13.3
Impact Factor
Theranostics 2017; 7(11):2775-2793. doi:10.7150/thno.19443 This issue Cite
Review
Advanced Gene Manipulation Methods for Stem Cell Theranostics
Department of Chemistry and Chemical Biology, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA
Received 2017-2-1; Accepted 2017-4-18; Published 2017-7-8
Abstract
In the field of tissue engineering, autologous cell sources are ideal to prevent adverse immune responses; however, stable and reliable cell sources are limited. To acquire more reliable cell sources, the harvesting and differentiation of stem cells from patients is becoming more and more common. To this end, the need to control the fate of these stem cells before transplantation for therapeutic purposes is urgent. Since transcription factors orchestrate all of the gene activities inside of a cell, researchers have developed engineered and synthetic transcription factors to precisely control the fate of stem cells allowing for safer and more effective cell sources. Engineered transcription factors, mutant fusion proteins of naturally occurring proteins, comprise the three main domains of natural transcription factors including DNA binding domains, transcriptional activation domains, and a linker domain. Several key advancements of engineered zinc finger proteins, transcriptional activator-like effectors, and deficient cas9 proteins have revolutionized the field of engineered transcription factors allowing for precise control of gene regulation. Synthetic transcription factors are chemically made transcription factor mimics that use small molecule based moieties to replicate the main functions of natural transcription factors. These include hairpin polyamides, triple helix forming oligonucleotides, and nanoparticle-based methods. Synthetic transcription factors allow for non-viral delivery and greater spatiotemporal control of gene expression. The developments in engineered and synthetic transcription factors have lowered the risk of tumorigenicity and improved differentiation capability of stem cells, as well as facilitated many key discoveries in the fields of cancer and stem cell biology, thus providing a stepping stone to advance regenerative medicine in the clinic for cell replacement therapies.
Keywords: Gene Regulation, Transcription Factors, Cellular Reprogramming, Stem Cells, Regenerative Medicine, Nanotechnology.
Introduction
Although stem cell therapy and cellular reprogramming hold great potential for regenerative medicine, precise control of stem cell differentiation and reprogramming fate is one of the most critical issues to be addressed before their therapeutic applications can be fully realized. In particular, using human patient-derived stem cells, such as human induced pluripotent stem cells (hiPSCs), autologous adult stem cells, and reprogramming patient-derived somatic cells can offer a great opportunity for the treatment of multiple devastating injuries and neuro-degenerative diseases such as Parkinson's and Alzheimer's diseases, and spinal cord injury. However, several hurdles, including selective and effective control of cell fate and a better understanding of the cellular reprogramming and differentiation mechanisms, must be surmounted before innovative clinical approaches can be developed.
To this end, regulating cellular reprogramming and stem cell differentiation using the ectopic expression of key transcription factors (TFs) not only has high impact in the field of regenerative medicine, but can also provide new insight into the fundamental mechanisms of cell state transitions. TF proteins are master regulators of gene expression that function to activate or deactivate specific gene networks within cells, which alter cellular functions and fate such as cell differentiation and cellular reprogramming. TFs can sense and respond to various stimuli within cells and in turn initiate various signaling cascades and alter the genetic circuitry of the cell [1]. While TFs are complex proteins that involve several different interactions, they are made up of three basic components: i) a nuclear localization domain that allows for the protein to enter the nucleus; ii) a DNA binding domain that can locate and bind to specific DNA sequences; and iii) an activation domain that can recruit RNA polymerase and the transcriptional preinitiation complex to the desired DNA promoter region and begin transcription. While TFs has been well known to regulate the gene expression profile of cells for decades, the pioneering work by Yamanaka and colleagues, generating induced pluripotent stem cells, has sparked a huge rise in the study and engineering of transcription factors to alter the genetic programming of cells [2, 3]. Forced expression of transcription factors has been clearly demonstrated to induce differentiation, cellular reprogramming, and apoptosis. Extensive research has gone into utilizing this ability of transcription factors to directly convert from one cell type to another. Transdifferentiation (lineage reprogramming, direct conversion, and direct reprogramming) occurs when a cell is converted from one cell type to another without entering a stem state. The ability to convert from one cell type directly to another has many advantages in the field of regenerative medicine as it would allow for a larger cell source and eliminate immune rejection. Through forced expression of various transcription factors, scientists have shown induced myogenic transdifferentiation [4-6], cardiogenic transdifferentiation [7-14], and neurogenic transdifferentiation [15-18]. In addition, transcription factors are involved in almost every cellular process and can be related to several disorders. Thus, the ectopic expression of transcription factors has proven to be a very valuable tool in studying the progression and expression pattern of diseases such as cancer [19-21]. While the most common method of delivery for transcription factors is viral vector expression, several research groups have developed delivery methods including electroporation, polymer nanocarriers, protein nanocapsules, lipid capsules, oligonucleotide nanoparticles, and other nanoparticle delivery vehicles to deliver whole transcription factors into the cell. However these methods are normally hindered by low delivery efficiency and protein degradation [22-29]. Because of these limitations, new technologies are needed to improve current strategies for gene expression manipulation. These approaches include the use of engineered transcription factors that can target any specific location within the genome and provide efficient activation of endogenous genes, however the use of viral-based delivery methods impede their translational potentials. Although small molecule based transcription factors provide the advantage of non-viral delivery, often times they suffer from low solubility and nuclear permeability. A novel platform, NanoScript, was designed to provide enhanced cellular uptake and non-viral delivery through a nanoparticle-based transcription factor mimic.
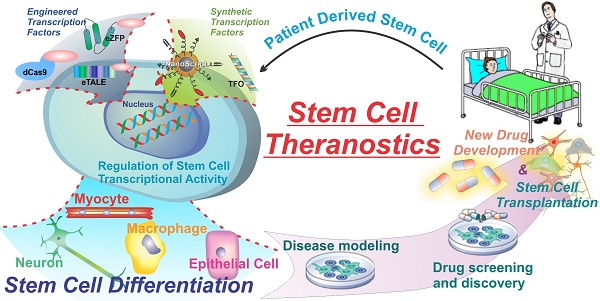
Stem cells, specifically, have great potential for therapeutic strategies. They can differentiate into many different cell types in the body, they can sense inflammation and migrate to sites of injury and disease, and they can be grown and differentiated in culture for the creation of disease models and diagnostics. However, one of the major pitfalls of current stem cell therapies is the lack of control over stem cell fate. Unguided stem cell therapies can lead to devastating adverse effects such as teratoma formation when implanted. The major current hurdles of using transcription factors to control cell fate are the limited number of gene targets and the lack of spatial and temporal control. Nevertheless, transcription factors are one of the most powerful proteins in the body due to their far-reaching impact, and therefore are the focus of much study and research to develop the next generation of transcription factor technologies. New technologies such as engineered and synthetic transcription factors hold the key to further the field of stem cell theranostics and regenerative medicine. By delivering engineered and synthetic transcription factors into cells we can specifically control cell fate and function. With greater control over stem cell fate researchers can develop more accurate disease models, more advanced drug screening methods. This in turn can lead to more advanced and effective drug development as well as furthering stem cell-based therapies (Figure 1).
Engineered Transcription Factors
While expression of natural transcription factors (TFs) is very alluring and has several therapeutic and research applications, the designs of natural TFs are restricted to a limited number of targets, exogenously expressed factors, or integrative viral delivery which limits clinical application. In addition, there are few cases that can be spatiotemporally regulated to control gene expression. To address some of these issues, several types of engineered transcription factors have been developed to specifically target any gene and impart several other key advantages over natural transcription factors. These engineered transcription factors normally consist of a DNA-binding domain fused to a transcriptional activation domain. The DNA binding domain can target specific DNA sequences while the effector domains can act to either activate or repress genes by modulating the transcriptional machinery or altering the epigenetic state. Three examples of this are outlined in figure 2. The next section of this review will highlight some of the main types of engineered transcription factors including Zinc-finger proteins, Transcriptional activator-like effectors, and Cas9 based transcription factors fused to various types of effector domains for the regulation of transcription and the control of cellular behavior (Figure 2) [30].
Engineered Zinc Finger Proteins (eZFPs)
Zinc Finger Proteins (ZFPs) are the most typical class of DNA binding domains that are found on naturally occurring transcription factors. Since the development of polydactyl zinc finger proteins, first discovered by the study of TFIIA, a general transcription factor [31], the feasibility of using zinc fingers as therapeutic agents has been achieved due to their ability to bind to longer stretches of DNA allowing them to be unique to one location in the genome [32].
Developing the Technology
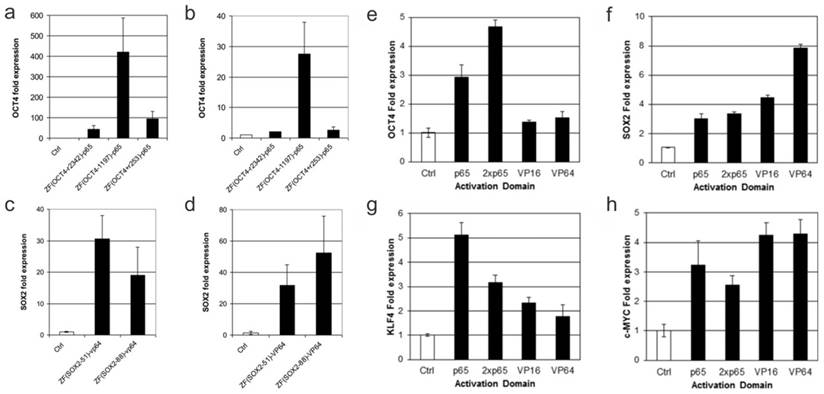
The first use of engineered ZFPs (eZFPs) was described by Beerli et al. in 2000. eZFPs that targeted the untranslated region of the proto-oncogenes EERB-2 and EERB-3 were designed. Then, activation or repression domains were fused to the eZFPs and the engineered transcription factors were introduced into the cells. Beerli el al. were able to show regulation of the target genes and in addition treatment of the engineered transcription factors in SKBR3 breast cancer cells resulted in inhibition of the cell-cycle and accumulation in the G1 phase. This demonstration showed zinc finger proteins can potentially be used as a therapeutic strategy [33]. As a study to show the robustness of eZFP-TFs, Ji et al. constructed ZF-TFs for OCT4, SOX2, KLF4, and c-MYC, the four factor important in cellular reprograming. ZFPs were used to target the promoter, distal enhancer, proximal enhancer, as well as downstream from the proximal promoter. In addition, various activation domains including VP16, VP64, and p65 were also examined. Ji et al. were able to find the optimal binding sites for the various genes as well as show expression with each of the different activation domains in HEK293T cells as well as fibroblast cells. Figure 3 shows that when using the zinc finger to target the promoter region of the OCT4 gene binding 1197 base pairs upstream of the transcription start site elicited the highest expression of OCT4 using the activation domain p65. In addition, it was shown that binding 51 base pairs upstream using the VP64 activation domain gave the highest expression of SOX2 in K562 cells whereas binding 84 base pairs upstream gave the highest expression of SOX2 in fibroblasts. Ji and colleagues also tested the relative efficiency of different activation domains for the 4 genes tested. They showed that for different genes different activation domains worked the best to increase transcription (Figure 3). This suggests the design of the eZFP-TF may be more complex than originally thought [34].
Engineered and Synthetic transcription factors can modulate gene expression and alter cell behavior to initiate cellular processes such as differentiation. Engineered Transcription factors are mutations of naturally occurring transcription factors used to gain key advantages such as target specificity while synthetic transcription factors are chemically derived transcription factor mimics.
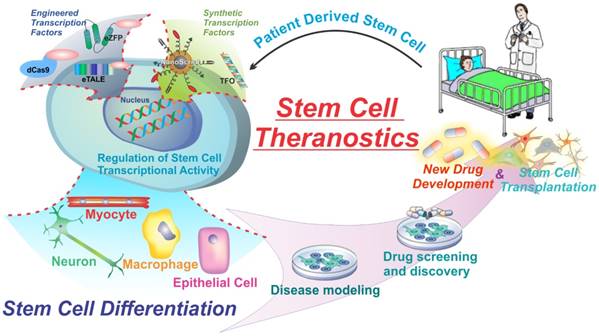
Commonly used engineered transcription factors for gene manipulation. Zinc finger proteins (a) TALEs (b) and dCas9 (c) are used as DNA binding domains to target specific gene sequences. They are then fused to effector domains to activate or repress transcription, or alter the epigenetic state of the histones. Crystal structures are of zinc finger proteins, TALEs, and spCas9 (nuclease active) not fused to effector domains. Figure adapted from Thakore et al. (2016) Nature Methods (30). Example crystal structures generated from PDB files 2I13, 3UGM, and 4OO8.
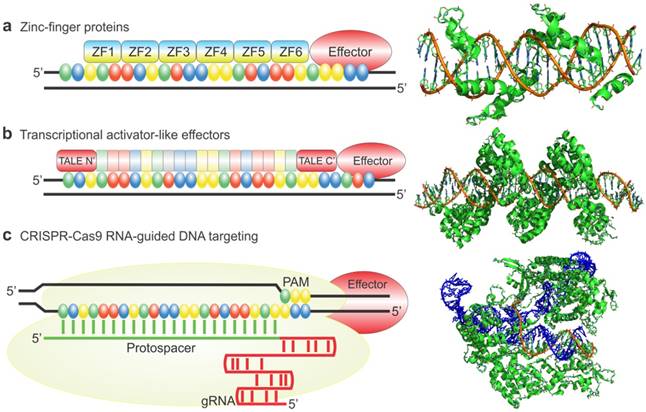
Engineered Zinc Finger proteins fused to p65 and VP64 activation domains upregulates OCT4, SOX2 respectively, in K562 (a and c) and BJ fibroblasts (b and d) at various lengths upstream of the transcriptional start site (a-d). Relative activation of genes using ZFP-TF fused to various activation domains including p65, 2xp65, VP16 and VP64 (e-h). Modified from Ji et al. (34) https://creativecommons.org/licenses/by/3.0/legalcode
Epigenetic Targeting
In addition to the use of standard activation domains, groups have recently investigated the use of epigenetic modifiers in conjunction with the ZFP system. Snowden et al. showed that engineered ZFPs can be used as repressors for VEGF-A by fusing the eZFP to the ligand binding domain of thyroid hormone receptor. The fusion protein was able to deacetylate the histones H3 and H4 and cause repression of the target genes. In addition, the eZFP was able to reduce expression of VEGF by greater than 20 fold dropping to levels of non-angiogenic tumors making it a viable strategy for cancer therapy [35]. Cui et al. demonstrated the fusion of the DNA methyltransferase dnmt3a to an eZFP targeting the P16 promoter region. P16 is a gene that is heavily involved in cancer growth and metastasis and, when inhibited, has been shown to lead to a worse prognosis. This study was performed to test whether DNA methylation could play a role in P16 under expression. The ZFP-dnmt3a fusion repressed the P16 gene approximately 2.5 fold and was able to confer an increased migration and invasion in both transfected GES-1 cells as well as BGC823 cells in both a transwell system as well as an in vivo mouse metastasis model [36]. Li et al. that Dnmt3a and Dnmt3b could be fused to different DNA binding domains including GAL4 and an engineered Cys2His2 zinc finger protein to target both cellular and viral promoters. They were able to show an 18 fold reduction of the titer of Herpes Simplex Virus type 1 using a methyltransferase fused to a ZFP targeting the Herpes Simplex Virus type 1 gene IE175K [37]. Rivenbark et al. demonstrated the use of DNMT3a fused to eZFP to suppress MAPSIN and SOX2 in a human breast cancer cell line. Through this they were able to demonstrate heritable gene silencing through DNA methylation [38].
Therapeutic Applications
Dai et al. fused the p65 activation domain from the nf-kb transcription factor to an eZFP targeting VEGF to upregulate its expression in mice that experienced femoral artery ligation. Treated groups received eZFP treatment to the ischemic muscle and showed a significant increase in VEGF mRNA expression as well as increased capillary density and cell proliferation compared to untreated groups. In addition, blood flow ratio of the ischemic limb compared to nonischemic were significantly higher in the treated groups. Dai et al. showed that the eZFP could activate VEGF expression in vivo and can potentially be used as a treatment for peripheral artery disease [39]. Graslund et al. used eZFPs conjugated to VP64 domains to probe the targeting sites near known proximal regulatory regions to induce y-GLOBIN expression in K562 cells. Optimal binding site and induce expression of y-GLOBIN were found to be showing a 16-fold increase over native K562 cells. This activation could potentially be used as a treatment for sickle cell disease and thalassemic diseases showing the therapeutic potential of eZFPs [40].
Engineered Transcription Activator-Like Effectors (eTALE)
A second class of DNA binding domains, transcription activator-like effectors (TALEs), was shown to have a unique DNA binding domain that is made of tandem repeats of 34 amino acid residues. By tuning the repeating variable residues located at the 12th and 13th position of each repeat, the recognition sequence of the TALE can be tailored [41-45]. Since this discovery, many groups have utilized TALEs to engineer transcription factors with a high degree of specificity with ease.
Developing the Technology
Morbitzer et al. showed that TALE could be designed to target user-specific genes and targeted the promoters of BS4, EGL3, or KNAT1. In all three cases, the desired gene was upregulated and showed that a desired location in the genome can be efficiently targeted with the TALE system [46]. Zhang et al. utilized a novel technique to manufacture TALEs which uses type II restriction enzymes to ligate linkers between the orthogonal repeated domains allowing for a much more facile generation of TALEs. This technique enhances the practical use of TALEs in generating custom DNA-binding domains. Zhang then tested 17 different TALEs to recognize specific DNA binding sites including SOX2 and KLF4 causing an increase in mCherry reporter expression. It is also of interest to note that expression seemed to be inversely related to the number of base pair mismatches from the predicted DNA sequence [47]. Gao et al. utilized designer TALE TFs to induce expression of OCT4 in mouse embryonic fibroblasts. The OCT4-TALE, greatly increases OCT4 expression in the fibroblasts and can replace exogenously expressed OCT4 when reprogramming fibroblasts into induced pluripotent stem cells, highlighting the incredible capability of TALE-TFs to induce reprogramming of fibroblasts to stem cells [48]. Perez-Pinera et al. and Maeder et al simultaneously utilized a combinatorial technique where they transfected multiple TALE-TFs that targeted several areas in the promoter region and showed the delivery of multiple TALE-TFs targeting the same gene at different sites gave rise to a marked increase in expression compared to expression of a singular TALE-TF. A representative scheme of the TALE-TF is shown in figure 4 showing the fusion of the VP64 domain onto the TALE DNA binding domain, with an NLS to facilitate nuclear localization. The location of binding of each TALE-TF with respect to the transcription start site of the gene of interest is highlighted in Figure 4b. Relative expression induced by each TALE-TF treated individually to cells as well as when co-expressed showing the significant enhancement of gene expression when TALE-TFs are multiplexed (Figure 4). Moreover, genes that are in a silenced state can be robustly activated by this combinatorial approach [49, 50].
Schematic diagram of eTALE-TF (a). Location of binding sites for various eTALEs in various genes including IL1RN, KLK3, CEACAM5, and ERBB2 (b). Relative luciferase activity detected for the various genes tested in a promoter reporter assay (c-f). Relative mRNA expression in transfected cells for various genes tested (g-j). Protein expression detected using Elisa (k-l) or western blot (m-n). Adapted by permission from Macmillan Publishers Ltd: Nature Methods (49) copyright (2013)
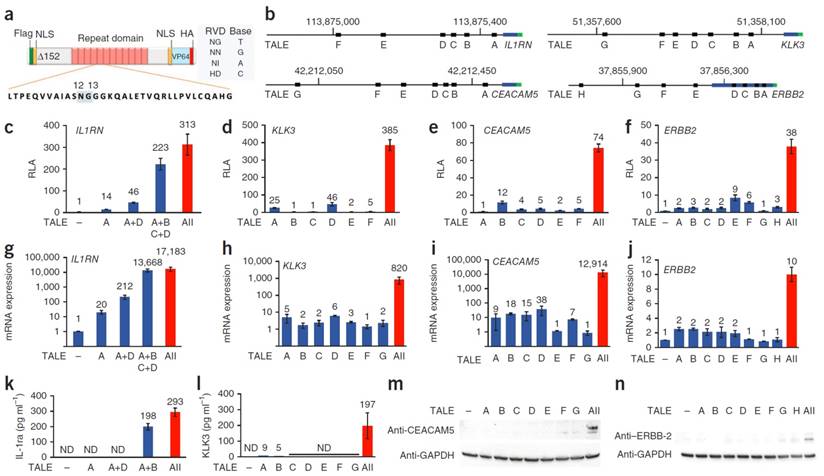
Epigenetic Targeting
Bultmann et al. studied the epigenetic effect of histone states on the efficiency of TALE-mediated gene regulation. While eTALEs were able to efficiently upregulate the pluripotency gene Oct4 in mouse embryonic stem cells where the Oct4 promoter would be active, very little upregulation of Oct4 in was found in mouse neural stem cells where the Oct4 promoter is silenced. However, the activation of Oct4 in neural stem cells was able to be rescued using epigenetic modulators such as valproic acid or 5-aza-2'-deoxycytidine which activated the Oct4 promoter. This demonstration showed the dependence of TALE-based gene regulation on the epigenetic state of the promoters as well as the synergistic effect epigenetic modulators when used in combination with TALE-TFs. In addition, Tale based TFs can be used to alter the transcriptome of various types of stem cells found in the body, demonstrating their potential for therapeutic use [51]. Maeder et al. also used TALE fusions to add a TET1 hydroxylase catalytic domain onto the TAL effector. By doing so, specific CpG methylation marks could be removed from specific genes that are silenced in the genome. This allows for the study of the effect of specific methylation marks on the animal genome and development as well as the activation of endogenous genes by the deletion of silencing marks [50]. Bernstein et al. used a TALE DNMT fusion targeted at the CDKN2A locus that encodes the cyclin-dependent kinase inhibitor P16. Using this system they were able to show a two-fold repression of P16 mRNA in fibroblasts. This in turn increased the replication of the fibroblasts by permitting entry into the cell cycle [52]. Hu et al. studied the effect of TALE-TFs on the expression of OCT4, a stringently silenced gene, in human and mouse somatic cells. It was found that TALE-VP64 had the most efficient expression -120 to -80 bp upstream of the transcription start site. In addition, a combination approach, as previously described by Perez-Pinera, showed to achieve upregulation of 30-fold expression. Furthermore, p300, a histone acetyltransferase, further enhanced the gene expression and protein production of OCT4 in normally silenced cells [53].
Therapeutic Applications
Tremblay et al. utilized this technology to treat Friedreich ataxia, a disorder with reduced expression of the FRATAXIN gene. In order to treat the disorder, 12 different TALE-TFs were generated to target different regions of the FRATAXIN gene. These TALE-VP64 fusions were transfected into 293FT cells, and the optimized conditions showed a two to three-fold increase in the FRATAXIN gene expression in 293FT cells, however, protein expression was not characterized. This demonstration showed the potential of eTALE for therapeutic use for treating disorders such as Friedreich ataxia, however it suffers since the therapy would have to be administered systemically, which is challenging [54]. Very recently, Barbon et al. showed the potential of TALE-TFs for treating diseases in a coagulation factor VII (FVII) deficient human hepatocyte model. The TALE-TF was designed to bind to a location in the promoter between the two known point mutations that cause the disorder. The TALE-TF was cotransfected with both disease-mimic and wild-type reporters and showed a 100 fold increase in disease-mimic reporter expression. In addition, the TALE-TF was transfected into HepG2 cells and Hep10 (liver cells from 10 patients) cells and showed upregulation of gene expression as well as an increase in F7 protein expression. These demonstrations showed the therapeutic potential of TALE-TFs [55].
Summary of key results from engineered transcription factors being applied to endogenous genes in mammalian cells.
| DBD | Effector Domain | Cell Tested | Gene target | Multiplexing | Fold Activation/Repression | Reference |
|---|---|---|---|---|---|---|
| eZFP | KRAB | A431 | erB-2 | No | Expression abolished | 33 |
| K562 | y-Globin | No | 10 fold repression | 40 | ||
| VP64 | A431 | erB-2 | No | 8 fold activation | 33 | |
| K562 | y-Globin | No | 16 fold activation | 40 | ||
| K562 | SOX2 | No | 30 fold activation | 34 | ||
| BJ Fibroblast | SOX2 | No | ~50 fold activation | 34 | ||
| vErbA | U87MG | VEGF-A | No | 20 fold repression | 35 | |
| p65 | Ischemic rabbit muscle | VEGF | No | 15 fold activation | 39 | |
| K562 | OCT4 | No | ~400 fold activation | 34 | ||
| BJ Fibroblast | OCT4 | No | ~30 fold activation | 34 | ||
| dnmt3a | HEK293T | P16 | No | 2.5 fold repression | 36 | |
| eTALE | KRAB | ESCs | Oct4 | No | 10 fold repression | 48 |
| VP64 | 293FT | SOX2 | No | 5.5 fold activation | 47 | |
| KLF4 | No | 2.2 fold activation | 47 | |||
| 293FT | FRATAXIN | No | 3.1 fold activation | 54 | ||
| 293T | IL1RN | Yes | 17,183 fold activation | 49 | ||
| KLK3 | Yes | 820 fold activation | 49 | |||
| CEACAM5 | Yes | 12,914 fold activation | 49 | |||
| ERBB2 | Yes | 10 fold activation | 49 | |||
| 293T | VEGF-A | Yes | 50 fold activation | 50 | ||
| miRNA302a | Yes | ~250 fold activation | 50 | |||
| NIH3T3 | Oct4 | Yes | 30 fold activation | 53 | ||
| 293T | OCT4 | Yes | 20 fold activation | 53 | ||
| MEFs | Oct4 | No | 20 fold activation | 48 | ||
| HepG2 | F7 | No | 4 fold activation | 55 | ||
| Hep10 | F7 | No | 4 fold activation | 55 | ||
| VP16 | ESCs | oct4 | No | 3-4 fold activation | 51 | |
| ES-NSC | oct4 | No | No activation | 51 | ||
| p65 | 293T | VEGF-A | Yes | 35 fold activation | 50 | |
| miRNA302a | Yes | ~100 fold activation | 50 | |||
| TET1 | 293 | RHOXF2 | No | 14,000 fold activation | 50 | |
| HBB | No | 40,000 fold activation | 50 | |||
| DNMT3a-3L | HFFs | CDKN2A | No | 2.5 fold repression | 52 | |
| dCas9 | KRAB | HeLa | CD71 | No | ~4 fold repression | 58 |
| CXCR4 | No | ~5 fold repression | 58 | |||
| VP64 | ESC | SOX17 | No | 287 fold activation | 70 | |
| 293T | ASCL1 | Yes | 249 fold activation | 59 | ||
| NANOG | Yes | 13 fold activation | 59 | |||
| HBG1 | Yes | 134 fold activation | 59 | |||
| MYOD1 | Yes | 47 fold activation | 59 | |||
| VEGFA | Yes | 2 fold activation | 59 | |||
| TERT | Yes | 2 fold activation | 59 | |||
| IL1B | Yes | 10 fold activation | 59 | |||
| IL1R2 | Yes | 19 fold activation | 59 | |||
| NTF3 | Yes | 25 fold activation | 60 | |||
| VP160 | 293T | IL1RN | Yes | 6.5 fold activation | 62 | |
| SOX2 | Yes | 9.5 fold activation | 62 | |||
| OCT4 | Yes | 8 fold activation | 62 | |||
| VP192 | 293 | OCT4 | Yes | 70 fold activation | 71 | |
| hESC | FOXA2 | Yes | ~10 fold activation | 71 | ||
| SOX17 | Yes | ~500 fold activation | 71 | |||
| PDX1 | Yes | ~60 fold activation | 71 | |||
| NKX6.1 | Yes | ~8 fold activation | 71 | |||
| SAM system | 293FT | IL1B | No | ~20,000 fold activation | 63 | |
| HBG1 | No | ~5,000 fold activation | 63 | |||
| ZFP42 | No | ~800 fold activation | 63 | |||
| ASCL1 | No | ~500 fold activation | 63 | |||
| NANOG | No | ~300 fold activation | 63 | |||
| LIN28A | No | ~300 fold activation | 63 | |||
| MYOD1 | No | ~300 fold activation | 63 | |||
| IL1R2 | No | ~300 fold activation | 63 | |||
| POU5F1 | No | ~80 fold activation | 63 | |||
| KLF4 | No | ~10 fold activation | 63 | |||
| P300 Core | 293T | IL1RN | Yes | 9,920 fold activation | 66 | |
| MYOD | Yes | 50 fold activation | 66 | |||
| OCT4 | Yes | 32 fold activation | 66 | |||
| VPR | 293T | MIAT | Yes | 280 fold activation | 64 | |
| NEUROD1 | Yes | 87 fold activation | 64 | |||
| ASCL1 | Yes | 4,600 fold activation | 64 | |||
| RHOXF2 | Yes | 18,000 fold activation | 64 | |||
| TTN | Yes | 20,000 fold activation | 64 | |||
| ACTC1 | Yes | 330 fold activation | 64 | |||
| Casilio | 293T | OCT4 | Yes | 100 fold activation | 65 | |
| DNMT3A | 293 | IL6ST | Yes | ~2 fold repression | 67 | |
| BACH2 | Yes | ~2 fold repression | 67 | |||
| CDKN2A | Yes | 39% decrease | 68 | |||
| DNMT3a-3l | Skov-3 | EpCAM | Yes | 2 fold repression | 69 |
Inactive/deficient Cas9 (dCas9)
Lastly, a third class of DNA binding domains has been developed, the CRISPR/Cas9 system. This system has been engineered to rely on the Cas9 protein recognizing and binding to guide RNA (gRNA) that is delivered into the cells. This system, discovered in prokaryotes, provides resistance against viruses in most bacteria [56]. A deficient Cas9 (dCas9) version of the protein was engineered with mutations in the nuclease portion of the protein ablating the cleavage of DNA while maintaining its ability to bind to gRNAs. The new deficient system was then used for transcriptional interference and was able to achieve up to 300-fold repression of a given gene in bacteria while only a modest repression in mammalian cells [57].
Developing the Technology
Gilbert et al took the technology one-step further and attempted to improve the repression in mammalian cells by fusing a repressor domain, KRAB, onto the dCas9. By doing so, a 15-fold repression of expression in mammalian cells was achieved. In addition, Gilbert fused activation domains to the dCas9 and was able to show up to 25 fold activation of a reporter plasmid [58]. Perez-Pinera et al. demonstrated that this dCas9 fused with a VP64 activation domain was able to target the promoter region of the endogenous IL1RN gene and showed induction of the gene. In addition, upregulation was seen in eight other genes including ASCL1, NANOG, HBG1, HBG2, MYOD1, BEGFA, TERT, IL1B, and IL1R2. In some cases, a single gRNA was sufficient to induce expression of the genes where in other cases 4 gRNAs were necessary to induce expression [59] Concurrently Maeder et al. performed similar work using a dCas9-VP64 fusion to express VEGFA and NTF3 yielding very similar results [60]. Farzadfard et al. demonstrated the activation of genes using orthogonal gRNAs in both natural and synthetic promoters. In addition, small molecule triggered activation was achieved by modifying the pRPR1 promoter with an anhydrotetracycline (aTc) inducible site which would be stimulated in the presence of aTc. Therefore, in the presence of aTc, there was a 20-fold increase in blue fluorescent protein expression compared to aTc null controls [61]. Cheng et al. delivered multiple gRNAs for multiple genes and demonstrated the ability for multiplexed endogenous gene activation when dCas9 was delivered with multiple gRNAs for different genes. In addition, by altering the ratios of gRNA, the relative levels of expression were tuned. It was shown that the dCas9-TF system can work in vivo when injected into the cytoplasm of mouse zygotes along with an EGFP reporter construct containing the promoter region of Nanog [62].
Increasing Activation
Konermann et al. were able to use the dCas9-VP64 system with protein interacting aptamers fused to the sgRNA to facilitate the recruitment of effector domains. Plasmids for both dCas9-VP64 as well as MS2-p65 were transfected into HEK293 and A375 cells. This allowed for multiple distinct effectors to be bound to dCas9, modeled after the natural transcription activation process. A screen of potential gRNAs to impart resistance to BRAF inhibitors, drugs designed to inhibit the mutated form of B-raf found in many cancers, was also achieved and the top hits were able to confer resistance to BRAF inhibitors in cell lines as well as patient-derived samples, showing the ability of Cas9 as a therapeutic strategy for stem cell-based therapies [63]. Later, Chavez et al. demonstrated the rational design of a chimeric activation domain composed of the VP64 activation domain fused to p65 and Rta activation domains connected by glycine serine linkers. The new activation domain, termed VPR, showed significantly higher activation, up to 320 fold, compared to VP64 alone. This chimeric protein is a step toward solving the low activation efficiency seen by typical dCas9-VP64 transcription factors. Chavez was able to then demonstrate the usefulness of this technology by increasing the differentiation of induced pluripotent stem cells, an attractive candidate for stem cell transplantation, into induced neurons by activating NGN2 and NEUROD1 using dCas9-VPR [64]. Lastly, Cheng et al. developed a Casilio system that is based on the RNA binding protein Pumilio. Using this system they were able to achieve multiplexing and multimerization of several different types of effector domains including: activation domains, epigenetic modifiers, and fluorescent proteins. Using this system, they were able to achieve multimerization in a much easier manner, as well as, achieve robust gene expression compared to traditional fusion protein [65].
Epigenetic Targeting
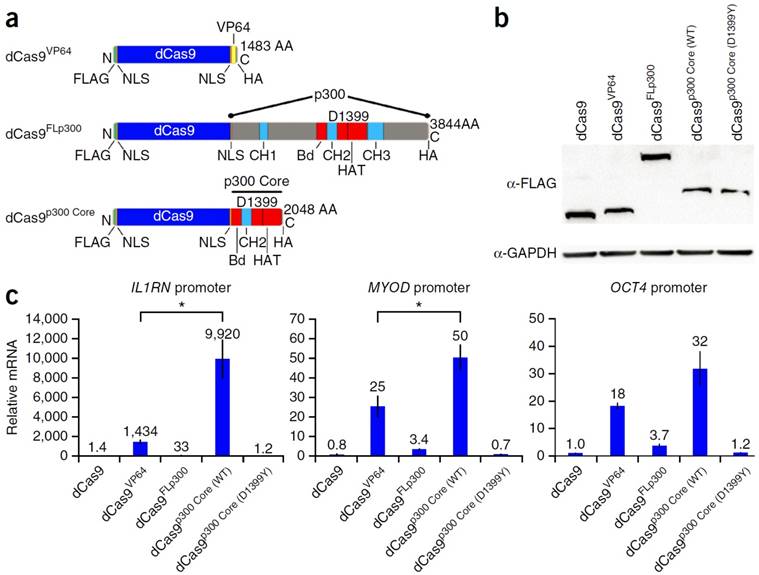
Hilton et al. demonstrated the ability to fuse the catalytic core of the p300 histone acetyl transferase to the dCas9. This allowed for the site-specific acetylation of the H3K27 and thus the transactivation of the gene. Compared to the typical dCas9-VP64, the dCas9-p300core was able to show higher upregulation transcription, demonstrating the efficiency of the p300 core mediating gene activation in the promoter of multiple genes. Figure 5 outlines the work done by Hilton et al. detailing the scheme of the different dCas9 systems tested, including: Cas9-VP64, Cas9-P300, and Cas9-P300core. RT-PCR analysis showed that the p300 core upregulated the IL1RN gene 9,920 fold, the MYOD gene 50 fold, and the OCT4 gene 32 fold. This is significantly higher than the Cas9-VP64 which upregulated the genes by 1,434, 25, and 18 fold respectively (Figure 5) [66]. Vojta et al. utilized the dCas9 system fused to the DNMT3A DNA methyltransferase catalytic domain. By doing so, they were able to use the gRNA to direct site-specific DNA methylation to control gene transcription. This system was able to increase the methylation in the promoter region of the IL6ST gene as well as the BACH2 gene. Along with the increase in DNA methylation, there was a repression of gene expression as measured by qPCR. IL6ST and BACH2 are known to be involved in inflammation and autoimmune diseases, and therefore this novel Cas9-DNMT system can potentially have implications for the study of treatment of these diseases [67]. McDonald et al. utilized a Cas9 DNMT3a fusion to target the CpG island at the CDKN2A promoter. The tumor suppressor gene CDKN2A is one of the most frequently hypermethylated regions in cancer, thus inhibiting the tumor suppressor gene. When transfected into HEK293T the methylation of the promoter region of CDKN2A was increased by 43%. The mRNA expression was reduced by 30% and the protein expression was reduced by 26% most likely due to Cas9 based inhibition. This was a proof of concept paper that showed dCas9 fused to DNA methyltransferase domains could be used to study DNA methylation of endogenous genes involved in cancer development [68]. Stepper et al. utilized a more potent epigenetic effector domain consisting of a fusion of the DNMT3a C-terminal domain with the C-terminal domains of DNMT3l. They showed that the fusion protein was able to increase methylation up to 4.6 times higher compared to DNMT3a alone fused to the dCas9. In addition, this increase in DNA methylation led to a 2 fold reduction of mRNA when targeted to the promoter region of the EpCAM gene in Skov-3 cells and CXCR4 and TFRC genes in HEK 293T cells [69].
Therapeutic & Differentiation Applications
Kearns et al. utilized dCas9 to study its effect on the differentiation of both embryonic stem cell cultures and induced pluripotent stem cells. dCas9 was fused to either the VP64 activation domain or the KRAB repression domain to control gene expression. Guide RNAs for the gene SOX17, a gene involved in endoderm differentiation, were treated with the dCas9-VP64 fusion to embryonic stem cells. An increase in the SOX17 gene was observed as well as an accumulation of the SOX17 protein shown by immunofluorescence. Next, the dCas9-KRAB system was treated to pluripotent stem cells with gRNA targeting OCT4, a gene involved in maintaining pluripotency. When the dCas9-KRAB system was treated, OCT4 was significantly repressed. In addition, several markers for pluripotency could not be detected using immunofluorescence indicating that the pluripotent stem cells had begun differentiating [70]. Balboa et al. also tested the differentiation capabilities of the dCas9 system. First, they delivered a dCas9-VP192 construct targeted to the OCT4 promoter in fibroblast cells using electroporation to test whether this system could replace ectopically expressed OCT4 in fibroblast reprogramming. In addition, they delivered plasmids to ectopically express KLF4, SOX2, and c-MYC. They were able to show that the dCas9 system was able to generate IPSC colonies as efficiently as the ectopically expressed OCT4. Next, they used dCas9-VP192 to activate FOXA2, SOX17, PDX1, and NKX6.1, all genes that are involved in endoderm and pancreatic differentiation, in IPSCs. They were able to differentiate IPSC colonies into cells expressing pancreatic progenitor cells. This demonstration showed the ability of dCas9 to induce directed differentiation as de-differentiation [71]. Using the VPR technology, Lin et al. demonstrated the use of dCas9-VPR in Drosophila cells as well as in Drosophila in vivo. This was done by generating transgenic flies that expressed the dCas9-VPR as well 2 distinct sgRNA. The dCas9-TF was used to activate both Twist and Snail in vitro. Wg was activated in vivo to physiologically relevant levels by generating transgenic flies expressing transgenic 10X-UAS:3xFLAG-Cas9-VP64 and VPR constructs. This created partial duplication of the wing pouch and other patterning abnormalities caused by Wg overexpression, thus showing dCas9-VPR's applicability in multicellular organisms [72].
Conclusions
While eZFPs, TALEs, and dCas9 all have been developed to alter the genome or control gene expression they each have their own unique advantages. While many believe that dCas9 is the most advanced system and will replace eZFPs and TALEs all together, this may not be the case. Studies, such as those done by Gao et al., show that there may be more nuanced details that need to be explored. They showed that in their system the dCas9 was less efficient at reprogramming somatic cells to iPSCs compared the TALE system [73]. However, it is indeed clear that the advantages of the dCas9 system are remarkable including: its ability to multiplex, its ease of scalability, as well as its cost efficiency making it arguably the most promising future for the field of engineered transcription factors.
Schematic diagram of dCas9 DNA binding domain fused to activation domain, p300, and p300 core (a). Western blot in cells transfected with the dCas9 fusion proteins (b). Relative mRNA expression of IL1RN, MYOD, and OCT4 in HEK293T cells (c). Adapted by permission from Macmillan Publishers Ltd: Nature Biotechnology (66) copyright (2015)
These examples of engineered transcription factors demonstrate the wide array of tools that have been developed to control cellular fate. Engineered transcription factor research provides the framework for future advancements that can drastically change the field of stem cell replacement therapy. By having control over stem cells, researchers and clinicians can significantly mitigate issues such as tumorigenesis and unguided differentiation. In addition, engineered transcription factor has the ability to create a virtually unlimited cell sources where patient's skin or stem cells can be harvested and be differentiated or transdifferentiated into the desired cell phenotype to treat a variety of diseases.
Stimuli-Responsive Transcription Factors
While current methods of inducing gene expression are extraordinary, one limitation of these techniques is that lack of spatial and temporal control of gene regulation after induction. To gain spatial-temporal control over the delivery and activation of engineered transcription factor, scientists and bioengineers have developed stimuli-responsive transcription factors to achieve such feat. This portion of the review will focus on the design and generation of light inducible transcription factors to alter gene expression.
One of the first methods used to optogenetically control gene expression inside of a cell was proposed by Ye et al. in 2011. This research group took advantage of the fact that the transcription factor nuclear factor of activated T cells (NFAT) relied on calcineurin to activate the transcription factor. Cells were co-transfected with a melanopsin expression vector and the luciferase reporter construct for NFAT. Melanopsin is known to trigger a calcium response inside the cell when exposed to blue light and therefore is thought to induce NFAT activity when stimulated. After cells were transfected, they were exposed for 24 hours to blue-pulse light with 5 seconds on and 10 seconds off at a power of 1.5*10^8 photons s-1m-2 [74]. As a proof of concept study, secreted alkaline phosphatase (SEAP) expression vectors were used and their cells were exposed to blue light for 48 hours with 5 seconds on and 10 seconds off. By 24 hours the levels of SEAP had already reached maximum levels. This proved that the system was able to work well and efficiently activate the target gene. In addition, the amount of SEAP could be tuned by irradiating the sample for varying amounts of time. To validate the therapeutic use of their design, the group also tested the system in mice with glucagon-like peptide 1, a glucose homeostasis peptide. This was achieved by subcutaneously injecting HEK293 cells, which constitutively expressed melanopsin and had a NFAT driven glucagon-like peptide 1 expression vector, in both healthy and diabetic mice. Significant increase in the glucagon-like peptide was observed in both healthy and diabetic mice. The mice were exposed to blue light pulses for 48 hours and showed an improvement in glucose homeostasis measured by increased insulin levels as well as a lower glycemic excursion when the mice were injected with glucose [74]. Binder et al demonstrated the light-based control of gene expression using a photocaged IPTG the inducer of expression in a T7 RNA polymerase expression system. When exposed to light the IPTG would be uncaged and could induce expression in the E. coli. This can be advantageous as it can be used to trigger a gradual response to get specific dose-dependent expression. In addition, using this photocaged IPTG, a light responsive bacterial expression system was made that could be demonstrated at the single cell level using microfluidics [75].
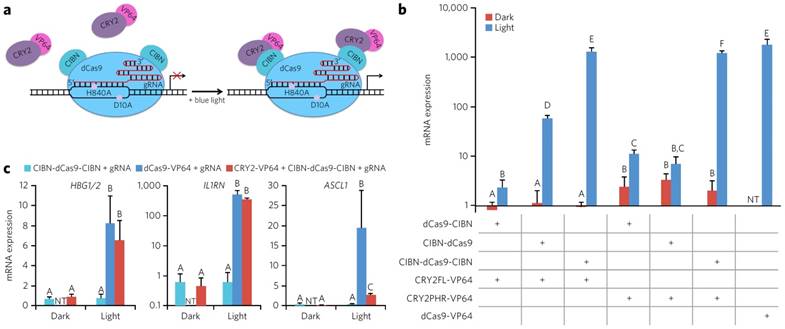
The other main design for the use of optically controlled transcription factors is the use of fusion proteins. In this strategy, a DNA binding protein is fused to one-half of an optogenetic pair, while an activation of transcription domain is fused to the other half. The creation of mutant fusion proteins is feasible using standard molecular biology techniques and cloning the fusion protein into an expression vector and expressing the protein inside of a cell. This design allows for the homo- and hetero-dimerization of the optogenetic pairs after induction with blue light bringing the activation domain and the DNA binding domain together, allowing the construct to initiate transcription. One of the first demonstrations of optogenetic gene switch was by Yazawa et al. Briefly, they fused Flavin-binding kelch repeat (FKF1) to a VP16 activation domain, and fused Gigantea (GI) to the Gal4 DNA binding domain. When irradiated with blue light, the GI and FKF1 heterodimerized bringing Gal4 and VP16 together thus allowing transcription. The design was shown to be able to induce a 5-fold increase in luciferase transcription [76]. In a similar approach, Konermann et al. developed a CRY2-CIB1 interaction that is found in A. thaliana into an artificial transcription factor. CRY2 protein was fused to the C-terminus of a TALE [44, 77]. The fused system was transfected into primary cortical neurons to target various genes. As much as a 7-fold increase when illuminating with blue light compared to the control was observed. In addition, this group was able to create a mutation in the NLS portion of the TALE which significantly reduced the background expression of NGN2 compared to controls with the NLS. The high level of gene activation and low level of background made the system a very robust and highly effective tool. Furthermore, epigenetic modifiers were attached to the fusion protein to either methylate or deacetylate the histone at the target location. Using this system, up to a two-and-a-half-fold increase in methylation on H4K20me3 and H3K27me3 and sixty percent reduction of H4K8Ac were observed. This demonstration offers a new way to control the expression of genes using an epigenetic approach rather than transcriptional to alter the histone structure and epigenetic landscape of the genome. Experiments were carried out primarily in primary mouse neurons as well as in vivo highlighting the ability to utilize optogenetically active TFs in primary cells for therapeutic purposes [77]. A slightly different approach proposed by Wang et al. involves the VVD protein from Neurospora crassa. Wang proposed a fusion of half the Gal4 DNA binding domain, termed Gal4 (64), with the light oxygen voltage sensing domain (LOV domain) of VVD, and the p65 activation domain. When irradiated with blue light, the LOV domain of VVD homodimerized bringing the Gal4 (64) pieces together which then also dimerized and allowed DNA binding. After transfecting to HEK 293 cells with the fusion protein as well as a Gaussia princeps luciferase reporter plasmid, a 13 and 81-fold increase in luciferase gene expression was when exposed to light for .5 and 1 hour respectively compared to samples that were not exposed to light [78]. Motta-Mena et al. developed a method to reduce the time required for the optogenetic switch to be shut off. Using the protein EL222, a transcription factor found in bacteria, as well as a VP16 domain, allowed for a light inducible switch that shut off in less than a minute after irradiation was stopped. EL222 contains a DNA binding domain as well as a LOV domain. Under exposure to blue light, the EL222 protein dimerizes and allows for DNA recognition. When illuminated, this domain was able to have a greater than 100 fold increase in Renilla luciferase gene activation from a reporter plasmid. However, one of the main important points of this demonstration was the ability to have a relatively short time scale of activation and deactivation kinetics. Activation was observed in less than 10 seconds, and deactivation was observed in less than 50 seconds. The EL222 based TF provides much greater temporal control over the activation of transcription compared to previous designs. A T cell-derived Jurkat splicing line 1 (JSL1) cell line was created that stably expressed both the EL222 protein as well as a flag-tagged CUPGB Elav-like family 2 (CELF2) plasmid controlled by a EL222 promoter. In the dark there was no expression of CELF2, however, in the light there was a moderate expression of the protein. The system was also tested in zebrafish embryos, where the EL222 TF mRNA and an EL222 mCherry reporter plasmid were injected into the embryo at a single cell stage. When exposed to light the developing zebrafish expressed mCherry, whereas when left in the dark no mCherry was observed. In short, this group was able to create a platform that was nontoxic and had much greater temporal control of transcription compared to other designs [79]. Jayaraman et al. described the use of a photoactive EL222 DNA binding domain. This technique showed a minimal expression in the dark state with a 5 fold increase in expression in the light state in an E. Coli system [80]. Muller et al. created an On/Off switch system using the phytochrome B and phytochrome interacting factor 6(PIF6) from A. thaliana. PIF6 was fused to a TETR DNA binding domain and a VP16 activation domain was fused to PhyB. This allowed an On/Off switch of the transcription factor virtually instantaneously when exposed to different wavelengths of light. In addition, this technique uses low-energy light which reduces concerns about protein stability as well as the health of the cells both in vitro and in vivo. In addition, the longer wavelength light has a much greater penetration depth, and therefore is more suited for in vivo application where UV and blue light may not be able to reach. The longer wavelength light allows stem cell based therapies to have temporal control of gene expression in vivo [81]. Polstein et al. have engineered a CRISPR-Cas9 that can be light activated as well. Using the CRY2-CIB1 binding partners, they fused activation domains to the catalytically inactive dCas9. This allowed for significantly enhanced gene expression when exposed to blue light for multiple genes in HEK 293 cells. A schematic diagram of dCas9 binding to DNA, as well as CIBN recruiting CRY2 when exposed to blue light is depicted in Figure 6a. In addition, various orientation and truncations of the Cas9-CIBN-CRY2 system were tested and treated to cells targeting the IL1RN promoter under both light and dark conditions. The mRNA was studied using qPCR and presented in Figure 6b. Lastly, two other genes were tested including HGB1/2 and ASCL1. For both HGB1/2 and IL1RN the Cas9-CIBN-CRY2 system, when exposed to light, worked almost as well as a constitutively active Cas9-VP64 fusion targeting the same genomic locus. In addition, low background signal was detected for the samples kept in the dark (Figure 6) [82].
Much of this information has been previously reviewed by Muller et al. as well as a more in-depth review of the use of optogenetic proteins by Tischer et al. [83, 84].
Schematic diagram of dCas9 DNA binding domain fused to the CIBN optogenetic fusion protein and the VP64 activation domain fused to the CRY2 fusion protein. Under blue light irradiation the binding partners fuse and bring the dCas9 and VP64 together to initiate transcription. (a). Relative mRNA expression of IL1RN in HEK293 cells under various fusion protein conditions under exposure to blue light or being kept in dark. Variations were made by changing the binding of the C or N terminus as well as using a mutated CRY2 fusion protein (b). Relative mRNA expression of various genes tested with our without the CRY2 binding partner and comparing it to dCas9-VP64 control (c). Adapted by permission from Macmillan Publishers Ltd: Nature Chemical Biology (82) copyright (2015)
Using these light responsive engineered transcription factors, complex cellular patterns can be made as well as enhanced spatial and temporal control of cellular behavior both in vitro and in vivo. In many therapeutic approaches, temporal control of protein expressions is crucial. Light responsive transcription factors are one avenue to gain greater temporal control of gene activation post-delivery of stem cells for therapeutic strategies such as cancer therapy or regenerative medicine.
Synthetic Transcription Factors
The use of engineered transcription factors has drastically changed the way that researchers can study and manipulate the human genome. However, as a therapeutic strategy, engineered transcription factors are limited by commonly used viral-based delivery methods. Viral based delivery, currently, is not approved for therapeutic application due to the risks associated with genomic integration, mutagenesis, and general cytotoxicity. To overcome these critical challenges associated with viral delivery, synthetic transcription factors have been developed to promote the translational potential of transcription factor-based therapies. Since synthetic transcription factors are small molecule or nanoparticle-based, they can bypass safety and other concerns that are raised by viral-based delivery methods by not integrating into the DNA and having low cytotoxicity through the use of biocompatible nanomaterials.
Small Molecule Mimics
One of the early examples of synthetic transcription factor mimics was pyrrole-imidazole polyamides. These small molecules can recognize different base pair sequences in the minor groove of DNA by altering the pyrrole-imidazole order in the polyamide [85]. In 1997, Gottesfeld et al. demonstrated the use of pyrrole-imidazole polyamides to inhibit gene expression of endogenous genes by binding to the TFIIIA binding site to interfere gene expression. While this methodology does not mimic the exact mechanism of transcription factors, by using this competitive binding, Gottesfeld was able to alter the gene expression in the same way repressive transcription factor would [86]. Mapp and colleagues then fused a hairpin polyamide to the amphipathic helix, a different peptide shown capable of activating genes [87-89]. By doing this, they were able to target the promoter region of a template plasmid and were able to show a 13 fold enhancement of activated transcription over basal levels of the template plasmid [90]. Kwon et al. utilized a hairpin polyamide to function as a DNA-binding domain and conjugated this to Wrenchnolol, which is a non-peptidic activation domain known to bind to the Sur-2 subunit of the human mediator complex [91]. While this molecule did not have that high a cell permeability, it did demonstrate that a fully non-peptidic structure can be used to make an artificial transcription factor [92]. Xiao et al. demonstrated the use of a hairpin polyamide fused to a new activation peptoid termed carboxyfluoresceinated peptoid 3. This combination structure was able to enter cells with a high efficiency as well as initiate the transcription of a reporter plasmid that has a luciferase reporter gene. A dose-dependent increase in luciferase expression was observed with a maximum of a fivefold increase [93]. More recently, Pandian et al. demonstrated that pyrrole-imidazole polyamides can be adapted with epigenetic modifying agents such as SAHA. One such compound was termed δ. This compound was able to efficiently upregulate genes such as Oct 3/4 and Nanog as well as alter Rex1, Cdh1, Zeb2 and other genes involved in the mesenchymal to epithelial transition of cells [94].
Alternatively, Kuznetsova et al. designed a triplex-forming oligonucleotide (TFO) that was then fused to the VP16 activation domain to induce transcription [95]. TFOs are oligonucleotides that can bind to the major groove of DNA and are generally used to inhibit transcription similar to the earlier described polyamide [96-100]. After HOBT ester activation of the TFO, Kuznetsova reported a 3-fold increase in luciferase expression demonstrating an artificial transcription factor can be synthesized to upregulate gene expression [95]. On the other hand, Stanojevic and colleagues developed another artificial transcription factor that combined the use of a TFO conjugated to a synthetic fragment of the VP16 activation domain through the use of a bifunctional linker. This peptide TFO conjugate was able to show a 30-fold enhancement of the template plasmid in HeLa and BHK-21 cell lines proving the ability of artificial transcription factor to upregulate genes when delivered to cells in vitro even without the use of transfection agents such as lipofectamine [101].
Small molecule transcription factors, while being non-viral and clearly defined, suffer from disadvantages such as relatively low cell permeability and gene enhancement. To overcome these concerns, nanoparticle-based transcription factors offer a unique approach and have gained significant traction. The goal of these nanoparticle constructs is to mimic the function of transcription factors but improve cellular and nuclear uptake in a non-viral manner to increase synthetic transcription factors delivery and efficacy as well. This nanoparticle-based transcription factor mimic has been termed, NanoScript.
NanoScript
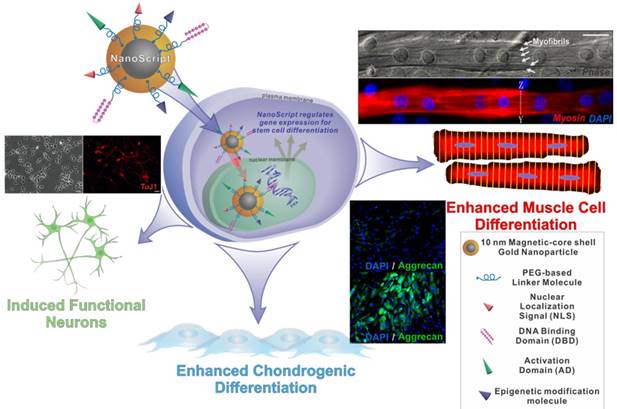
NanoScript is a next generation artificial nanoparticle-based synthetic transcription factor. The goal of NanoScript is to mimic both the structure and function of natural transcription factors by incorporating the key domains that are found in natural transcription factors using a nanoparticle as a linker domain. Hairpin polyamides were used as DNA binding domain, while a tandem repeat of VP16 was used as an activation domain, and the peptide NLS from the SV40 large T-antigen was used as a nuclear localization signal. Patel et al. first synthesized 10 nm gold nanoparticles. Mercaptoundecanoic acid was coated onto the nanoparticle, and EDC/NHS coupling was used to conjugate the biomolecules onto the surface of the nanoparticles. Once fully functionalized, the NanoScript nanoparticle system was constructed. The NanoScript system can translocate into the nucleus and directly alter gene expression levels. NanoScript has been demonstrated to induce differentiation into muscle cells, chondrocytes, and functional neurons (Figure 7).
Schematic diagram of NanoScript entering the cell and translocating into the nucleus where it can alter gene expression. NanoScript is composed of a magnetic core gold shell nanoparticle conjugated to the three effector domains, nuclear localization signals, DNA binding domains, and Activation domains. NanoScript has been used to differentiate stem cells into muscle cells, chondrocytes, as well as neurons.
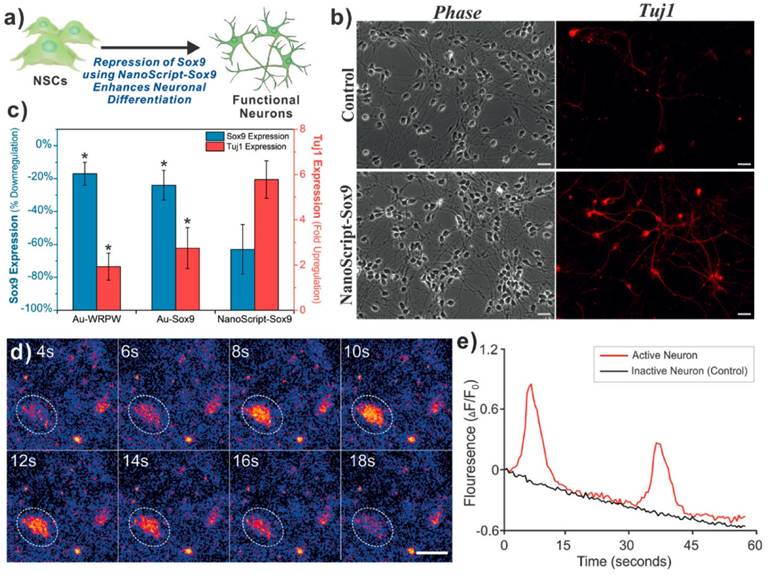
NanoScript was then transfected into cells along with a reporter plasmid. Robust cellular and nuclear localization of NanoScript was observed using 3D-SIM microscopy. In addition, a 15-fold enhancement of reporter expression was seen when transfected with NanoScript compared to control conditions [102]. This demonstration showed that NanoScript can upregulate gene expression of reporters within cells. To further expand their technology, Patel et al. then designed the polyamide to target the consensus sequence of several myogenic genes such as MYOD, MYOGENIN, MYF5, and MRF. This new version of NanoScript was used to activate endogenous genes in stem cells to initiate their differentiation into muscle cells. After 7 days of induction using NanoScript, a significant increase in myogenic gene expression of up to 28-fold increase in MYH1 was observed. This correlated with an increase in the protein expression of myosin and myogenin and the formation of myofibrils quantified using immunostaining. In addition, NanoScript based gene activation was compared to the delivery of MyoD protein using commercially available cationic-based lipid delivery vehicles where a significantly higher increase in gene expression using NanoScript was observed [103]. This demonstration further confirmed the potential of NanoScript to upregulate endogenous genes in a non-viral manner. Additionally, NanoScript has demonstrated its ability to not only able to upregulate gene expression, but also repress gene expression as many natural transcription factors can. A short repressor peptide WRPW was chosen to prevent the formation of the basal transcription machinery at the transcription initiation site and recruit Groucho and TLE family of proteins to inhibit gene expression [104]. With this new repression domain taking the place of the activation domain, the new NanoScript was designed to knockdown the SOX9 gene which is known to be a key neuronal switch gene. When treated with NanoScript, SOX9 was repressed by 60%. In correlation, Tuj1 expression had a 6--fold increase showing the inhibition of SOX9 led to the further differentiation of neural stem/progenitor cells into the neuronal lineage. Calcium imaging was then performed to show the functionality of the induced neurons and the potency of the NanoScript platform to induce differentiation through transcriptional repression [105] (Figure 8).
In one demonstration NanoScript-Sox9 was used to differentiate neural stem cells into functional neurons. Gene expression was determined for NanoScript compared to nanoparticles with just the DNA binding domain, or just the repressor peptide (c). Fluorescent microscopy is used to show TUJ1 staining (b). Calcium imaging was used to show functionality of the induced neurons (d,e). Adapted with permission from Wiley: Angewandte Chemie. (105) Copyright 2015
Finally, to enhance the gene-manipulating efficiency of NanoScript, the researchers added small molecules that can modify the chromatin landscape of the genome making it more accessible to initiate transcription of genes. With the addition of an epigenetic modulator onto NanoScript, the efficiency of the platform was improved. A modified version of the histone acetyltransferase activator, CTB, was synthesized to be conjugated onto the nanoparticle. CTB, known to activate the p300 pathway, has been shown to influence the SOX9 activation in chondrogenesis. When delivered to adipose-derived stem cells (ADSCs), CTB modified NanoScript increased the HAT activity of the cells by 50%. In addition, the CTB loaded NanoScript increased the gene expression of AGGRECAN, SOX9, and COLLAGEN II after 7 days of differentiation, measured by qPCR, as well as increased the level of immunostaining signal [106]. This demonstration showed that the inclusion of an epigenetic modulator has a drastic effect on the efficiency of NanoScript and opens the door for the use of other molecules such as SAHA or 5-azacytadine. NanoScript, an example of next generation synthetic transcription factors having high biocompatibility, high efficiency, and being non-viral, can potentially be translated for further therapeutic applications.
Current Limitations
While there have been significant advancements in the field of engineered and synthetic transcription factors, major hurdles still remain to be overcome before the widespread use of these technologies. First and foremost, the concern over immunogenicity of many of the components of these systems and the delivery vehicles that are used is critical [107]. Another potential pitfall of these technologies, specifically those that fuse a catalytically active domain to a DNA binding domain, is the off-target effects [108]. Catalytically active domains, such as p300, can alter the epigenetic state at locations not targeted by the DNA binding domains. In addition, repressive marks from KRAB domains have been shown to spread past the targeted DNA region. Although it has been shown that these off-target effects are possible through CHiP-seq and epigenome assays, remarkably, transcriptome analysis shows a high specificity in many cases with few or no off-target effects [109, 110]. In addition, one potential solution that is being explored is developing effector domains that are inactive until DNA binding. This will greatly limit the off-target effects of these effector domains.
Conclusions and Future Directions
The field of regenerative medicine is constantly growing. With the growing need for cells, tissues, and organs to treat a variety of diseases, a need for advanced tools to control the growth and differentiation of cells and tissues is abundantly clear. To this end, engineered and synthetic transcription factors can help provide the tools necessary to solve many of these problems. Engineered and synthetic transcription factors provide a platform for the future studies of diseases and disorders as well as the discovery and development of therapeutic strategies. The ability to control any gene or gene network in the body opens up unimaginable opportunity in the fields of regenerative medicine, drug discovery, systems biology, and genetics. As the field of engineered and synthetic transcription factors advances, the ability to completely control cell fate and function will become more of a reality. Advances in the field can lead to a virtually unlimited supply of tailor-made cells for autologous cell transplantation for cell replacement therapy, thus greatly impact the field of stem cell-based regenerative medicine.
As the field of engineered and synthetic transcription factors evolves, we can expect to see major advances in regenerative medicine, specifically stem cell theranostics. Patient cells will be harvested, engineered, and transplanted back to treat devastating diseases such as Alzheimer's, Parkinson's, cancer, and many more. Skin cells will be able to be harvested and converted into iPSCs and grown into organoids to guide patient-specific therapies. Stem cells will be engineered to become fluorescent when they are near sites of disease or tumors. All of these can be made possible by the enhanced control of cellular behavior and phenotype imparted by engineered and synthetic transcription factors. Eventually, the biomedical community will be able to mimic the embryonic development of tissues and organs by controlling the gene expression network of cells to stably and reproducibly generate any cell type in the human body. With all the opportunities, one is certain; the advancement of engineered and synthetic transcription factors can help address the healthcare needs of today and the future.
Abbreviations
DNA: deoxyribonucleic acid; hIPSC: human induced pluripotent stem cell; TFs: Transcription Factors; RNA: ribonucleic acid; eZFP: engineered zinc finger protein; TFIIA: Transcription factor 2 A; erbB-2: Receptor tyrosine-protein kinase; Oct4: octamer-binding transcription factor 4; Sox2: (sex determining region Y)-box 2; Klf4: Kruppel-like factor 4; c-Myc: avian myelocytomatosis viral oncogene homolog; VP16: Herpes simplex virus protein vmw65; VP64: 4 tandem repeats of VP16; p65: Activation domain of the transcription factor p65; VEGF: Vascular endothelial growth factor; DNMT: DNA methyltransferase; TALE: transcription activator like effector; Bs4: Lycopersicon esculentum bacterial spot disease resistance protein 4; EGL3: Enhancer of glabra 3; KNAT: homeobox protein knotted-1-like 1; TET1: Ten-eleven translocation methylcytosine dioxygenase 1; CDKN2A: Cyclin Dependent Kinase Inhibitor 2A; p300: Histone acetyltransferase p300; dCas9: deficient Cas9; gRNA: guide ribonucleic acid; KRAB: The Krüppel associated box; IL1RN: interleukin-1 receptor antagonist; ASCL1: Achaete-Scute Family BHLH Transcription Factor 1; HBG1: Hemoglobin subunit gamma-1; MyoD1: Myogenic Differentiation 1; BEGF: biotinylated epidermal growth factor; TERT: Telomerase reverse transcriptase; IL1B: Interleukin 1 Beta; NTF3: Neurotrophin-3; PRPR1: protease polyprotein ArgI; Atc: anhydrotetracycline; EGFP: enhanced green fluorescent protein; VPR: VP64-p65-rta; NGN2: Neurogenin 2; NeuroD1: Neuronal Differentiation 1; IL6st: Interleukin 6 Signal Transducer; BACH2: BTB Domain And CNC Homolog 2; qPCR: Quantitative polymerase chain reaction; EpCAM: Epithelial cell adhesion molecule; CxCR4: C-X-C chemokine receptor type 4; TFRC: Transferrin receptor protein 1; SOX17: (sex determining region Y)-box 17; FOXA2: Forkhead Box A2; PDX1: insulin promoter factor 1; NKX6.1: NK6 homeobox 1; Wg: Wingless; NFAT: Nuclear factor of activated T-cells; SEAP: secreted alkaline phospatase; FkF1: flavin-binding kelch repeat F box protein; GI: Gigantea; CRY2: Cryptochrome Circadian Clock 2; CIB1: Calcium and integrin-binding protein 1; NLS: Nuclear localization signal; VVD: Vivid PAS protein; LOV: light oxygen voltage domain; JSL1: Jurkat splicing line 1; CELF2: CUPGB Elav-like family 2; PIF6: phytochrome interacting factor 6; TETR: Tetracycline repressor protein; PhyB: Phytochrome B; Rex1: zinc finger protein 42; CDH1: cadherin 1; Zeb2: Zinc Finger E-Box Binding Homeobox 2; TFO: Triplex forming oligonucleotide; HOBT: Hydroxybenzotriazole; SAHA: Suberoylanilide Hydroxamic Acid; MYF5: myogenic factor 5; MRF: myogenic regulatory factor; MYH1: Myosin Heavy Chain 1; TLE: Transducin-like enhancer protein; SOX9: (sex determining region Y)-box 9; CTB: N-(4-Chloro-3-trifluoromethyl-phenyl)-2-ethoxy-benzamide; ADSCs: Adipose derived mesenchymal stem cells; ChIP-seq: chromatin immunoprecipitation-sequencing.
Acknowledgements
This work has been partially financially supported the NIH Director's Innovator Award (1DP20D006462-01), NIH R21 (1R21NS085569-01), New Jersey Commission on Spinal Cord (CSR13ERG005), NSF CHE-1429062, CBET-1236508, American Chemical Society New Directions Award (PRF# 55869-ND10), and the University City Science Center's QED Award. NIH T32 GM008339.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425-32
2. Mali P, Cheng L. Concise review: Human cell engineering: cellular reprogramming and genome editing. Stem cells. 2012;30:75-81
3. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-76
4. Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987-1000
5. Schafer BW, Blakely BT, Darlington GJ, Blau HM. Effect of cell history on response to helix-loop-helix family of myogenic regulators. Nature. 1990;344:454-8
6. Liu Z, Fan H, Li Y, Zheng SG. Experimental Studies on the Differentiation of Fibroblasts into Myoblasts induced by MyoD Genes in vitro. International Journal of Biomedical Science: IJBS. 2008;4:14-9
7. Blankesteijn WM, Creemers E, Lutgens E, Cleutjens JP, Daemen MJ, Smits JF. Dynamics of cardiac wound healing following myocardial infarction: observations in genetically altered mice. Acta physiologica Scandinavica. 2001;173:75-82
8. Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708-11
9. Chen JX, Krane M, Deutsch MA, Wang L, Rav-Acha M, Gregoire S. et al. Inefficient reprogramming of fibroblasts into cardiomyocytes using Gata4, Mef2c, and Tbx5. Circ Res. 2012;111:50-5
10. Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G. et al. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13:215-22
11. Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L. et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593-8
12. Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN. et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599-604
13. Islas JF, Liu Y, Weng KC, Robertson MJ, Zhang S, Prejusa A. et al. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc Natl Acad Sci U S A. 2012;109:13016-21
14. Fu JD, Stone NR, Liu L, Spencer CI, Qian L, Hayashi Y. et al. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem cell reports. 2013;1:235-47
15. Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A. et al. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A. 2011;108:10343-8
16. Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ. et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220-3
17. Caiazzo M, Dell'Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D. et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224-7
18. Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035-41
19. Leong DT, Lim J, Goh X, Pratap J, Pereira BP, Kwok HS. et al. Cancer-related ectopic expression of the bone-related transcription factor RUNX2 in non-osseous metastatic tumor cells is linked to cell proliferation and motility. Breast cancer research: BCR. 2010;12:R89
20. Deniaud E, Baguet J, Chalard R, Blanquier B, Brinza L, Meunier J. et al. Overexpression of transcription factor Sp1 leads to gene expression perturbations and cell cycle inhibition. PloS one. 2009;4:e7035
21. Chen C, Breslin MB, Lan MS. Ectopic expression of a small cell lung cancer transcription factor, INSM1 impairs alveologenesis in lung development. BMC pulmonary medicine. 2016;16:49
22. Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AK, Han MS, Mirkin CA. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027-30
23. Carter PJ. Introduction to current and future protein therapeutics: a protein engineering perspective. Experimental cell research. 2011;317:1261-9
24. Bale SS, Kwon SJ, Shah DA, Banerjee A, Dordick JS, Kane RS. Nanoparticle-mediated cytoplasmic delivery of proteins to target cellular machinery. ACS nano. 2010;4:1493-500
25. Gu Z, Biswas A, Zhao M, Tang Y. Tailoring nanocarriers for intracellular protein delivery. Chemical Society reviews. 2011;40:3638-55
26. Tang R, Kim CS, Solfiell DJ, Rana S, Mout R, Velazquez-Delgado EM. et al. Direct delivery of functional proteins and enzymes to the cytosol using nanoparticle-stabilized nanocapsules. ACS nano. 2013;7:6667-73
27. Choi SO, Kim YC, Lee JW, Park JH, Prausnitz MR, Allen MG. Intracellular protein delivery and gene transfection by electroporation using a microneedle electrode array. Small. 2012;8:1081-91
28. Yan M, Du J, Gu Z, Liang M, Hu Y, Zhang W. et al. A novel intracellular protein delivery platform based on single-protein nanocapsules. Nature nanotechnology. 2010;5:48-53
29. Liu Y, Wang H, Kamei K, Yan M, Chen KJ, Yuan Q. et al. Delivery of intact transcription factor by using self-assembled supramolecular nanoparticles. Angew Chem Int Ed Engl. 2011;50:3058-62
30. Thakore PI, Black JB, Hilton IB, Gersbach CA. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nature methods. 2016;13:127-37
31. Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. The EMBO journal. 1985;4:1609-14
32. Beerli RR, Barbas CF 3rd. Engineering polydactyl zinc-finger transcription factors. Nature biotechnology. 2002;20:135-41
33. Beerli RR, Dreier B, Barbas CF 3rd. Positive and negative regulation of endogenous genes by designed transcription factors. Proc Natl Acad Sci U S A. 2000;97:1495-500
34. Ji Q, Fischer AL, Brown CR, Eastlund ER, Dvash T, Zhong B. et al. Engineered zinc-finger transcription factors activate OCT4 (POU5F1), SOX2, KLF4, c-MYC (MYC) and miR302/367. Nucleic acids research. 2014;42:6158-67
35. Snowden AW, Zhang L, Urnov F, Dent C, Jouvenot Y, Zhong XH. et al. Repression of vascular endothelial growth factor A in glioblastoma cells using engineered zinc finger transcription factors. Cancer Res. 2003;63:8968-76
36. Cui C, Gan Y, Gu L, Wilson J, Liu Z, Zhang B. et al. P16-specific DNA methylation by engineered zinc finger methyltransferase inactivates gene transcription and promotes cancer metastasis. Genome Biol. 2015;16:252
37. Li F, Papworth M, Minczuk M, Rohde C, Zhang Y, Ragozin S. et al. Chimeric DNA methyltransferases target DNA methylation to specific DNA sequences and repress expression of target genes. Nucleic acids research. 2007;35:100-12
38. Rivenbark AG, Stolzenburg S, Beltran AS, Yuan X, Rots MG, Strahl BD. et al. Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics. 2012;7:350-60
39. Dai Q, Huang J, Klitzman B, Dong C, Goldschmidt-Clermont PJ, March KL. et al. Engineered zinc finger-activating vascular endothelial growth factor transcription factor plasmid DNA induces therapeutic angiogenesis in rabbits with hindlimb ischemia. Circulation. 2004;110:2467-75
40. Graslund T, Li X, Magnenat L, Popkov M, Barbas CF 3rd. Exploring strategies for the design of artificial transcription factors: targeting sites proximal to known regulatory regions for the induction of gamma-globin expression and the treatment of sickle cell disease. The Journal of biological chemistry. 2005;280:3707-14
41. Kay S, Hahn S, Marois E, Hause G, Bonas U. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science. 2007;318:648-51
42. Romer P, Recht S, Strauss T, Elsaesser J, Schornack S, Boch J. et al. Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. The New phytologist. 2010;187:1048-57
43. Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S. et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509-12
44. Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501
45. Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annual review of phytopathology. 2010;48:419-36
46. Morbitzer R, Romer P, Boch J, Lahaye T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc Natl Acad Sci U S A. 2010;107:21617-22
47. Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nature biotechnology. 2011;29:149-53
48. Gao X, Yang J, Tsang JC, Ooi J, Wu D, Liu P. Reprogramming to pluripotency using designer TALE transcription factors targeting enhancers. Stem cell reports. 2013;1:183-97
49. Perez-Pinera P, Ousterout DG, Brunger JM, Farin AM, Glass KA, Guilak F. et al. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nature methods. 2013;10:239-42
50. Maeder ML, Linder SJ, Reyon D, Angstman JF, Fu Y, Sander JD. et al. Robust, synergistic regulation of human gene expression using TALE activators. Nature methods. 2013;10:243-5
51. Bultmann S, Morbitzer R, Schmidt CS, Thanisch K, Spada F, Elsaesser J. et al. Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic acids research. 2012;40:5368-77
52. Bernstein DL, Le Lay JE, Ruano EG, Kaestner KH. TALE-mediated epigenetic suppression of CDKN2A increases replication in human fibroblasts. J Clin Invest. 2015;125:1998-2006
53. Hu J, Lei Y, Wong WK, Liu S, Lee KC, He X. et al. Direct activation of human and mouse Oct4 genes using engineered TALE and Cas9 transcription factors. Nucleic acids research. 2014;42:4375-90
54. Tremblay JP, Chapdelaine P, Coulombe Z, Rousseau J. Transcription activator-like effector proteins induce the expression of the frataxin gene. Human gene therapy. 2012;23:883-90
55. Barbon E, Pignani S, Branchini A, Bernardi F, Pinotti M, Bovolenta M. An engineered tale-transcription factor rescues transcription of factor VII impaired by promoter mutations and enhances its endogenous expression in hepatocytes. Scientific reports. 2016;6:28304
56. Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709-12
57. Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173-83
58. Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442-51
59. Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR. et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nature methods. 2013;10:973-6
60. Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nature methods. 2013;10:977-9
61. Farzadfard F, Perli SD, Lu TK. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS synthetic biology. 2013;2:604-13
62. Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW. et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell research. 2013;23:1163-71
63. Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583-8
64. Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, E PRI. et al. Highly efficient Cas9-mediated transcriptional programming. Nature methods. 2015;12:326-8
65. Cheng AW, Jillette N, Lee P, Plaskon D, Fujiwara Y, Wang W. et al. Casilio: a versatile CRISPR-Cas9-Pumilio hybrid for gene regulation and genomic labeling. Cell research. 2016;26:254-7
66. Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE. et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nature biotechnology. 2015;33:510-7
67. Vojta A, Dobrinic P, Tadic V, Bockor L, Korac P, Julg B. et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic acids research. 2016;44:5615-28
68. McDonald JI, Celik H, Rois LE, Fishberger G, Fowler T, Rees R. et al. Reprogrammable CRISPR/Cas9-based system for inducing site-specific DNA methylation. Biol Open. 2016;5:866-74
69. Stepper P, Kungulovski G, Jurkowska RZ, Chandra T, Krueger F, Reinhardt R. et al. Efficient targeted DNA methylation with chimeric dCas9-Dnmt3a-Dnmt3L methyltransferase. Nucleic acids research. 2016;45:1703-13
70. Kearns NA, Genga RM, Enuameh MS, Garber M, Wolfe SA, Maehr R. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141:219-23
71. Balboa D, Weltner J, Eurola S, Trokovic R, Wartiovaara K, Otonkoski T. Conditionally Stabilized dCas9 Activator for Controlling Gene Expression in Human Cell Reprogramming and Differentiation. Stem cell reports. 2015;5:448-59
72. Lin S, Ewen-Campen B, Ni X, Housden BE, Perrimon N. In Vivo Transcriptional Activation Using CRISPR/Cas9 in Drosophila. Genetics. 2015;201:433-42
73. Gao X, Tsang JC, Gaba F, Wu D, Lu L, Liu P. Comparison of TALE designer transcription factors and the CRISPR/dCas9 in regulation of gene expression by targeting enhancers. Nucleic acids research. 2014;42:e155
74. Ye H, Daoud-El Baba M, Peng RW, Fussenegger M. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science. 2011;332:1565-8
75. Binder D, Grunberger A, Loeschcke A, Probst C, Bier C, Pietruszka J. et al. Light-responsive control of bacterial gene expression: precise triggering of the lac promoter activity using photocaged IPTG. Integrative biology: quantitative biosciences from nano to macro. 2014;6:755-65
76. Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Induction of protein-protein interactions in live cells using light. Nature biotechnology. 2009;27:941-5
77. Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L. et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472-6
78. Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nature methods. 2012;9:266-9
79. Motta-Mena LB, Reade A, Mallory MJ, Glantz S, Weiner OD, Lynch KW. et al. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nature chemical biology. 2014;10:196-202
80. Jayaraman P, Devarajan K, Chua TK, Zhang H, Gunawan E, Poh CL. Blue light-mediated transcriptional activation and repression of gene expression in bacteria. Nucleic acids research. 2016;44:6994-7005
81. Muller K, Zurbriggen MD, Weber W. Control of gene expression using a red- and far-red light-responsive bi-stable toggle switch. Nat Protoc. 2014;9:622-32
82. Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nature chemical biology. 2015;11:198-200
83. Müller K, Naumann S, Weber W, Zurbriggen Matias D. Optogenetics for gene expression in mammalian cells. Biological Chemistry. 2015 p. 145
84. Tischer D, Weiner OD. Illuminating cell signalling with optogenetic tools. Nature reviews Molecular cell biology. 2014;15:551-8
85. Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Current opinion in structural biology. 2003;13:284-99
86. Gottesfeld JM, Neely L, Trauger JW, Baird EE, Dervan PB. Regulation of gene expression by small molecules. Nature. 1997;387:202-5
87. Ma J, Ptashne M. A new class of yeast transcriptional activators. Cell. 1987;51:113-9
88. Gerber HP, Seipel K, Georgiev O, Hofferer M, Hug M, Rusconi S. et al. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 1994;263:808-11
89. Giniger E, Ptashne M. Transcription in yeast activated by a putative amphipathic alpha helix linked to a DNA binding unit. Nature. 1987;330:670-2
90. Mapp AK, Ansari AZ, Ptashne M, Dervan PB. Activation of gene expression by small molecule transcription factors. Proc Natl Acad Sci U S A. 2000;97:3930-5
91. Uesugi M. Synthetic molecules that modulate transcription and differentiation: Hints for future drug discovery. Comb Chem High T Scr. 2004;7:653-9
92. Kwon Y, Arndt HD, Mao Q, Choi Y, Kawazoe Y, Dervan PB. et al. Small molecule transcription factor mimic. J Am Chem Soc. 2004;126:15940-1
93. Xiao X, Yu P, Lim HS, Sikder D, Kodadek T. A cell-permeable synthetic transcription factor mimic. Angew Chem Int Ed Engl. 2007;46:2865-8
94. Pandian GN, Nakano Y, Sato S, Morinaga H, Bando T, Nagase H. et al. A synthetic small molecule for rapid induction of multiple pluripotency genes in mouse embryonic fibroblasts. Scientific reports. 2012;2:544
95. Kuznetsova S, Ait-Si-Ali S, Nagibneva I, Troalen F, Le Villain JP, Harel-Bellan A. et al. Gene activation by triplex-forming oligonucleotide coupled to the activating domain of protein VP16. Nucleic acids research. 1999;27:3995-4000
96. Maher LJ. Prospects for the therapeutic use of antigene oligonucleotides. Cancer Invest. 1996;14:66-82
97. Giovannangeli C, Diviacco S, Labrousse V, Gryaznov S, Charneau P, Helene C. Accessibility of nuclear DNA to triplex-forming oligonucleotides: The integrated HIV-1 provirus as a target. P Natl Acad Sci USA. 1997;94:79-84
98. Strobel SA, Doucette-Stamm LA, Riba L, Housman DE, Dervan PB. Site-specific cleavage of human chromosome 4 mediated by triple-helix formation. Science. 1991;254:1639-42
99. Strobel SA, Dervan PB. Single-site enzymatic cleavage of yeast genomic DNA mediated by triple helix formation. Nature. 1991;350:172-4
100. Grigoriev M, Praseuth D, Guieysse AL, Robin P, Thuong NT, Helene C. et al. Inhibition of Gene-Expression by Triple Helix-Directed DNA Cross-Linking at Specific Sites. P Natl Acad Sci USA. 1993;90:3501-5
101. Stanojevic D, Young RA. A highly potent artificial transcription factor. Biochemistry. 2002;41:7209-16
102. Patel S, Jung D, Yin PT, Carlton P, Yamamoto M, Bando T. et al. NanoScript: a nanoparticle-based artificial transcription factor for effective gene regulation. ACS nano. 2014;8:8959-67
103. Patel S, Yin PT, Sugiyama H, Lee KB. Inducing Stem Cell Myogenesis Using NanoScript. ACS nano. 2015;9:6909-17
104. Fisher AL, Ohsako S, Caudy M. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol Cell Biol. 1996;16:2670-7
105. Patel S, Chueng ST, Yin PT, Dardir K, Song Z, Pasquale N. et al. Induction of stem-cell-derived functional neurons by NanoScript-based gene repression. Angew Chem Int Ed Engl. 2015;54:11983-8
106. Patel S, Pongkulapa T, Yin PT, Pandian GN, Rathnam C, Bando T. et al. Integrating epigenetic modulators into NanoScript for enhanced chondrogenesis of stem cells. J Am Chem Soc. 2015;137:4598-601
107. Gaj T, Gersbach CA, Barbas CF 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397-405
108. Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nature biotechnology. 2014;32:677-83
109. Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nature reviews Molecular cell biology. 2016;17:5-15
110. Polstein LR, Perez-Pinera P, Kocak DD, Vockley CM, Bledsoe P, Song L. et al. Genome-wide specificity of DNA binding, gene regulation, and chromatin remodeling by TALE- and CRISPR/Cas9-based transcriptional activators. Genome Res. 2015;25:1158-69
Author contact
![]() Corresponding author: Ki-Bum Lee, Department of Chemistry and Chemical Biology, Rutgers, The State University of New Jersey, Piscataway, NJ 08854 (USA) Email: kbleeedu Website: http://kblee.rutgers.edu/
Corresponding author: Ki-Bum Lee, Department of Chemistry and Chemical Biology, Rutgers, The State University of New Jersey, Piscataway, NJ 08854 (USA) Email: kbleeedu Website: http://kblee.rutgers.edu/
 Global reach, higher impact
Global reach, higher impact