13.3
Impact Factor
Theranostics 2017; 7(10):2634-2648. doi:10.7150/thno.17853 This issue Cite
Research Paper
Co-Activation of PKC-δ by CRIF1 Modulates Oxidative Stress in Bone Marrow Multipotent Mesenchymal Stromal Cells after Irradiation by Phosphorylating NRF2 Ser40
1. Department of Blood Transfusion, Radiation Biology Laboratory, The Second Affiliated Hospital, Third Military Medical University, Chongqing 400038, China
2. General Hospital of Xinjiang Military Area Command, Urumqi 830000, China
* Equal contribution to this work
Received 2016-10-8; Accepted 2017-4-19; Published 2017-6-25
Abstract
The high mortality associated with pancytopenia and multi-organ failure resulting from hematopoietic disorders of acute radiation syndrome (h-ARS) creates an urgent need for developing more effective treatment strategies. Here, we showed that bone marrow multipotent mesenchymal stromal cells (BMMSCs) effectively regulate oxidative stress following radiative injury, which might be on account of irradiation-induced elevation of protein levels of CR6-interacting factor 1(CRIF1) and nuclear factor E2-related factor 2(NRF2). Crif1-knockdown BMMSCs presented increased oxidative stress and apoptosis after irradiation, which were partially due to a suppressed antioxidant response mediated by decreased NRF2 nuclear translocation. Co-immunoprecipitation (Co-IP) experiments indicated that CRIF1 interacted with protein kinase C-δ (PKC-δ). NRF2 Ser40 phosphorylation was inhibited in Crif1-deficient BMMSCs even in the presence of three kinds of PKC agonists, suggesting that CRIF1 might co-activate PKC-δ to phosphorylate NRF2 Ser40. After radiative injury, the supporting effect of BMMSCs for the colony forming ability of HSCs in vitro was reduced, and the deficiency of CRIF1 aggravated such damage. Thus, CRIF1 plays an essential role in PKC-δ/NRF2 pathway modulation to alleviate oxidative stress in BMMSCs after irradiative injury, and at some level it may maintain the HSCs-supporting effect of BMMSCs after radiative injuries.
Keywords: h-ARS, BMMSCs, Oxidative Stress, Irradiation, CRIF1, NRF2, PKC-δ.
Introduction
Acute radiation syndrome (ARS) results in severe hematopoietic (1-9Gy), gastrointestinal (9-20Gy), and cerebrovascular syndromes (>20Gy) [1]. Patients with hematopoietic syndromes resulting from ARS (h-ARS) at low doses (1-3Gy) may recover with effective supportive treatment, including intravenous cytokines, antiemetics, analgesics, antibiotics, and blood transfusions [2]. However, after exposure to higher radiation levels (6-9Gy), a hematopoietic stem cells (HSCs) transplant may be required [3], because HSCs are extremely sensitive to radiation [4]. However, previous studies indicated that this treatment did not significantly improve survival [5]. Transplanted HSCs might effectively replenish HSC quantity, but these cells do not effectively exert the hematopoietic function in a radiation-destroyed hematopoietic microenvironment. Therefore, the destruction of the hematopoietic microenvironment in the bone marrow upon radiation might be the main cause for HSC transplantation failure [6].
Bone marrow multipotent mesenchymal stromal cells (BMMSCs) are a major component of the hematopoietic microenvironment, providing physical support and secreting a variety of cytokines for regulating HSC self-renewal, differentiation, and retention [7-9]. Several recent studies reported that BMMSCs were more resistant to radiation than HSCs [10-12], and their relative radiation resistance was an important contributing factor in their ability to rescue hematopoiesis after radiation damage [13]. The mechanisms of BMMSCs radioresistance involve efficient DNA damage recognition, double-strand break repair, and apoptosis evasion [14]. However, the molecular mechanisms underlying BMMSCs radiation tolerance have not been fully elucidated. Upon exposure to ionizing radiation, intracellular reactive oxygen species (ROS) attack cellular macromolecules, and DNA damage represents a critical cellular injury with many pathogenetic consequences [15]. Nuclear factor E2-related factor 2 (NRF2) is a major stress-response transcription factor that activates a variety of cytoprotective and antioxidant genes, playing a crucial role in defense against ROS-mediated cellular injury [16-18].Under normal conditions, NRF2 is constantly ubiquitinated by the Kelch-like erythroid cell-derived-associated protein 1 (KEAP1)-cullin 3 (Cul3) ubiquitin ligase E3 complex, resulting in proteasomal degradation in the cytoplasm [19]. External stresses induce the dissociation of NRF2 from KEAP1, resulting inNRF2 nuclear translocation and activation of its downstream target genes through binding to antioxidant response element (ARE) motifs [20]. Previous studies showed that oxidative stress-induced covalent modification of KEAP1 and PKC-mediated phosphorylation of serine 40 (Ser40) in NRF2 contributed to the dissociation of NRF2 from KEAP1 [21, 22]. Therefore, NRF2 may be important for BMMSCs radiation resistance because of its potent regulatory effect during oxidative stress. However, few studies clarified the relationship between NRF2 and BMMSCs radiation resistance.
CR6-interacting factor 1 (CRIF1) is a multifunctional protein that was first identified as a regulator of cell cycle and growth [23, 24]. This protein is also known to interact with some transcription factors [25-27] and is a constitutive protein of the large mitoribosomal subunit [28]. CRIF1 deficiency leads to various defects, including the deterioration of mitochondrial function, which results in high levels of mitochondrial ROS [29, 30]. Here, we showed that CRIF1 played an important role in modulating BMMSCs oxidative stress after irradiation, acting as an NRF2 regulator by facilitating the phosphorylation of NRF2 Ser40 through interacting with PKC-δ.
Materials and Methods
Cell culture and reagents
Primary human BMMSCs derived from bone marrow samples obtained from healthy donors (ages from 18-31) at the Department of Hematology at the Second Affiliated Hospital, Third Military Medical University, Chongqing, China. All donors gave their informed consent, and the study was approved by the Third Military Medical University Institutional Ethics Committee. The isolated cells were purified and characterized as previously described [31], and cultured in human mesenchymal stem cell growth medium (HUXMX-90011, Cyagen, USA) at 37°C in a humidified atmosphere containing 5% CO2. In this study, BMMSCs were defined using criteria proposed by the International Society for Cellular Therapy (ISCT). Expression of CD73, CD90, and CD105, and lack of CD14, CD34, and CD45 were assessed using flow cytometry. Adipogenic, osteogenic, and cartilage differentiation of BMMSCs were also verified (Figure S1) as previously described [31]. In all in vitro experiments, cells were utilized from passage 3 to passage 5.
Bone marrow mononuclear cells (BMMNCs) for colony forming assay were collected from bone marrow samples after informed consent of a healthy donor (18 years old). Mononuclear cells (MNCs) were isolated by density-gradient centrifugation with Percoll Plus (GE Healthcare, USA) and treated with ammonium chloride for red blood cells lysing.
Phorbol 12-myristate 13-acetate (PMA), puromycin and tert-butylhydroquinone (t-BHQ) were purchased from Sigma. Prostratin and oleic acid were purchased from Santa Cruz. For the experiments, cells were treated with 40 nmol/L PMA, 20 μmol/L oleic acid, 1 μmol/L prostratin or 100 mmol/L t-BHQ.
Mice
Nrf2+/+ and Nrf2-/- mice from the C57BL/6 parental strain were kindly provided by Prof. HT Zheng (Third Military Medical University) [32]. The animal experimental protocols were approved by the Institutional Animal Care and Use Committee of the Third Military Medical University.
Irradiation of cells and mice
When cultured cells reached 80% confluence, they were irradiated with 9 Gy by Co-60 at a dose rate of 700 cGy/min. Nrf2+/+ and Nrf2-/- mice at 8 weeks of age were exposed to total body irradiation (TBI) at 5 Gy according to previous studies [33, 34]. Control groups of cells and mice were placed in the same place but not exposed to irradiation.
Transfection of BMMSCs with Crif1 shRNA lentiviral vector, Crif1 overexpression lentiviral vector, and Nrf2 overexpression plasmid vector
A Crif1 shRNA (or overexpression) lentiviral vector was constructed by inserting the Crif1 interference sequence (or overexpression sequence) into a pGreenPuro shRNA vector and pSIH1-H1-Puro shRNA vector (SystemBiosciences, USA) (See Table S1 for Crif1 interference sequences; Table S2 for sequences of amplification primers used for Crif1 overexpression). Lentiviral vectors were generated by transfecting the 293FT packaging cell line with the Crif1 shRNA (or overexpression) vectors (pGreenPuro-shCrif1 and pSIH1-H1-Puro-shCrif1, or pGreenPuro-oeCrif1). A group of control cells was transfected with empty vectors (pLentis-CMV-SFFV-GTP and pLentis-CMV-HI-Puro) as a control. Lentiviral vectors were added into BMMSCs culture medium. After 48 h, infected cells were selected using 3 μg/mL puromycin and were maintained in 1.5 μg/mL puromycin (Sigma). BMMSCs containing Crif1 shRNA vectors (Crif1-SI), Crif1 overexpression vectors (Crif1-OE) and empty vectors control (Con-EV) were used for experiments such as irradiation treatment.
The recombinant expression (or nonspecific control) plasmid vector pcDNA3.1-3xFlag harbored a full-length Nrf2 cDNA (GenBANK No: NM_006164) (or nonspecific sequence) were constructed by YouBio biotechnology company (Changsha, China). Then 1 × 104 BMMSCs were transfected with 5μg vectors using Lipofectamine® 3000 reagent (Thermo Fisher Scientific, USA) followed the manufacturer's instructions.
Western blot analysis
Cells were washed once in PBS and lysed in RIPA lysis buffer (P0013B; Beyotime, Shanghai, China) at 4°C. Proteins were denatured by boiling. Protein concentrations were determined using the Enhanced BCA Protein Assay kit (P0010S; Beyotime). Protein samples were separated in a 12% SDS-polyacrylamide gel and transferred to PVDF membranes (Immobilon-P membranes, Millipore, USA). Membranes were blocked with Tris-buffered saline/Tween 20 (TBST, 0.1% Tween 20) containing 5% bovine serum albumin (BSA) for 1 h and then incubated with the appropriate primary antibody overnight at 4°C. After three 10-min washes with TBST, membranes were incubated for 1 h at 24°C with the appropriate horseradish peroxidase-conjugated secondary antibody. After extensive washing, immunoreactive bands were detected by the BeyoECL Plus reagent (P0018; Beyotime) using a Photo-Image System (Molecular Dynamics, Sunnyvale, CA, USA). Immunoblotting was performed with the following primary antibodies: phosphorylated NRF2 Ser40 (pNRF2 Ser40) (ab76026; Abcam), NRF2 (ab62352; Abcam), GCLC (ab53179; Abcam), GGT1 (ab175384; Abcam), NQO1 (ab34173; Abcam), HO1 (ab13243; Abcam), Retinoblastoma (RB) (ab24; Abcam), p53 (ab1101; Abcam), p21 (ab109520; Abcam), p16 (ab51243; Abcam), CRIF1 (sc-134882; Santa Cruz), PKC-δ (sc-213-G; Santa Cruz), and β-actin (sc-8432; Santa Cruz).
RNA isolation, cDNA synthesis, and gene expression detection
Total RNA was harvested from BMMSCs using TRIzol reagent (15596-026; Invitrogen, USA) according to the manufacturer's protocol. The RNA was used to synthesize complementary DNA (cDNA) using the PrimeScript RT Reagent kit (RR047A; TaKaRa, Japan). Real-time quantitative PCR (qPCR) was used to analyze the relative expression of specified mRNAs in selected samples. Triplicate qPCR was performed by Real-Time PCR Systems (StepOnePlus; ABI, USA) in 20-μL reactions containing FastStart Universal SYBR Green Master Mix (04913850001; Roche, USA) and 0.3 pM primers (See Table S3 for sequences of primers). Quantitation of gene expression relative to β-actin was determined using the 2-ΔΔCT method [35].
Immunocytochemistry
After treatment, cells were washed twice in PBS and fixed in 4% formaldehyde for 20 min at 24°C. Cells were washed again in PBS, permeabilized for 10 min in 0.2% Triton X-100, and incubated in blocking solution containing 5% BSA in PBS. Cells were incubated with anti-CRIF1 (1:200) and anti-NRF2 (1:200) overnight at 4°C. Cells were washed three times for 10 min each in PBS and incubated with secondary antibodies conjugated with Alexa Fluor 647 and Cy3 (Beyotime) for 1 h at 24°C. Cells were incubated with DAPI for nuclear staining. Fluorescence images were obtained using laser confocal microscopy (Leica SP5, Germany).
Immunoprecipitation and co-immunoprecipitation
The lysis buffer was used for both immunoprecipitation (IP) and co-immunoprecipitation (Co-IP) experiments. Using IP for NRF2 ubiquitination assay, total cellular proteins from BMMSCs were extracted and incubated with 1 μg of NRF2 primary antibody (ab62352; Abcam) at 4°C on a rocker overnight. Twenty microliters of resuspended Protein G PLUS Agarose (sc-2002; Santa Cruz) was added to the samples, followed by incubation at 4°C for 2 h. Immunoprecipitates were collected by centrifugation at 1,000×g for 5 min at 4°C, and the sediments were washed three times in lysis buffer, resuspended in 40 μL of 1× electrophoresis sample buffer, and boiled for 5 min. The eluted proteins were analyzed by standard western blot procedures with an anti-ubiquitin antibody (sc-8017; Santa Cruz). To analyze the interaction between CRIF1 and PKC-δ using Co-IP, total cellular proteins were extracted and processed in the same way as IP experiments. Normal rabbit/goat IgG (negative control), anti-CRIF1 (sc-134882; Santa Cruz), and anti-PKC-δ (sc-213-G; Santa Cruz) were used for immunoprecipitation. The eluted proteins were analyzed by western blot with antibodies against PKC-δ and CRIF1, respectively.
Cell apoptosis
Cells (1 × 106) were collected and stained with the Annexin-V-FLUOS Staining kit for apoptosis analysis by flow cytometry according to the manufacturer's instructions (11988549001; Roche). Flow cytometry was performed on a FACSAria cell sorter (BD Biosciences, USA), and the results were analyzed using FACSDiva software (version 6.1.3; BD Biosciences). For analysis of cell caspase-3 activity, a Caspase-3 Activity Assay kit (C1115; Beyotime) was used according to the manufacturer's instructions.
Senescence-associated β-galactosidase (SA-β-Gal) staining
For SA-β-Gal staining, 1 × 104 cells were seeded in 6-well plates for 24 h followed by 9 Gy irradiation. At 72 h, the irradiated cells were washed once in PBS and subjected to SA-β-Gal staining with a Cell Senescence SA-β-Gal Staining kit (C0602; Beyotime). The percentage of senescent cells was calculated by the number of stained cells (blue) out of at least 500 cells in different microscope fields.
Measurement of intracellular ROS
The levels of intracellular ROS were quantified using the DCFH-DA (S0033; Beyotime) and dihydroethidium (DHE) probes (S0063; Beyotime). Cells were washed three times in PBS. DCFH-DA or DHE probes diluted to a final concentration of 10 mM were added and incubated for 30 min at 37ºC in the dark. After being washed three times in PBS, the relative levels of fluorescence were quantified by flow cytometry with an appropriate detection spectrum (535 nm emission for DCFH-DA and 610 nm emission for DHE).
Determination of glutathione (GSH)
After treatment, the harvested cell pellets were frozen, thawed three times, and centrifuged at 12,000 ×g for 5 min at 4°C. Supernatants were collected for use. The clear supernatant was used to evaluate GSH, which was determined using a GSH Assay kit (S0053; Beyotime).
Indirect fluorescence-activated cell sorting (FACS) analysis in quadri-color-labeled mouse BMMSCs
Cells isolated from the bone marrow of mice with or without TBI were incubated for 20 min at 4°C with antibodies conjugated to phycoerythrin (PE), allophycocyanin (APC), peridinin chlorophyll protein (PerCP), and APC-Cy7 against mouse Sca-1, CD29, CD45, and CD11b (Sigma). Acquisitions were performed on a flow cytometer for quantitative analysis of BMMSCs with Sca-1(+), CD29(+), CD45(-), and CD11b(-). For ROS analysis, quadri-color-labeled cells were incubated with DCFH-DA probes for 30 min at 4°C, while for indirect FACS analysis of CRIF1, quadri-color-labeled cells were incubated with a CRIF1 anti-rabbit antibody for 1 h at 24°C and mixed with the appropriate FITC-labeled goat anti-rabbit IgG (H+L) for 30 min at 24°C. Cells were washed twice in PBS, and acquisitions were performed on a flow cytometer.
Colony-forming unit (CFU) assay
The CFU assay was performed to assess the colony-forming ability of the BMMNCs co-cultured with irradiated BMMSCs. Four hours after 9 Gy irradiation, BMMSCs were co-cultured with BMMNCs for 48h. BMMNCs were re-suspended in Iscove's modified Dulbecco medium (IMDM) with 2% fetal bovine serum (FBS) at a concentration of 2×105/mL, and 0.3 mL of the cell suspension was added to 3 mL methylcellulose medium (MethoCult GF+ H4435; StemCell Technologies, Canada). The mixture of 1.1 mL was planted in triplicate into 35-mm tissue culture dishes. The dishes were incubated at 37ºC in a humidified atmosphere containing 5% CO2. After 14 days of culture, colonies belonging to burst-forming unit-erythroid (BFU-E), colony-forming unit-granulocyte, macrophage (CFU-GM), and colony-forming unit-granulocyte, erythrocyte, macrophage, megakaryocyte (CFU-GEMM) consisting of 40 or more cells were scored by trained personnel under an inverted phase-contrast microscope. The results were reported as the mean colony count per 104 nucleated cells.
Statistical analysis
Data are presented as the means ± standard error of mean (SEM). Statistical significance was evaluated using a Student's t-test for comparison between two data sets and one-way ANOVA followed by Tukey's honestly significant difference (HSD) test for analysis of three or more sets of data in SPSS19.0.
Results
Radiation increased oxidative stress in BMMSCs
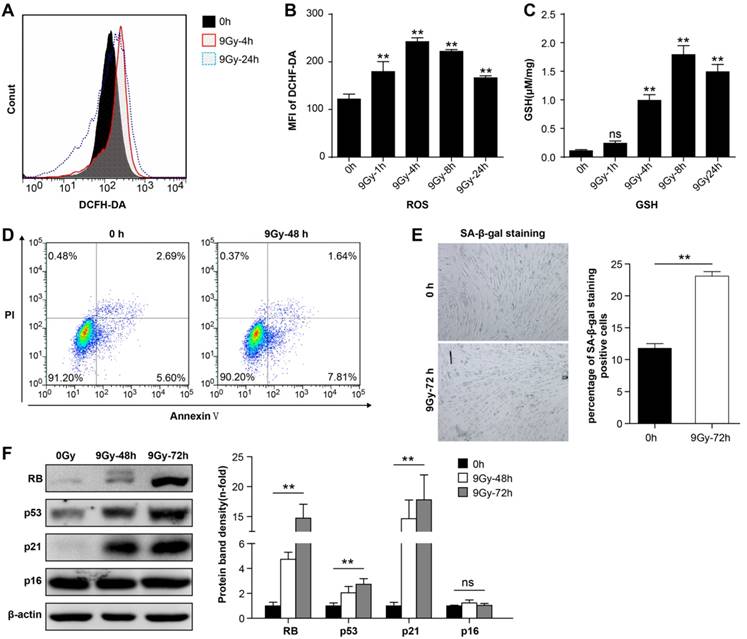
BMMSCs were irradiated with a single dose of 9 Gy [31], and flow cytometry analysis showed that intracellular ROS levels increased immediately after irradiation (Figure 1A and 1B). ROS levels reached the peak within 4 h after irradiation, and 24h later levels recovered to a certain degree but remained elevated compared to the normal state. Meanwhile, 9 Gy of irradiation remarkably elevated GSH level in BMMSCs (Figure 1C). These results suggested that oxidative stress in BMMSCs significantly increased at the early phase after high-dose irradiation. To determine the detrimental effect of oxidative stress in BMMSCs, cellular apoptosis and senescence were analyzed before and after irradiation. The apoptotic cell rate increased from 8.29% to 9.45% after irradiation as revealed by Annexin V and PI staining (Figure 1D), suggesting that BMMSCs were relatively radioresistant. However, SA-β-Gal staining showed that 9 Gy of irradiation significantly increased BMMSCs senescence (Figure 1E). Several proteins related to cell senescence were detected by western blot after irradiation at 48 and 72 h (Figure 1F). The results showed that protein levels of RB, p53, and p21 significantly increased, confirming that cell senescence was induced by irradiation, while no remarkable change of p16 was observed.
Radiation-induced oxidative stress in BMMSCs (a) Fluorescence-activated cell sorting (FACS) results and (b) quantitation of ROS levels in BMMSCs at the indicated time points after irradiation (9 Gy) as determined using DC H-DA probes. (c) The GSH content of BMMSCs was determined at the indicated time points in irradiated BMMSCs. (d) BMMSCs apoptosis 48 h after irradiation was examined using an Annexin V assay. (e) BMMSCs senescence 72 h after irradiation was examined using SA-β-Gal staining (left panel) and quantification (right panel). (f) Proteins (RB, p53, p21, and p16) related to cell senescence were detected by western blot after irradiation at 48 and 72 h. The representative immunoblots (left panel) and relative levels of proteins normalized to β-actin (right panel) are shown. All experiments were performed in triplicate in the same cell line, and representative images are shown. Data represent the mean ± SEM, *p< 0.05, **p< 0.01, ns (not significant), compared to the control (or 0 h group)
Irradiation not only elevated protein levels of NRF2 and CRIF1 but also induced their nuclear accumulation
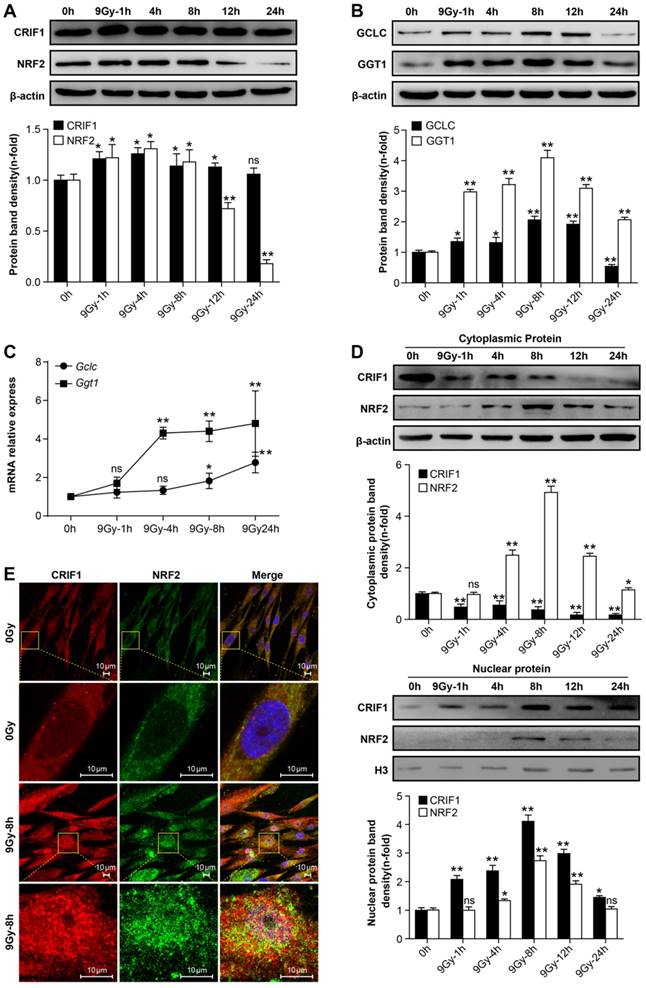
Western blot was used to quantify NRF2 and CRIF1 protein levels in BMMSCs. The results showed that NRF2 and CRIF1 protein levels increased significantly at early phases following irradiation, especially 4-8 h after irradiation. NRF2 returned to baseline levels at 12 h; however, there was no obvious decrease of CRIF1 up to 24 h after radiation (Figure 2A). We also observed that NRF2 downstream target proteins and mRNAs such as Gclc and Ggt1 showed similar expression patterns as NRF2 in BMMSCs after irradiation (Figure 2B and 2C). Nuclear translocation of NRF2 is crucial for activating its downstream target genes [36]. To assess the effect of irradiation on nuclear translocation of NRF2 and CRIF1, the cytoplasm and nuclear fraction proteins were extracted and detected by western blot (Figure 2D), and subcellular localization was observed using immunocytochemistry (Figure 2E). The western blot results and fluorescence images showed that the nuclear translocation of NRF2 and CRIF1 markedly increased upon irradiation, and these two proteins were co-localized in BMMSCs nuclei after irradiation. These data indicated that CRIF1 was likely involved in NRF2 regulation because of its protein level and nuclear translocation in BMMSCs after irradiation.
Knockdown of Crif1 exacerbated BMMSCs oxidative stress after irradiation
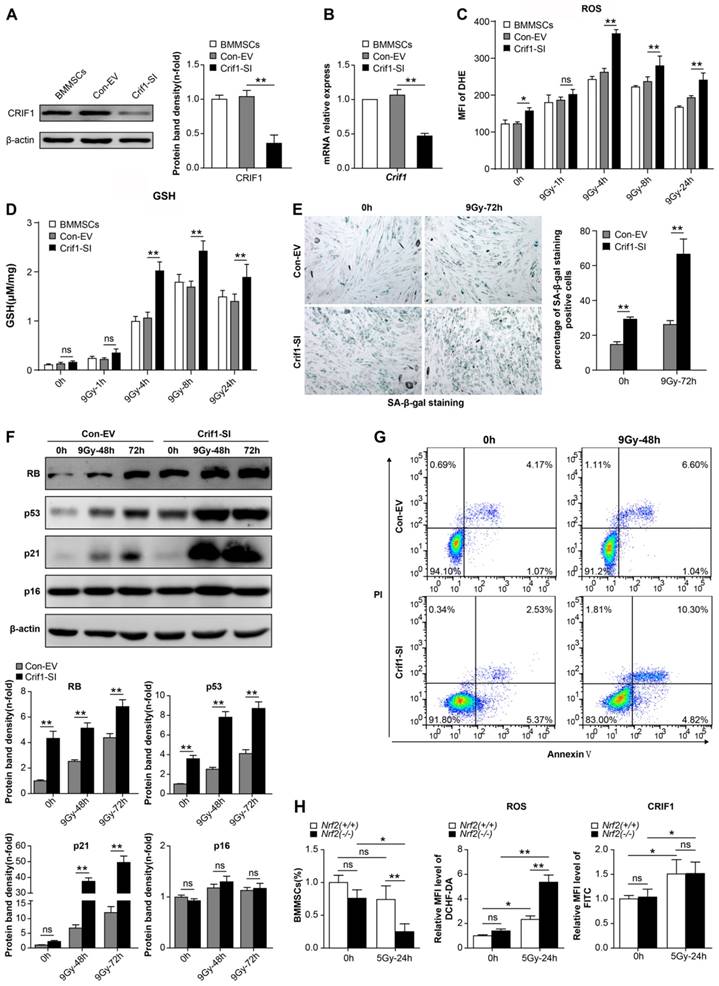
To further confirm whether CRIF1 played an important role in the anti-oxidative response after radiative injury in BMMSCs, we knocked down Crif1 expression with lentiviral particles harboring Crif1 shRNA, given the high level of CRIF1 protein expression in BMMSCs in physiological and pathological conditions. Both protein and mRNA levels of CRIF1 were effectively inhibited by shRNA vector transfection (Figure 3A and 3B). Crif1-knockdown BMMSCs presented higher intracellular ROS levels after irradiation than those of the control cells at all time points, especially at 4h, and even 24h after irradiation the ROS levels were still notably higher than the resting state levels (Figure 3C). Additionally, GSH content was higher after irradiation in Crif1-deficient BMMSCs than that in the control cells (Figure 3D). Furthermore, the proportion of senescent cells (Figure 3E) significantly increased in Crif1-deficient BMMSCs after irradiation compared to the control cells. The levels of proteins (RB, p53, and p21) involved in cell senescence significantly increased in Crif1-deficient BMMSCs compared to the control cells after irradiation, unlike p16 (Figure 3F). Moreover, the apoptotic analysis using Annexin V and PI staining (Figure 3G) and caspase-3 activity (Figure S2) indicated that the apoptosis significantly increased in Crif1-deficient BMMSCs after irradiation compared to the control cells. To further explore the regulation effect of CRIF1 on NRF2, we used an Nrf2-/- mice model to evaluate the relationship between NRF2 and CRIF1 in BMMSCs after 5 Gy TBI. The results agreed with our hypothesis that quadri-color-labeled BMMSCs decreased in Nrf2-/- mice compared to the control mice, while ROS levels in these cells were significantly elevated in Nrf2-knockout mice. However, the results of indirect fluorescent staining showed that CRIF1 expression increased in BMMSCs after 5 Gy TBI whether mice were deficient in NRF2 or not (Figure 3H). When activating NRF2 through tert-butylhydroquinone (t-BHQ) or Nrf2 overexpression vectors in BMMSCs before irradiation, western blot results showed that the protein level of NRF2 increased, and the cell senescence induced by irradiation was in remission. In Crif1-deficient BMMSCs, the same treatment did not significantly increase NRF2 protein levels and not reduce the level of cell senescence (Figure S3). These results demonstrated that Crif1-knockdown aggravated oxidative stress in BMMSCs after irradiation and influenced stem cell death and senescence, probably associated to the NRF2 pathway.
Knockdown of Crif1 resulted in reduced NRF2 protein level and nuclear translocation in BMMSCs after radiative injury
To further elucidate the relationship between CRIF1 and NRF2 protein expression, we analyzed the effect of Crif1-knockdown on NRF2 expression in BMMSCs. Western blot results showed that NRF2 protein level decreased in Crif1-deficient BMMSCs independently of the irradiation (Figure 4A). However, qPCR results indicated that the relative mRNA expression of Nrf2 did not significantly change in Crif1-deficient BMMSCs (Figure 4B), suggesting that Crif1-knockdown affected NRF2 at the protein level. After irradiation, NRF2 protein levels in Crif1-deficient BMMSCs significant decreased than NRF2 levels in control cells at peak time points (8 h and 12 h). Moreover, the peak time point of NRF2 expression appeared at 4 h instead at 8 h in the control cells (Figure 4C). The cytoplasmic and nuclear protein levels determined by western blot and the subcellular localization observed using immunocytochemistry consistently indicated that NRF2 nuclear translocation was markedly reduced in Crif1-deficient BMMSCs after irradiation compared to nuclear translocation in the control cells (Figure 4D and 4E). Furthermore, the increased levels of NRF2-dependent proteins (HO1, GGT1, and GCLC) expression in Crif1-deficient BMMSCs decreased after irradiation, and the levels of NRF2 target genes such as Ho1, Ggt1, and Gclc also decreased (Figure 4F and 4G).
Protein levels and nuclear translocation of NRF2 and CRIF1 after irradiation (a) CRIF1 and NRF2 protein expression was assessed by western blot in BMMSCs at the indicated time points after an irradiation dose of 9 Gy. The representative immunoblots (upper panel) and relative levels of proteins normalized to β-actin (lower panel) are shown. (b) Western blot results of target proteins of NRF2 (GCLC and GGT1) in BMMSCs at the indicated time points after irradiation. The representative immunoblots (upper panel) and relative levels of proteins normalized to β-actin (lower panel) are shown. (c) qPCR results of Gclc and Ggt1 in BMMSCs at the indicated time points after irradiation. (d) The cytoplasmic and nuclear proteins were detected at the indicated time points in irradiated BMMSCs. The representative immunoblots (upper panel) and relative levels of cytoplasmic (or nuclear) proteins normalized to β-actin (or histone H3) (lower panel) are shown. H3 was used as nuclear protein-loading control. (e) Immunofluorescence analysis of CRIF1 and NRF2 8 h after irradiation. All experiments were performed in triplicate in the same cell line, and representative images are shown. Data represent the mean ± SEM. *p< 0.05, **p< 0.01, ns (not significant), compared to the control
Effect of Crif1-knockdown on BMMSC oxidative stress after irradiation (a) Western blot results of CRIF1 in Crif1-deficient BMMSCs (Crif1-SI) and control cells (Con-EV). The representative immunoblots (left panel) and relative levels of proteins normalized to β-actin (right panel) are shown. BMMSCs transfected with Crif1 shRNA are referred as Crif1-SI, and BMMSCs transfected with empty control vectors are referred as Con-EV. (b) qPCR analysis of Crif1 in Crif1-deficient BMMSCs (Crif1-SI) and control cells (Con-EV). (c) ROS levels were quantified in Crif1-deficient BMMSCs (Crif1-SI) and control cells (Con-EV) at the indicated time points after an irradiation dose of 9 Gy. (d) The GSH content of Crif1-deficient BMMSCs (Crif1-SI) and control cells (Con-EV) was determined at the indicated time points after irradiation. (e) Senescence was examined in Crif1-deficient BMMSCs (Crif1-SI) and control cells (Con-EV) 72 h after irradiation using SA-β-Gal staining (left panel) and quantification (right panel). (f) Proteins (RB, p53, p21, and p16) related to cell senescence were detected by western blot in Crif1-deficient BMMSCs (Crif1-SI) and control cells (Con-EV) after irradiation at 48 and 72 h. The representative immunoblots (upper panel) and relative levels of proteins normalized to β-actin (lower panel) are shown. (g) Apoptosis of Crif1-deficient BMMSCs (Crif1-SI) and control cells (Con-EV) at 48 h after irradiation was examined. BMMSCs were transfected with GFP-free lentiviral Crif1 shRNA and empty control vectors.(h) FACS analysis of the BMMSCs amount, ROS levels, and CRIF1 protein expression levels of Sca-1(+), CD29(+), CD45(-), and CD11b(-) labeled cells from bone marrow cells in Nrf2-/- and Nrf2+/+ mice (n=3) before 5 Gy total body irradiation (TBI) and 24 h after irradiation. All experiments were performed in triplicate in the same cell line, and representative images are shown. Data represent the mean ± SEM. *p< 0.05, **p< 0.01, ns (not significant), compared to the control
Effect of Crif1-knockdown on NRF2 protein level and nuclear translocation in BMMSCs after irradiation. (a) Western blot results of NRF2 in Crif1-deficient BMMSCs (Crif1-SI) and control cells (Con-EV). The representative immunoblots (upper panel) and relative levels of proteins normalized to β-actin (lower panel) are shown. BMMSCs transfected with Crif1 shRNA are referred as Crif1-SI, and BMMSCs transfected with empty control vectors are referred as Con-EV. (b) qPCR analysis of Nrf2 in Crif1-deficient BMMSCs (Crif1-SI) and control cells (Con-EV). (c) Western blot analysis of NRF2 expression in irradiated Crif1-knockdown (Crif1-SI) and control BMMSCs (Con-EV) at the indicated time points after an irradiation dose of 9 Gy. The representative immunoblots (upper panel) and relative levels of proteins normalized to β-actin (lower panel) are shown. (d) Cytoplasmic and nuclear fraction proteins CRIF1 and NRF2 in Crif1-deficient BMMSCs (Crif1-SI) and control cells (Con-EV) 8 h after irradiation were detected by western blot. The representative immunoblots (upper panel) and relative levels of proteins normalized to β-actin (lower panel) are shown. (e) Immunofluorescence analysis of CRIF1 and NRF2 in Crif1-deficient BMMSCs (Crif1-SI) and control cells (Con-EV) 8h after irradiation. (f) Western blot of target proteins of NRF2 in Crif1-deficent BMMSCs (Crif1-SI) and control cells (Con-EV) at the indicated time points after an irradiation dose of 9 Gy. The representative immunoblots (upper panel) and relative levels of proteins normalized to β-actin (lower panel) are shown. (g) qPCR results of NRF2 target genes (Ho1, Ggt1, and Gclc) in Crif1-deficent BMMSCs (Crif1-SI) and control cells (Con-EV) at the indicated time points after an irradiation dose of 9 Gy. All experiments were performed in triplicate in the same cell line, and representative images are shown. Data represent the mean ± standard error of mean (SEM). *p< 0.05, **p< 0.01, ns (not significant), compared to the control
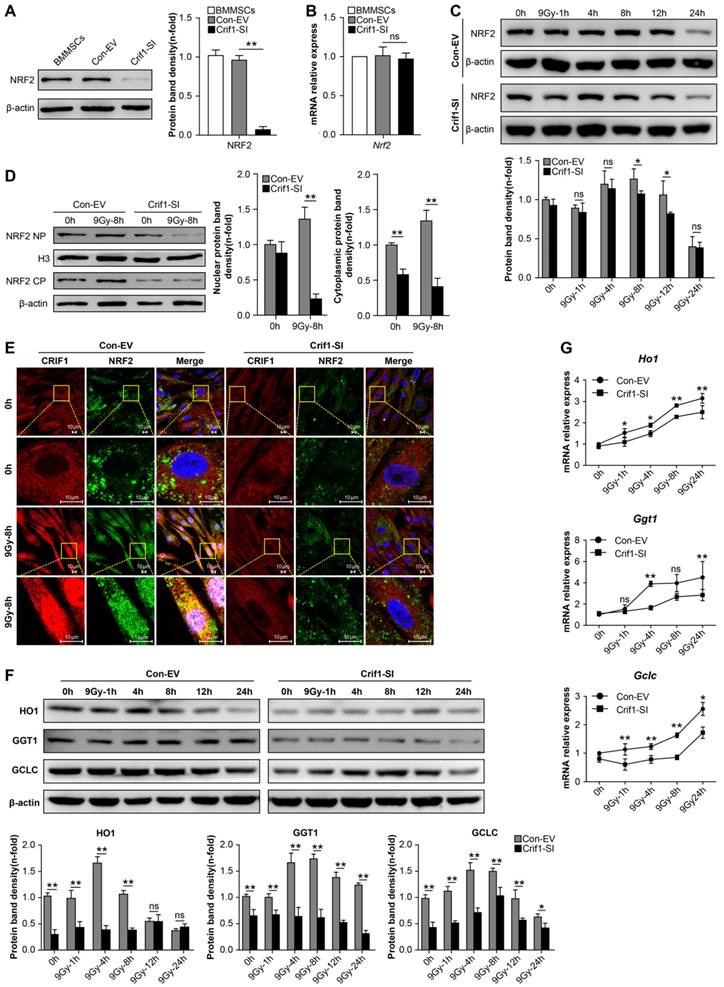
Interaction between CRIF1and PKC-δ contributed to NRF2 Ser40 phosphorylation and NRF2 stability
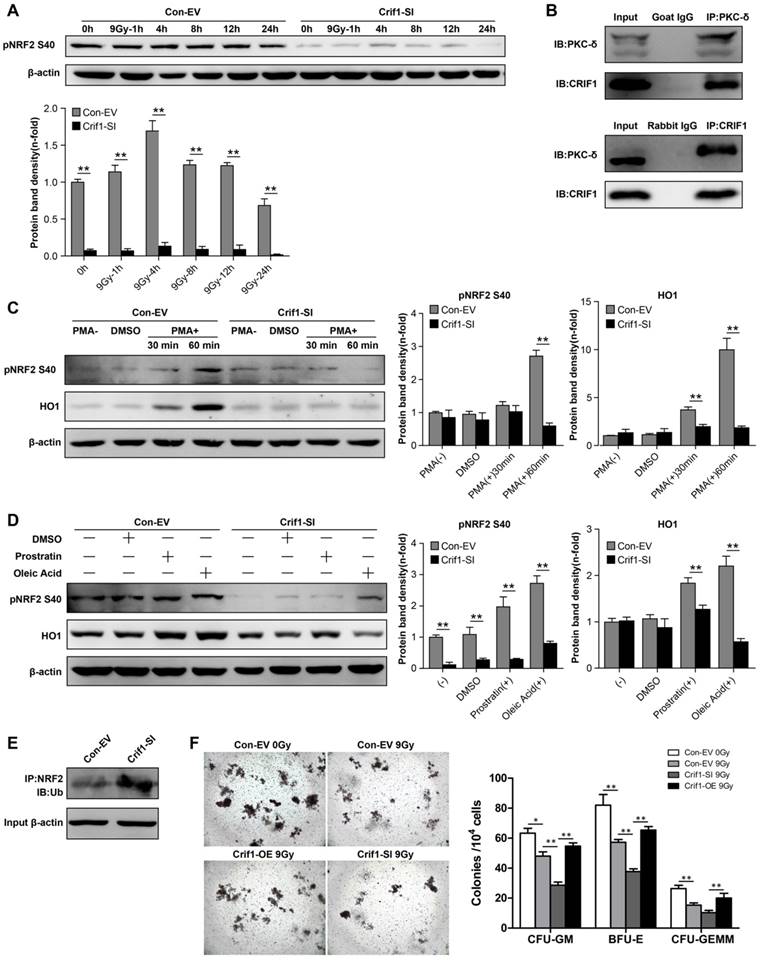
The phosphorylation of NRF2 Ser40 by PKC-δ is a critical mechanism for releasing NRF2 from KEAP1 and translocating it into the nucleus [37, 38]. To determine the mechanism underlying CRIF1-mediated regulation of protein expression level and subcellular localization of NRF2, we determined the level of NRF2 Ser40 phosphorylation after irradiation. As expected, NRF2 Ser40 phosphorylation was significantly reduced in Crif1-deficient BMMSCs compared to the control cells without irradiation. Similarly, NRF2 Ser40 phosphorylation in Crif1-deficient BMMSCs significantly decreased compared to the phosphorylation in the control cells at all time points after irradiation (Figure 5A). Co-IP experiments demonstrated that PKC-δ and CRIF1 interacted with each other (Figure 5B). To verify the role of CRIF1 in PKC-δ activity, three PKC agonists were utilized. PMA was used as a potent agonist, and prostratin and oleic acidwere used as moderately efficient agonists. After agonist exposure, we found that NRF2 Ser40 phosphorylation significantly increased in the control cells, while the phosphorylation was suppressed in Crif1-deficient BMMSCs. The expression of HO1 followed the same pattern (Figure 5C and 5D). Considering that NRF2 is degraded by the proteasome, we analyzed the ubiquitination level of NRF2 by immunoprecipitation, and the results showed a higher level of NRF2 ubiquitination in Crif1-deficient BMMSCs, indicating that NRF2 was less stable upon Crif1-knockdown than it was in the control cells (Figure 5E).
The CFU assay using the BMMNCs, which represented the HSCs in vitro, was carried out to verify the effect of CRIF1 in the HSCs-supporting function of irradiated BMMSCs. The results suggested that the BMMNCs co-cultured with irradiated BMMSCs formed less colony units than the control BMMSCs without irradiation, and the BMMNCs co-cultured with irradiated Crif1-deficient BMMSCs presented less clone-forming rate than the irradiated control BMMSCs. Furthermore, when co-cultured with irradiated Crif1-overexpress BMMSCs, the BMMNCs ability of clone formation was restored (Figure 5F). We may deduce that CRIF1 deficiency in BMMSCs may result in a reduced HSCs-supporting function of BMMSCs after irradiation.
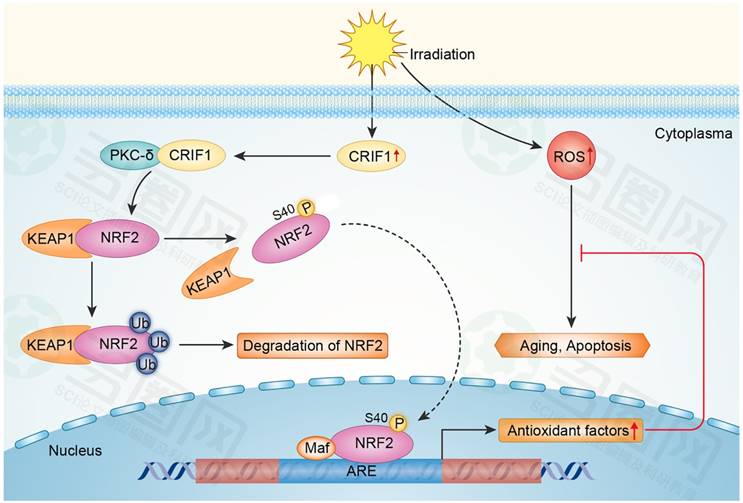
Our results suggested that CRIF1 likely co-activated PKC-δ and regulated oxidative stress of BMMSCs after irradiation mainly through the PKC-δ/NRF2 pathway (Figure 6).
Discussion
ROS generated by irradiation damage both HSCs and MSCs in bone marrow, but the radiation sensitivity of these two kinds of cells is not the same. Previous studies showed the radioresistance of BMMSCs relative to HSCs, and the association of the irradiation with reduced BMMSCs function [38, 39]. The radioresistant phenotype of BMMSCs may be potentially useful for treating radiation-induced tissue lesions, especially h-ARS. In animal experiments, MSC infusion successfully reconstituted hematopoiesis destroyed by radiation [40]. The potent ROS-scavenging ability of BMMSCs upon irradiation is a key mechanism for their radioresistance [41].Our results indicated that ROS levels peaked 4 h after irradiation but returned to baseline levels at 24 h. Meanwhile, BMMSCs seemed to prefer senescence rather than apoptosis after irradiation. This phenotype of BMMSCs is similar to previously described phenotypes that at high (20 Gy) [42] or low (40 mGy) [43] radiation doses preferentially entered senescence rather than apoptosis. The accumulation and scavenging of ROS in BMMSCs after irradiation may be associated with cell senescence. However, the molecular mechanism underlying this ROS-scavenging ability is not fully understood. A recent study revealed that CRIF1 deficiency induced oxidative stress in human umbilical vein endothelial cells (HUVECs) [44]. Here, we presented several results in support of the role of CRIF1 in modulating oxidative stress in BMMSCs after irradiation. CRIF1 deficiency in BMMSCs resulted in increased intracellular ROS levels after irradiation, and these Crif1-deficient cells showed accelerated cellular apoptosis and senescence compared to wild-type cells. In addition, our results showed that p16 levels did not significantly change after irradiation, regardless of CRIF1 status in BMMSCs. This evidence agrees with the results of a previous report that indicated that accelerated senescence triggered by irradiation was accompanied by the accumulation of p21 but did not correlate with p16 in BMMSCs [45]. Furthermore, some studies showed that p21 levels significantly increased within 24 h after irradiation in BMMSCs, and within 3-6 days they began to decline, whereas p16 level increased when p21 level was decreasing. Since we observed the p16 expression within 48 and 72 h after irradiation, we did not see any difference in p16 levels [42, 46, 47].
Contribution of the interaction between CRIF1 and PKC-δ to NRF2 Ser40 phosphorylation and NRF2 stability. (a) Western blot results of NRF2 Ser40 phosphorylation levels in Crif1-knockdown (Crif1-SI) and control BMMSCs (Con-EV) at the indicated time points after an irradiation dose of 9 Gy. The representative immunoblots (upper panel) and relative levels of proteins normalized to β-actin (lower panel) are shown. BMMSCs transfected with Crif1 shRNA are referred as Crif1-SI, and BMMSCs transfected with empty control vectors are referred as Con-EV. (b) Co-immunoprecipitation (Co-IP) analysis to determine PKC-δ and CRIF1 interaction in BMMSCs. (c) Control (Con-EV) and Crif1-knockdown (Crif1-SI) BMMSCs were treated with PMA or (d) prostratin and oleic acid separately at the indicated time points. NRF2 Ser40 phosphorylation levels and HO1 protein levels were examined by western blot. The representative immunoblots (upper panel) and relative levels of proteins normalized to β-actin (lower panel) are shown. (e) Analysis of NRF2 ubiquitination by IP in Crif1-knockdown (Crif1-SI) and control BMMSCs (Con-EV). β-actin levels were used as a loading control (lower panel). (f) Colony-forming unit (CFU) assay of bone marrow mononuclear cells (BMMNCs) co-cultured with BMMSCs. The representative morphology of colonies (×25 magnification) (left panel) and the amount of colonies per 104 BMMNCs (right panel) are shown. BMMSCs transfected with Crif1 overexpression vectors are referred as Crif1-OE. IB: immunoblot; IP: immunoprecipitation. All experiments were performed in triplicate in the same cell line, and representative images are shown. Data represent the mean ± standard error of mean (SEM). *p< 0.05, **p< 0.01, ns (not significant), compared to the control
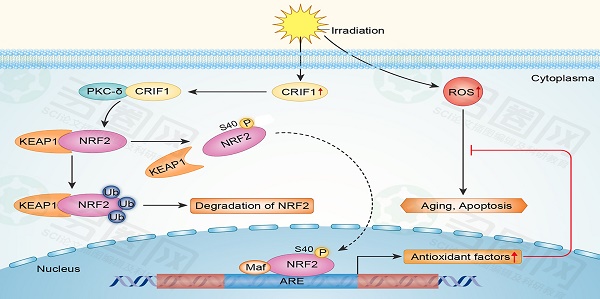
Proposed mechanism for CRIF1 modulation of the PKC-δ / NRF2 signaling pathway to facilitate redox homeostasis in BMMSCs after irradiation. CRIF1 co-activates KC-δ to phosphorylate NRF2 at Ser40 and promotes the dissociation of NRF2 from KEAP1. More stable NRF2 translocates into nucleus to activate its downstream target genes through binding to antioxidant response element (ARE) motifs, resulting in alleviation of oxidative stress in BMMSCs induced by radiative injury.
Mechanistically, our results suggested that CRIF1 positively regulated the NRF2/ARE antioxidant pathway in BMMSCs. NRF2 is one of the most crucial transcription factors for cytoprotection. In vitro and in vivo studies verified that NRF2-mediated transcription protected cells and tissues from the pathogenic effects of irradiation [18, 48, 49]. Consistent with the above-mentioned results, our study showed that populations of BMMSCs in Nrf2-/- mice decreased compared to cells from control mice 24 h after treatment with 5 Gy TBI, while ROS levels were higher in BMMSCs of Nrf2-/- mice compared to the control mice. Therefore, NRF2 had a protective effect on BMMSCs upon radiation injury, and this protein may be involved in the pathogenesis of myelosuppression induced by radiative injury. Our in vivo and in vitro results confirmed that CRIF1 regulation oxidative stress in BMMSCs after irradiation occurred mainly through activating the NRF2 pathway. Besides, we noticed that NRF2 protein level increased at early phases following irradiation, however, it began to decline from 12 h and went below to baseline level at 24 h. This might since that NRF2 would be degradation by ubiquitination in the cytoplasm when finishing its task of transcriptional activation in the post-induction phase of oxidative stress [50, 51].
Increased NRF2 stability and its nuclear translocation were essential for the transcriptional activation of downstream antioxidant genes. Our results indicated that NRF2 transport into the nucleus increased 4 h after irradiation in BMMSCs, while the CRIF1 deficiency suppressed this process, but its molecular mechanism has not been fully elucidated. More recently, studies showed that PKC-mediated NRF2 Ser40 phosphorylation was necessary for NRF2 dissociation from KEAP1 and nuclear translocation [21, 22]. Niture et al. [52] identified PKC-δ as the major PKC isoform that phosphorylated NRF2 Ser40. Taking this into consideration, the present study further investigated this molecular mechanism. Co-IP results indicated that CRIF1 interacted with PKC-δ, and three PKC agonists with different mechanisms of action were employed to show that CRIF1 co-activated PKC-δ to phosphorylate NRF2 Ser40, increasing NRF2 stability, allowing it to be translocated into the nucleus to activate protective gene expression. In several previous studies, CRIF1 has been shown to co-activate several phosphatases, including protein kinase A (PKA) [31], p66shc [44], and Lck [53]. In a previous study, we showed that CRIF1 interacted with PKAα and assisted it in phosphorylating the cAMP response element-binding protein (CREB), promoting BMMSCs adipogenic differentiation after radiation damage [31]. Considering the results of our previous study and the current study, we provided several pieces of evidence indicating that CRIF1, which was highly expressed in BMMSCs, may have multiple functions in pathology and physiology after radiation damage. Several studies have described the HSCs-supporting function of BMMSCs [54-56]. According to our in vitro results of the CFU assay in BMMNCs co-cultured with irradiated BMMSCs, we speculated that CRIF1 may be helpful for the HSCs-supporting function of BMMSCs after irradiation. However, this hypothesis needs further in vivo experimental evidences.
A previous study [57] reported that CRIF1 interacted with both the N- and C-termini of NRF2 to promote the proteasome-mediated degradation of NRF2, which seemed to be contradictory with our hypothesis that CRIF1 may increase NRF2 stability. However, these conflicting results indicated a more complex regulation of NRF2 signaling by CRIF1. More recent studies revealed that uncontrolled constant nuclear accumulation of NRF2 had pathological consequences [58]. Therefore, NRF2 regulation is a complex and delicate process coordinating a variety of factors, including Bach1 and Fyn/GSK3, which activate/inactivate NRF2 transcriptional activation depending on if the stress signals subside or persist [59, 60]. Considering this, CRIF1 may play a dual role in NRF2-mediated transcriptional activation, where CRIF1 increases NRF2 stability when it exerts protective cell functions and may also promote ubiquitin-mediated NRF2 degradation when nuclear accumulation of NRF2 is excessive. However, the exact underlying mechanism involved in the regulation and balance of CRIF1 and NRF2 is not fully understood, needing further investigation.
Our study indicated that CRIF1 modulated PKC-δ/NRF2 signaling to facilitate redox homeostasis in BMMSCs after irradiation, and it may contribute to the supporting effect of BMMSCs for the colony forming ability of HSCs in vitro. Thereby, the present study increased the knowledge on CRIF1 biology, helped to elucidate the molecular mechanism underlying BMMSCs radiation resistance, and might get some implications to maintain the HSCs-supporting effect of BMMSCs after radiative injuries.
Abbreviations
APC: allophycocyanin; ARE: antioxidant-responsive element; ARS: acute radiation syndrome; Bach1: BTB domain and CNC homolog 1; BCA: bicinchoninic acid; BSA: bovine serum albumin; BMMNCs: Bone marrow mononuclear cells; BMMSCs: bone marrow multipotent mesenchymal stromal cells; Co-IP: Co-immunoprecipitation; CREB: cAMP-response element binding protein; CRIF1: CR6-interacting factor 1; DAPI: 4',6-diamidino-2-phenylindole; DHE: dihydroethidium; DCFH-DA: dichloro-dihydro-fluorescein diacetate; FACS: fluorescence activated cell sorter; FITC: fluorescein isothiocyanate; Fyn: FYN proto-oncogene, Src family tyrosine kinase; GCLC: catalytic subunit of glutamate-cysteine ligase; GGT1: gamma-glutamyltransferase 1; GSH: glutathione; GSK3: glycogen synthase kinase 3; h-ARS: hematopoietic disorders of acute radiation syndrome; HO1: heme oxygenase 1; HSC: hematopoietic stem cell; IP: immunoprecipitation; KEAP1: kelch-like ECH-associated protein 1; Lck: LCK Proto-Oncogene, Src Family Tyrosine Kinase; NQO1: NAD(P)H:quinoneoxidoreductase 1; NRF2: nuclear factor E2-related factor 2; PKA: protein kinase A; PKC-δ: protein kinase C delta; PBS: phosphate buffer saline; PE: phycoerythrin; PerCP: peridinin chlorophyll protein; PI: propidium iodide; PMA: phorbol 12-myristate 13-acetate; PVDF: polyvinylidene fluoride; RIPA: radio immunoprecipitation assay; ROS: reactive oxygen species; SA-β-Gal: senescence-associated β-galactosidase; SDS: sodium dodecyl sulfate; shRNA: short hairpin RNA; TBI: total body irradiation.
Acknowledgements
We thank Prof. HT Zheng (Third Military Medical University, Chongqing, China) for providing Nrf2+/+ and Nrf2-/- mice. This study was supported by the National Natural Science Foundation of China (No. 81472915), the National Natural Youth Science Foundation of China (No. 81602797), and the Youth Scientist Foundation of Chongqing (CSTC 2013JCYJJQ10001).
Author contribution
L.C. performed the experiments, analyzed the data, and wrote the manuscript; Q.R., Y.X., and L.X. performed the imaging experiments; L.C., F.J., C.W., and J.W. collection and assembly of data, data analysis and interpretation; Z.L. designed the experiments, reviewed the data, and edited the manuscript.
Supplementary Material
Supplementary figures and tables.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Eaton EB Jr, Varney TR. Mesenchymal stem cell therapy for acute radiation syndrome: innovative medical approaches in military medicine. Military Medical Research. 2015;2:2
2. Waselenko JK, MacVittie TJ, Blakely WF, Pesik N, Wiley AL, Dickerson WE. et al. Medical management of the acute radiation syndrome: recommendations of the Strategic National Stockpile Radiation Working Group. Annals of internal medicine. 2004;140:1037-51
3. Dainiak N. Rationale and recommendations for treatment of radiation injury with cytokines. Health physics. 2010;98:838-42
4. Wagemaker G. Heterogeneity of radiation sensitivity of hemopoietic stem cell subsets. Stem cells. 1995;13(Suppl 1):257-60
5. Fliedner TM, Chao NJ, Bader JL, Boettger A, Case C Jr, Chute J. et al. Stem cells, multiorgan failure in radiation emergency medical preparedness: a U.S./European Consultation Workshop. Stem cells. 2009;27:1205-11
6. Cao X, Wu X, Frassica D, Yu B, Pang L, Xian L. et al. Irradiation induces bone injury by damaging bone marrow microenvironment for stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1609-14
7. Wilson A, Trumpp A. Bone-marrow haematopoietic-stem-cell niches. Nature reviews Immunology. 2006;6:93-106
8. Wang LD, Wagers AJ. Dynamic niches in the origination and differentiation of haematopoietic stem cells. Nature reviews Molecular cell biology. 2011;12:643-55
9. Mercier FE, Ragu C, Scadden DT. The bone marrow at the crossroads of blood and immunity. Nature reviews Immunology. 2012;12:49-60
10. Chen MF, Lin CT, Chen WC, Yang CT, Chen CC, Liao SK. et al. The sensitivity of human mesenchymal stem cells to ionizing radiation. International journal of radiation oncology, biology, physics. 2006;66:244-53
11. Singh S, Kloss FR, Brunauer R, Schimke M, Jamnig A, Greiderer-Kleinlercher B. et al. Mesenchymal stem cells show radioresistance in vivo. Journal of cellular and molecular medicine. 2012;16:877-87
12. Nicolay NH, Liang Y, Lopez Perez R, Bostel T, Trinh T, Sisombath S. et al. Mesenchymal stem cells are resistant to carbon ion radiotherapy. Oncotarget. 2015;6:2076-87
13. Nicolay NH, Lopez Perez R, Debus J, Huber PE. Mesenchymal stem cells - A new hope for radiotherapy-induced tissue damage? Cancer letters. 2015;366:133-40
14. Nicolay NH, Lopez Perez R, Saffrich R, Huber PE. Radio-resistant mesenchymal stem cells: mechanisms of resistance and potential implications for the clinic. Oncotarget. 2015;6:19366-80
15. Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. The Biochemical journal. 2009;417:639-50
16. Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annual review of pharmacology and toxicology. 2007;47:89-116
17. Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends in biochemical sciences. 2014;39:199-218
18. Sekhar KR, Freeman ML. Nrf2 promotes survival following exposure to ionizing radiation. Free radical biology & medicine. 2015;88:268-74
19. Hong F, Sekhar KR, Freeman ML, Liebler DC. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. The Journal of biological chemistry. 2005;280:31768-75
20. Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y. et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochemical and biophysical research communications. 1997;236:313-22
21. Huang HC, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. The Journal of biological chemistry. 2002;277:42769-74
22. Bloom DA, Jaiswal AK. Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. The Journal of biological chemistry. 2003;278:44675-82
23. Chung HK, Yi YW, Jung NC, Kim D, Suh JM, Kim H. et al. CR6-interacting factor 1 interacts with Gadd45 family proteins and modulates the cell cycle. The Journal of biological chemistry. 2003;278:28079-88
24. Ran Q, Hao P, Xiao Y, Xiang L, Ye X, Deng X. et al. CRIF1 interacting with CDK2 regulates bone marrow microenvironment-induced G0/G1 arrest of leukemia cells. PloS one. 2014;9:e85328
25. Kwon MC, Koo BK, Moon JS, Kim YY, Park KC, Kim NS. et al. Crif1 is a novel transcriptional coactivator of STAT3. The EMBO journal. 2008;27:642-53
26. Park KC, Song KH, Chung HK, Kim H, Kim DW, Song JH. et al. CR6-interacting factor 1 interacts with orphan nuclear receptor Nur77 and inhibits its transactivation. Molecular endocrinology. 2005;19:12-24
27. Kwon MC, Koo BK, Kim YY, Lee SH, Kim NS, Kim JH. et al. Essential role of CR6-interacting factor 1 (Crif1) in E74-like factor 3 (ELF3)-mediated intestinal development. The Journal of biological chemistry. 2009;284:33634-41
28. Greber BJ, Bieri P, Leibundgut M, Leitner A, Aebersold R, Boehringer D. et al. Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science. 2015;348:303-8
29. Shin J, Lee SH, Kwon MC, Yang DK, Seo HR, Kim J. et al. Cardiomyocyte specific deletion of Crif1 causes mitochondrial cardiomyopathy in mice. PloS one. 2013;8:e53577
30. Byun J, Son SM, Cha MY, Shong M, Hwang YJ, Kim Y. et al. CR6-interacting factor 1 is a key regulator in Abeta-induced mitochondrial disruption and pathogenesis of Alzheimer's disease. Cell death and differentiation. 2015;22:959-73
31. Zhang X, Xiang L, Ran Q, Liu Y, Xiang Y, Xiao Y. et al. Crif1 Promotes Adipogenic Differentiation of Bone Marrow Mesenchymal Stem Cells After Irradiation by Modulating the PKA/CREB Signaling Pathway. Stem cells. 2015;33:1915-26
32. Long M, Rojo de la Vega M, Wen Q, Bharara M, Jiang T, Zhang R. et al. An Essential Role of NRF2 in Diabetic Wound Healing. Diabetes. 2016;65:780-93
33. Gan J, Meng F, Zhou X, Li C, He Y, Zeng X. et al. Hematopoietic recovery of acute radiation syndrome by human superoxide dismutase-expressing umbilical cord mesenchymal stromal cells. Cytotherapy. 2015;17:403-17
34. Hu KX, Sun QY, Guo M, Ai HS. The radiation protection and therapy effects of mesenchymal stem cells in mice with acute radiation injury. The British journal of radiology. 2010;83:52-8
35. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-8
36. Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes to cells: devoted to molecular & cellular mechanisms. 2011;16:123-40
37. Abdallah BM, Kassem M. New factors controlling the balance between osteoblastogenesis and adipogenesis. Bone. 2012;50:540-5
38. Misra J, Mohanty ST, Madan S, Fernandes JA, Hal Ebetino F, Russell RG. et al. Zoledronate Attenuates Accumulation of DNA Damage in Mesenchymal Stem Cells and Protects Their Function. Stem cells. 2016;34:756-67
39. Zhang Z, Zhang H, Liu F, Qiu M, Tong J. Effects of gamma radiation on bone-marrow stromal cells. Journal of toxicology and environmental health Part A. 2010;73:514-9
40. Yang X, Balakrishnan I, Torok-Storb B, Pillai MM. Marrow Stromal Cell Infusion Rescues Hematopoiesis in Lethally Irradiated Mice despite Rapid Clearance after Infusion. Advances in hematology. 2012;2012:142530
41. Hou J, Han ZP, Jing YY, Yang X, Zhang SS, Sun K. et al. Autophagy prevents irradiation injury and maintains stemness through decreasing ROS generation in mesenchymal stem cells. Cell death & disease. 2013;4:e844
42. Cmielova J, Havelek R, Soukup T, Jiroutova A, Visek B, Suchanek J. et al. Gamma radiation induces senescence in human adult mesenchymal stem cells from bone marrow and periodontal ligaments. International journal of radiation biology. 2012;88:393-404
43. Alessio N, Del Gaudio S, Capasso S, Di Bernardo G, Cappabianca S, Cipollaro M. et al. Low dose radiation induced senescence of human mesenchymal stromal cells and impaired the autophagy process. Oncotarget. 2015;6:8155-66
44. Nagar H, Jung SB, Kwon SK, Park JB, Shong M, Song HJ. et al. CRIF1 deficiency induces p66shc-mediated oxidative stress and endothelial activation. PloS one. 2014;9:e98670
45. Alessio N, Bohn W, Rauchberger V, Rizzolio F, Cipollaro M, Rosemann M. et al. Silencing of RB1 but not of RB2/P130 induces cellular senescence and impairs the differentiation potential of human mesenchymal stem cells. Cellular and molecular life sciences: CMLS. 2013;70:1637-51
46. Wang D, Jang DJ. Protein kinase CK2 regulates cytoskeletal reorganization during ionizing radiation-induced senescence of human mesenchymal stem cells. Cancer research. 2009;69:8200-7
47. Muthna D, Soukup T, Vavrova J, Mokry J, Cmielova J, Visek B. et al. Irradiation of adult human dental pulp stem cells provokes activation of p53, cell cycle arrest, and senescence but not apoptosis. Stem cells and development. 2010;19:1855-62
48. Kim SB, Pandita RK, Eskiocak U, Ly P, Kaisani A, Kumar R. et al. Targeting of Nrf2 induces DNA damage signaling and protects colonic epithelial cells from ionizing radiation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2949-55
49. Hseu YC, Lo HW, Korivi M, Tsai YC, Tang MJ, Yang HL. Dermato-protective properties of ergothioneine through induction of Nrf2/ARE-mediated antioxidant genes in UVA-irradiated Human keratinocytes. Free radical biology & medicine. 2015;86:102-17
50. Theodore M, Kawai Y, Yang J, Kleshchenko Y, Reddy SP, Villalta F. et al. Multiple nuclear localization signals function in the nuclear import of the transcription factor Nrf2. The Journal of biological chemistry. 2008;283:8984-94
51. Niture SK, Khatri R, Jaiswal AK. Regulation of Nrf2-an update. Free radical biology & medicine. 2014;66:36-44
52. Niture SK, Jain AK, Jaiswal AK. Antioxidant-induced modification of INrf2 cysteine 151 and PKC-delta-mediated phosphorylation of Nrf2 serine 40 are both required for stabilization and nuclear translocation of Nrf2 and increased drug resistance. Journal of cell science. 2009;122:4452-64
53. Vahedi S, Chueh FY, Chandran B, Yu CL. Lymphocyte-specific protein tyrosine kinase (Lck) interacts with CR6-interacting factor 1 (CRIF1) in mitochondria to repress oxidative phosphorylation. BMC cancer. 2015;15:551
54. Perdomo-Arciniegas AM, Vernot JP. Co-culture of hematopoietic stem cells with mesenchymal stem cells increases VCAM-1-dependent migration of primitive hematopoietic stem cells. International journal of hematology. 2011;94:525-32
55. Jing D, Fonseca AV, Alakel N, Fierro FA, Muller K, Bornhauser M. et al. Hematopoietic stem cells in co-culture with mesenchymal stromal cells-modeling the niche compartments in vitro. Haematologica. 2010;95:542-50
56. Huang X, Zhu B, Wang X, Xiao R, Wang C. Three-dimensional co-culture of mesenchymal stromal cells and differentiated osteoblasts on human bio-derived bone scaffolds supports active multi-lineage hematopoiesis in vitro: Functional implication of the biomimetic HSC niche. International journal of molecular medicine. 2016;38:1141-51
57. Kang HJ, Hong YB, Kim HJ, Bae I. CR6-interacting factor 1 (CRIF1) regulates NF-E2-related factor 2 (NRF2) protein stability by proteasome-mediated degradation. The Journal of biological chemistry. 2010;285:21258-68
58. Furfaro AL, Traverso N, Domenicotti C, Piras S, Moretta L, Marinari UM. et al. The Nrf2/HO-1 Axis in Cancer Cell Growth and Chemoresistance. Oxidative medicine and cellular longevity. 2016;2016:1958174
59. Lee OH, Jain AK, Papusha V, Jaiswal AK. An auto-regulatory loop between stress sensors INrf2 and Nrf2 controls their cellular abundance. The Journal of biological chemistry. 2007;282:36412-20
60. Niture SK, Jaiswal AK. Prothymosin-alpha mediates nuclear import of the INrf2/Cul3 Rbx1 complex to degrade nuclear Nrf2. The Journal of biological chemistry. 2009;284:13856-68
Author contact
![]() Corresponding author: E-mail address: zhongjunliedu.cn; johnneyusccom. Phone: +86-023-68755319, Fax: +86-023-68774776
Corresponding author: E-mail address: zhongjunliedu.cn; johnneyusccom. Phone: +86-023-68755319, Fax: +86-023-68774776
 Global reach, higher impact
Global reach, higher impact