13.3
Impact Factor
Theranostics 2017; 7(16):4057-4070. doi:10.7150/thno.20151 This issue Cite
Research Paper
Multiplexed Nucleic Acid Programmable Protein Arrays
1. State Key Laboratory of Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences (PHOENIX Center, Beijing), Beijing Institute of Radiation Medicine, Beijing, 102206, China;
2. The Virginia G. Piper Center for Personalized Diagnostics, Biodesign Institute, Arizona State University, Tempe, AZ 85287, USA;
3. Department of Microbial Pathogenesis and Immunology, College of Medicine, Texas A&M Health Science Center, College Station, TX 77843, USA;
4. Department of Medicine, Albert Einstein College of Medicine, NY 10461, USA; Department of Microbiology and Immunology, Albert Einstein College of Medicine, Bronx, NY 10461, USA.
* These authors contributed equally to this work.
Abstract
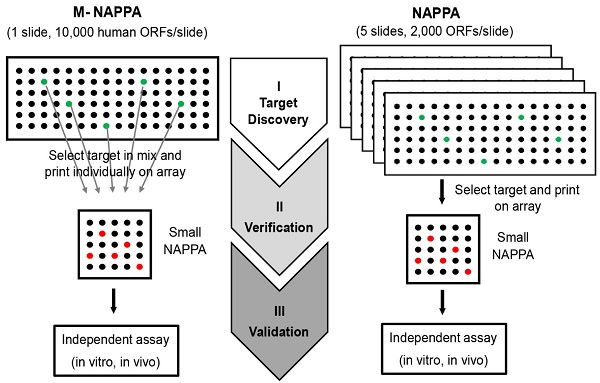
Rationale: Cell-free protein microarrays display naturally-folded proteins based on just-in-time in situ synthesis, and have made important contributions to basic and translational research. However, the risk of spot-to-spot cross-talk from protein diffusion during expression has limited the feature density of these arrays.
Methods: In this work, we developed the Multiplexed Nucleic Acid Programmable Protein Array (M-NAPPA), which significantly increases the number of displayed proteins by multiplexing as many as five different gene plasmids within a printed spot.
Results: Even when proteins of different sizes were displayed within the same feature, they were readily detected using protein-specific antibodies. Protein-protein interactions and serological antibody assays using human viral proteome microarrays demonstrated that comparable hits were detected by M-NAPPA and non-multiplexed NAPPA arrays. An ultra-high density proteome microarray displaying > 16k proteins on a single microscope slide was produced by combining M-NAPPA with a photolithography-based silicon nano-well platform. Finally, four new tuberculosis-related antigens in guinea pigs vaccinated with Bacillus Calmette-Guerin (BCG) were identified with M-NAPPA and validated with ELISA.
Conclusion: All data demonstrate that multiplexing features on a protein microarray offer a cost-effective fabrication approach and have the potential to facilitate high throughput translational research.
Keywords: Cell-free protein microarray, Proteomics, Protein-protein interaction, Antibody, Biomarker.
 Global reach, higher impact
Global reach, higher impact