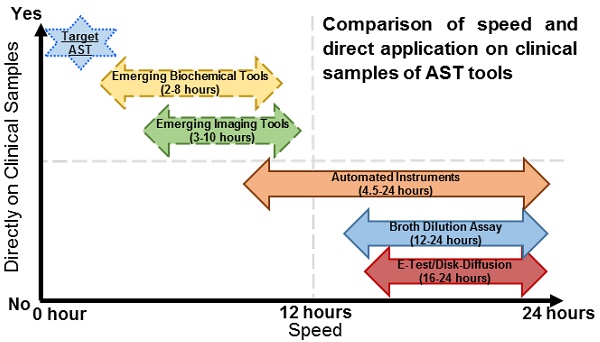
Summary of AST Technologies
| AST Technologies | Summary of Method | Time of AST | Direct on patient sample | Real MIC | FDA Approved | Reference |
|---|---|---|---|---|---|---|
| Current Technologies | ||||||
| Solid Media Cultures | ||||||
| 1. Agar Dilution Assay | Bacteria inoculated on agar plates with antibiotic discs of different concentrations | 16-24 Hours | No | Yes/No | Yes | [7] |
| 2. Disk Diffusion | Bacteria inoculated on agar plates with a single antibiotic disk | 16-24 Hours | No | Yes/No | Yes | [7, 12] |
| 3. E-test | Bacteria inoculated on agar plates with a graded antibiotic concentration strips | 16-24 Hours | No | Yes | Yes | [7, 12] |
| Liquid Media Cultures | ||||||
| 1. Broth Dilution Assay | Bacteria inoculated in liquid media with different antibiotics to monitor growth | 12-24 Hours | No | Yes | Yes | [7, 10] |
| 2. Automated Instruments | ||||||
| a) MicroScanWalkAway | Measure bacterial growth in the presence of antibiotics by recording bacterial turbidity using a photometer | 4.5-18 Hours | No | Yes | Yes | [8, 16] |
| b) Vitek-1/Vitek-2 | Measure bacterial growth in the presence of antibiotics by recording bacterial turbidity using a photometer | 6-11 Hours | No | Yes | Yes | [17 ,70] |
| c) BD Phoenix | Record bacterial growth in the presence of antibiotics by recording bacterial turbidity and colorimetric changes | 9-15 Hours | No | Yes | Yes | [71] |
| d) Sensititre | Record bacterial growth with antibiotics by measuring fluorescence | 18-24 Hours | No | Yes | Yes | [7] |
| Emerging Technologies Imaging Based Tools | ||||||
| 1. Multiplexed automated digital microscopy (MADM) | Image single bacteria growing into colonies with antibiotics and quantify growth rates | 3-5 Hours | Yes (Urine, Blood) | Yes | Yes | [31, 72, 73] |
| 2. Single-cell morphological analysis (SCMA) | Image single bacterial cell's morphology changes on antibiotic action | 3-4 Hours | Yes (Urine) | Yes | No | [32, 74] |
| 3. oCelloscope | Measure growth of bacterial cells using low resolution optical system | 1-4 Hours | Yes (Urine) | Yes | No | [33] |
| Non-Imaging Based Tools | ||||||
| 1. BacterioScan FLLS | Measures bacterial numbers and sizes on antibiotic action | 3-10 Hours | Yes (Urine) | Yes | No | [25, 43] |
| 2. LifeScaleMicochannel Resonator | Count bacterial cells and morphology changes on single cells post antibiotic action | > 3 Hours | No | Yes | No | [44] |
| 3. Genefluidics | Count 16s RNA increase as a proxy to bacterial growth | 4 Hours | Yes (Urine) | Yes/No | No | [45, 75] |
| 4. Smarticles | Bacteriophages which express luciferase on growing cells | - | - | - | No | [30] |
| Future Technologies | ||||||
| 1. AFM Cantilever | Measure cantilever fluctuations originating from bacterial motion as a proxy for metabolism | < 2 Hours | No | Yes | No | [22] |
| 2. PIT | Image and Quantify sub-nanometer motion of bacterial cells | < 2 Hours | Yes | Unknown | No | [59] |
| 3. Flow Cytometry | Count viable bacterial cells using dyes | 2-3 hours | No | Yes | No | [63] |
| 4. IMC | Heat signature of growing cells | 3-14 Hours | Yes | Yes | No | [68] |
Comparison of speed and direct application on clinical samples of current and emerging AST technologies
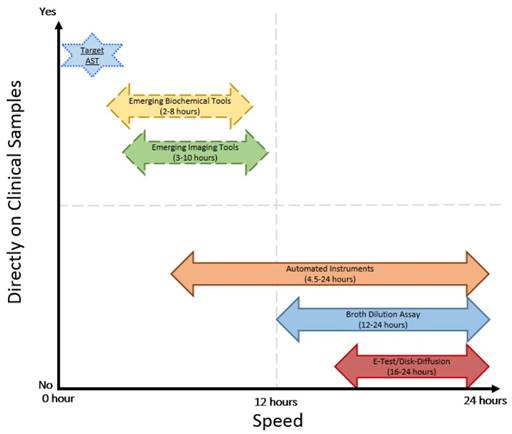
Isothermal micro calorimetry (IMC) is a novel technique that measures cumulative heat and generates heat curves of growing bacterial cells [67]. Heat curves of growing bacteria are similar to the growth curve measured by standard turbidity detection instruments. Since the lower limit of detection for IMC is ~ 104cfu/ml, the approach enables a faster AST [67]. IMC produces AST results within 3 hours using patient urine samples [68] and has been effective with various bacterial species, including Mycobacterium tuberculosis [69], E. coli, and S. aureus [67]. Although this new analytical tool uses a new signature of bacterial metabolism to perform AST, heat curves do not correlate with current standard techniques and do not shorten the time to generate MIC considerably due to dependence on culturing tools. Other discrepancies such as delays in onset of detectable heat due to insufficient bacterial numbers and lack of cellular level metabolic understanding of heat curves limit its current clinical use. While novel, sample preparation might involve purification of bacterial samples by overnight culturing to enable translation of this technique to complex clinical samples such as blood and sputum. This novel tool needs to be advanced further and studied in more to meet the expectations of current rapid AST.
Conclusions
Current manual and automated AST technologies are the backbone of today's clinical microbiology labs. Given their ease-of-use, relatively low cost to perform AST and prevalence across the globe, they will be indispensable in the immediate future. In the near future, we anticipate that the emerging and future innovative technologies, such as MADM, PIT and IMC, will lead the next wave of more powerful AST tools for rapid clinical diagnostics. Future tools to measure bacterial metabolic activity in real-time without culturing will be a quantum leap forward from the existing commercial AST technologies. These tools will enable a one-hour AST, within the time span of an outpatient clinical visit. Such rapid and real-time AST tools will not only help save lives [6], but also have the potential to enable accurate antibiotic treatment at disease onset, potentially slowing the evolution of antibiotic resistance and improving antibiotic stewardship. Given the ever-increasing spread of antibiotic resistance, we must develop innovative technologies which permit rapid AST within an hour, can be applied to fluids collected directly from the patient, and are applicable to slow-growing and non-cultivable microbes.
Acknowledgements
We thank Gordon and Betty Moore Foundation, and CNSF (#2137008 and #2137902) for financial support. We thank Rachael Liesman for providing picture of E-test and John Popovich for providing the picture of disk-diffusion assay.
Competing Interests
The authors have declared that no competing interest exists.
References
1. States U. Antibiotic resistance threats. Published Online First: 2013. http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf
2. Van Boeckel TP, Gandra S, Ashok A, Caudron Q, Grenfell BT, Levin SA. et al. Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. Lancet Infect Dis. 2014;14:742-750
3. Laxminarayan R, Duse A, Wattal C, Zaidi AKM, Wertheim HFL, Sumpradit N. et al. Antibiotic resistance — the need for global solutions. Lancet Infect Dis. 2013;3099:1057-1098
4. Zurek L, Ghosh A. Insects represent a link between food animal farms and the urban environment for antibiotic resistance traits. Appl Environ Microbiol. 2014;80:3562-3567
5. Daniels R. Surviving the first hours in sepsis: Getting the basics right (an intensivist's perspective). J Antimicrob Chemother. 2011;66:11-23
6. Barenfanger J, Drake C, Kacich G. Clinical and financial benefits of rapid bacterial identification and antimicrobial susceptibility testing. J Clin Microbiol. 1999;37:1415-8
7. Jorgensen JH, Ferraro MJ. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis. 2009;49:1749-55
8. Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3:163-75
9. Bauer KA, Perez KK, Forrest GN, Goff DA. Review of rapid diagnostic tests used by antimicrobial stewardship programs. Clin Infect Dis. 2014;59:S134-S145
10. Humphries RM, Hindler JA. Emerging Resistance, new antimicrobial agents... but no tests! the challenge of antimicrobial susceptibility testing in the current us regulatory landscape. Clin Infect Dis. 2016;63:83-88
11. Martínez JL, Coque TM, Baquero F. What is a resistance gene ? Ranking risk in resistomes. Nat Publ Gr. 2014;13:116-123
12. Balouiri M, Sadiki M, Ibnsouda SK. Methods for in vitro evaluating antimicrobial activity: A review. J Pharm Anal. 2016;6:71-79
13. Fluit ADC, Visser MR, Schmitz F. Molecular Detection of Antimicrobial Resistance. Clin Microbiol Rev. 2001;14:836-871
14. Fleming A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Br J Exp Pathol. 1929;10:226-336
15. Chotinantakul K, Suginta W, Schulte A. Advanced Amperometric Respiration Assay for Antimicrobial Susceptibility Testing. Anal Chem. 2014;86:10315-10322
16. McGregor A, Schio F, Beaton S, Boulton V, Perman M, Gilbert G. The MicroScan WalkAway diagnostic microbiology system-an evaluation. Pathology. 1995;27:172-6
17. Winstanley T, Courvalin P. Expert systems in clinical microbiology. Clin Microbiol Rev. 2011;24:515-556
18. Choi J, Yoo J, Lee M, Kim E, Lee JS, Lee S. et al. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci Transl Med. 2014;6:267ra174
19. Metzger S, Frobel R a, Dunne WM. Rapid simultaneous identification and quantitation of Staphylococcus aureus and Pseudomonas aeruginosa directly from bronchoalveolar lavage specimens using automated microscopy. Diagn Microbiol Infect Dis. 2014;79:160-165
20. Mohan R, Mukherjee A, Sevgen SE, Sanpitakseree C, Lee J, Schroeder CM. et al. A multiplexed microfluidic platform for rapid antibiotic susceptibility testing. Biosens Bioelectron. 2013;49:118-25
21. Godin M, Delgado FF, Son S, Grover WH, Bryan AK, Tzur A. et al. Using buoyant mass to measure the growth of single cells. Nat Methods. 2010;7:387-390
22. Longo G, Alonso-Sarduy L, Rio LM, Bizzini a, Trampuz a, Notz J. et al. Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors. Nat Nanotechnol. 2013;8:522-6
23. Etayash H, Khan MF, Kaur K, Thundat T, Farahi RH, Passian A. et al. Microfluidic cantilever detects bacteria and measures their susceptibility to antibiotics in small confined volumes. Nat Commun. 2016;7:12947
24. Sinn I, Albertson T, Kinnunen P, Breslauer DN, McNaughton BH, Burns M a. et al. Asynchronous magnetic bead rotation microviscometer for rapid, sensitive, and label-free studies of bacterial growth and drug sensitivity. Anal Chem. 2012;84:5250-6
25. Hayden RT, Clinton LK, Hewitt C, Koyamatsu T, Sun Y, Jamison G. et al. Rapid Antimicrobial Susceptibility Testing Using Forward Laser Light Scatter Technology. J Clin Microbiol. 2016;54:2701-2706
26. Price CS, Kon SE, Metzger S. Rapid antibiotic susceptibility phenotypic characterization of Staphylococcus aureus using automated microscopy of small numbers of cells. J Microbiol Methods. 2014;98:50-58
27. Kinnunen P, Sinn I, McNaughton BH, Newton DW, Burns M a, Kopelman R. Monitoring the growth and drug susceptibility of individual bacteria using asynchronous magnetic bead rotation sensors. Biosens Bioelectron. 2011;26:2751-5
28. MacH KE, Mohan R, Baron EJ, Shih MC, Gau V, Wong PK. et al. A biosensor platform for rapid antimicrobial susceptibility testing directly from clinical samples. J Urol. 2011;185:148-153
29. Ivančić V, Mastali M, Percy N, Gornbein J, Babbitt JT, Li Y. et al. Rapid antimicrobial susceptibility determination of uropathogens in clinical urine specimens by use of ATP bioluminescence. J Clin Microbiol. 2008;46:1213-1219
30. Listed] [No Author]. Roche gobbles Smarticles. Nat Biotechnol. 2015;33:1012-1012
31. Chantell C. Multiplexed Automated Digital Microscopy for Rapid Identification and Antimicrobial Susceptibility Testing of Bacteria and Yeast Directly from Clinical Samples. Clin Microbiol Newsl. 2015;37:161-167
32. Choi J, Jung Y-G, Kim J, Kim S, Jung U, Na H. et al. Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system. Lab Chip. 2012;13:280-287
33. Fredborg M, Andersen KR, Jørgensen E, Droce A, Olesen T, Jensen BB. et al. Real-time optical antimicrobial susceptibility testing. J Clin Microbiol. 2013;51:2047-53
34. Fredborg M, Rosenvinge FS, Spillum E, Kroghsbo S, Wang M, Sondergaard TE. Rapid antimicrobial susceptibility testing of clinical isolates by digital time-lapse microscopy. Eur J Clin Microbiol Infect Dis. 2015;34:2385-2394
35. Kim S, Cestellos-Blanco S, Inoue K, Zare R. Miniaturized Antimicrobial Susceptibility Test by Combining Concentration Gradient Generation and Rapid Cell Culturing. Antibiotics. 2015;4:455-466
36. Lu Y, Gao J, Zhang DD, Gau V, Liao JC, Wong PK. Single cell antimicrobial susceptibility testing by confined microchannels and electrokinetic loading. Anal Chem. 2013;85:3971-3976
37. Boedicker JQ, Li L, Kline TR, Ismagilov RF. Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab Chip. 2008;8:1265-1272
38. Chen CH, Lu Y, Sin MLY, Mach KE, Zhang DD, Gau V. et al. Antimicrobial Susceptibility Testing Using High Surface-to-Volume Ratio Microchannels. Anal Chem. 2010;82:1012-1019
39. Mohan R, Sanpitakseree C, Desai A V, Sevgen SE, Schroeder CM, Kenis PJA. A microfluidic approach to study the effect of bacterial interactions on antimicrobial susceptibility in polymicrobial cultures. RSC Adv. 2015;5:35211-35223
40. Quach DT, Sakoulas G, Nizet V, Pogliano J, Pogliano K. Bacterial Cytological Profiling (BCP) as a Rapid and Accurate Antimicrobial Susceptibility Testing Method for Staphylococcus aureus. EBIOM. 2016;4:95-103
41. Kadlec MW, You D, Liao JC, Wong PK. A Cell Phone-Based Microphotometric System for Rapid Antimicrobial Susceptibility Testing. J Lab Autom. 2013;19:258-266
42. Kinnunen P, McNaughton BH, Albertson T, Sinn I, Mofakham S, Elbez R. et al. Self-assembled magnetic bead biosensor for measuring bacterial growth and antimicrobial susceptibility testing. Small. 2012;8:2477-2482
43. Bugrysheva J V, Lascols C, Sue D, Weigel LM. Rapid Antimicrobial Susceptibility Testing of Bacillus anthracis, Yersinia pestis, and Burkholderia pseudomallei Using Laser Light Scattering Technology. J Clin Microbiol. 2016;5:1462-1471
44. Burg TP, Godin M, Knudsen SM, Shen W, Carlson G, Foster JS. et al. Weighing of biomolecules, single cells and single nanoparticles in fluid. Nature. 2007;446:1066-1069
45. Mohan R, Mach KE, Bercovici M, Pan Y, Dhulipala L, Wong PK. et al. Clinical Validation of Integrated Nucleic Acid and Protein Detection on an Electrochemical Biosensor Array for Urinary Tract Infection Diagnosis. PLoS One. 2011;6:e26846
46. Rolain JM, Mallet MN, Fournier PE, Raoult D. Real-time PCR for universal antibiotic susceptibility testing. J Antimicrob Chemother. 2004;54:538-541
47. Schoepp NG, Khorosheva EM, Schlappi TS, Curtis MS, Humphries RM, Hindler JA. et al. Digital Quantification of DNA Replication and Chromosome Segregation Enables Determination of Antimicrobial Susceptibility after only 15 Minutes of Antibiotic Exposure Zuschriften. Angew Chem Int Ed Engl. 2016;128:9709-9713
48. Buchan BW, Ledeboer NA. Emerging Technologies for the Clinical Microbiology Laboratory. Clin Microbiol Rev. 2014;27:783-822
49. Rogers GB, Marsh P, Stressmann AF, Allen CE. The exclusion of dead bacterial cells is essential for accurate molecular analysis of clinical samples. Eur Soc Clin Infect Dis. 2010;16:1656-1658
50. Clarridge JE. Impact of 16S rRNA Gene Sequence Analysis for Identification of Bacteria on Clinical Microbiology and Infectious Diseases. Clin Microbiol Rev. 2004;17:840-862
51. Martineau F, Picard OISJ, Lansac N, Me C, Roy PH, Ouellette M. et al. Correlation between the Resistance Genotype Determined by Multiplex PCR Assays and the Antibiotic Susceptibility Patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 2000;44:231-238
52. Besant JD, Sargent EH, Kelley SO. Rapid electrochemical phenotypic profiling of antibiotic-resistant bacteria. Lab Chip. 2015;15:2799-2807
53. Ertl P, Robello E, Battaglini F, Mikkelsen SR. Rapid antibiotic susceptibility testing via electrochemical measurement of ferricyanide reduction by Escherichia coli and Clostridium sporogenes. Anal Chem. 2000;72:4957-4964
54. Mann TS, Mikkelsen SR. Antibiotic susceptibility testing at a screen-printed carbon electrode array. Anal Chem. 2008;80:843-848
55. Aghayee S, Benadiba C, Notz J, Kasas S, Dietler G, Longo G. Combination of fluorescence microscopy and nanomotion detection to characterize bacteria. J Mol Recognit. 2013;26:590-5
56. Lissandrello C, Inci F, Francom M, Paul MR, Demirci U, Ekinci KL. Nanomechanical motion of Escherichia coli adhered to a surface. Appl Phys Lett. 2014:113701-5
57. Kasas S, Simone F, Benadiba C, Maillard C, Stupar P, Tournu H. Detecting nanoscale vibrations as signature of life. Proc Natl Acad Sci U S A. 2014;112:378-381
58. Syal K, Wang W, Shan X, Wang S, Chen HY, Tao N. Plasmonic imaging of protein interactions with single bacterial cells. Biosens Bioelectron. 2015;63:131-137
59. Syal K, Iriya R, Yang Y, Yu H, Wang S, Haydel SE. et al. Antimicrobial Susceptibility Test with Plasmonic Imaging and Tracking of Single Bacterial Motions on Nanometer Scale. ACS Nano. 2015;10:845-852
60. Yang Y, Yu H, Shan X, Wang W, Liu X, Wang S. et al. Label-Free Tracking of Single Organelle Transportation in Cells with Nanometer Precision Using a Plasmonic Imaging Technique. Small. 2015;11:2878-84
61. Shan X, Fang Y, Wang S, Guan Y, Chen HY, Tao N. Detection of charges and molecules with self-assembled nano-oscillators. Nano Lett. 2014;14:4151-4157
62. Yang Y, Yu H, Shan X, Wang W, Liu X, Wang S. et al. Label-Free Tracking of Single Organelle Transportation in Cells with Nanometer Precision Using a Plasmonic Imaging Technique. Small. 2015;11:2878-84
63. Álvarez-Barrientos A, Arroyo J, Cantón R, Nombela C, Sánchez-Pérez M. Applications of flow cytometry to clinical microbiology. Clin Microbiol Rev. 2000;13:167-195
64. Jepras RI, Paul FE, Pearson SC, Wilkinson MJ. Rapid assessment of antibiotic effects on Escherichia coli by bis-(1,3- dibutylbarbituric acid) trimethine oxonol and flow cytometry. Antimicrob Agents Chemother. 1997;41:2001-2005
65. Fredricks BA, DeCoster DJ, Kim Y, Sparks N, Callister SM, Schell RF. Rapid pyrazinamide susceptibility testing of Mycobacterium tuberculosis by flow cytometry. J Microbiol Methods. 2006;67:266-272
66. Van Belkum A, Dunne WM. Next-generation antimicrobial susceptibility testing. J Clin Microbiol. 2013;51:2018-2024
67. von Ah U, Wirz D, Daniels AU. Isothermal micro calorimetry-a new method for MIC determinations: results for 12 antibiotics and reference strains of E. coli and S. aureus. BMC Microbiol. 2009;9:106
68. Bonkat G, Braissant O, Widmer AF, Frei R, Rieken M, Wyler S. et al. Rapid detection of urinary tract pathogens using microcalorimetry: Principle, technique and first results. BJU Int. 2012;110:892-897
69. Howell M, Wirz D, Daniels AU, Braissant O. Application of a microcalorimetric method for determining drug susceptibility in Mycobacterium species. J Clin Microbiol. 2012;50:16-20
70. Eigner U, Schmid A, Wild U, Bertsch D, Fahr A. Analysis of the Comparative Workflow and Performance Characteristics of the VITEK 2 and Phoenix Systems Analysis of the Comparative Workflow and Performance Characteristics of the VITEK 2 and Phoenix Systems. J Clin Microbiol. 2005;43:3829-3834
71. Horstkotte MA, Knobloch JKM, Rohde H. et al. Evaluation of the BD PHOENIX Automated Microbiology System for Detection of Methicillin Resistance in Coagulase-Negative Staphylococci. J Clin Microbiol. 2004;42:5041-5046
72. Price CS, Kon SE, Metzger S. Rapid antibiotic susceptibility phenotypic characterization of Staphylococcus aureus using automated microscopy of small numbers of cells. J Microbiol Methods. 2014;98:50-8
73. Douglas IS, Price CS, Overdier KH, Wolken RF, Metzger SW, Hance KR. et al. Rapid automated microscopy for microbiological surveillance of ventilator-associated pneumonia. Am J Respir Crit Care Med. 2015;191:566-573
74. Choi J, Yoo J, Lee M, Kim E-G, Lee JS, Lee S. et al. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci Transl Med. 2014;6:267ra174
75. Sin MLY, Mach KE, Wong PK, Liao JC. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev Mol Diagn. 2014;14:225-44
Author contact
![]() Corresponding author: njtaoedu
Corresponding author: njtaoedu
 Global reach, higher impact
Global reach, higher impact