13.3
Impact Factor
Theranostics 2017; 7(17):4313-4321. doi:10.7150/thno.21804 This issue Cite
Review
Oral Microbiome: A New Biomarker Reservoir for Oral and Oropharyngeal Cancers
1. The School of Biomedical Sciences, Institute of Health and Biomedical Innovations, Queensland University of Technology, 60 Musk Avenue, Kelvin Grove, Brisbane, QLD 4059, Australia;
2. The Translational Research Institute, 37 Kent Street, Woolloongabba, Brisbane, QLD, 4102, Australia;
3. The University of Queensland Diamantina Institute, The University of Queensland, Translational Research Institute, 37 Kent Street, Woolloongabba, Brisbane, QLD, 4102, Australia.
Received 2017-7-7; Accepted 2017-8-23; Published 2017-9-26
Abstract
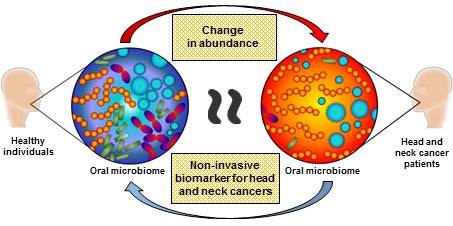
Current biomarkers (DNA, RNA and protein) for oral cavity and oropharyngeal cancers demonstrate biological variations between individuals, rendering them impractical for clinical translation. Whilst these biomarkers originate from the host, there is not much information in the literature about the influence of oral microbiota on cancer pathogenesis, especially in oral cancers. Oral microbiotas are known to participate in disease initiation and progression not only limited to the oral cavity, but also at other distant sites. Due to the close proximity of oral microbiota and oral cavity and oropharyngeal tumours, abundance changes in oral microbiota may provide useful information on tumourigenesis. This review aims to highlight information on the role of oral microbiota in oral cavity and oropharyngeal cancers. An in-depth analysis into the oral microbiota may provide a new avenue to diagnose and treat these patients.
Keywords: biomarker, oral and oropharyngeal cancers, oral microbiome.
Oral Microbiome
Oral microbiome, by definition, is the collective genomes of microorganisms that reside in the oral cavity [1-3]. Many researchers believe that the characterisation of oral microbiome is an essential step in understanding oral health and systemic diseases [2]. The oral cavity has densely populated microbial communities and has the largest core of commonly shared microbes among unrelated individuals [4]. As such, oral microbiome provides an ideal source for biomarker discoveries due to low inter- and intra- biological variations, in contrast to other tumour biomarkers originating from the host. The oral cavity and associated nasopharyngeal regions are also an ideal environment for the growth of microorganisms [5, 6]. The average normal oral temperature is 37°C without significant fluctuation, providing bacteria a stable habitat to thrive [5, 6]. In addition, saliva maintains a stable pH of 6.5 to 7.5, the preferred pH for most bacteria species [5, 6]. Saliva also keeps bacteria hydrated and acts as a medium to facilitate the transportation of nutrients to microorganisms [5, 6]. As such, the oral cavity harbors more than 700 bacterial species and is one of the most densely populated anatomical sites within the human body [5, 6].
There are two types of bacteria, aerobes (oxygen-dependent) and anaerobes (oxygen-independent) [7]. Together, they form multi-species communities, known as biofilms, which resist changes in their environment [7]. This symbiotic manner is known as coaggregation, where aerobes interact with oxygen and create a localised niche for the anaerobes to thrive [7]. Both types of organisms are therefore, essential to maintain the balance of the microbial ecosystem [5-7]. The bacteria in the oral cavity coevolve with each other (both pathogenic and mutualistic bacteria) to coexist in maintaining homeostasis. These bacteria have developed means to communicate with one another and thereby form successful multispecies organisations known as dental plaque [3]. Changes in environmental conditions are known to increase the potential for pathogenicity and promote oral diseases [6].
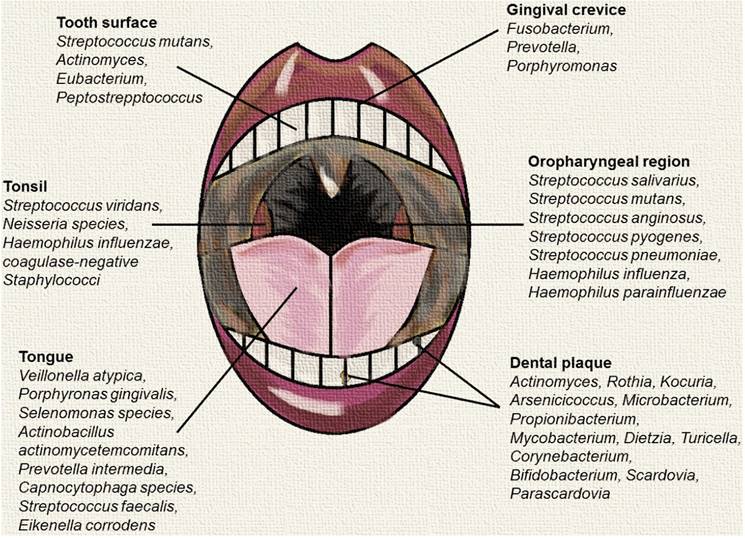
While the oral cavity includes several distinct microbial habitats such as periodontal pockets and the surface of teeth and cheeks, the tongue is the most populated niche and has a significant impact on other regions in the oral cavity [8]. Microbes in the tongue often travel around the oral cavity to colonise other regions, facilitated by saliva [8]. Microbes in the tongue include Veillonella atypica, Porphyromonas gingivalis, Selenomonas spp., Actinobacillus actinomycetemcomitans, Prevotella intermedia, Capnocytophaga spp. and many more [8, 9]. To facilitate food passage, the oropharynx is composed of cells that are able to produce mucus and is carpeted with cilia [10]. These distinct features attract numerous microbes that could not be found in the oral cavity [10]. Unique microbes residing in the oropharynx include Streptococcus pyogenes, Streptococcus pneumoniae, Haemophilus influenza and Haemophilus parainfluenzae [10]. Among the listed microbes, the latter three are pathogenic [10]. Inversely, there are also numerous microbes that could be found in the oral cavity, but not in the oropharynx such as Streptococcus faecalis, Eikenella corrodens, Enterobacteriaceae, Actinomyces, Lactobacilli, Veillonella and Treponema [10].
Oral microbiome in health and disease states
Physicians and dentists have been exploring the theory of focal infection in oral diseases that could potentially influence distant structures since the 19th century [12]. Some of the most heavily debated relationships between periodontal disease and systemic conditions include cardiovascular diseases, adverse pregnancy outcomes, diabetes mellitus and respiratory diseases [12]. The evolution of technology in molecular biology, microbiology, immunology and genetics enables medical researchers and clinicians to continue exploring the synergy between oral diseases and systemic conditions extensively. As a result, numerous mechanisms have been clarified with regards to the relationship between oral diseases and systemic conditions [13-17].
Predominant microbial communities within different sites of the oral and oropharyngeal region. This figure was created using data sources from Dewhirst et al., (2012), Hull et al., (2007) and Taylan et al., (2011) [2, 10, 11].
The association between oral diseases and systemic conditions has encouraged the exploration on the influence of oral dysbiosis and the pathogenesis of oral cancers. In recent years, studies have shown that chronic periodontitis is associated with increased risk of developing oral premalignant lesions, ultimately leading to head and neck squamous cell carcinoma (HNSCC) - mainly oral squamous cell carcinoma (OSCC) and oropharyngeal squamous cell carcinoma (OPSCC) [18-21]. Tezal et al., (2005) were able to associate chronic periodontitis to HNSCC based on an objective and quantitative measure of periodontitis history [18]. According to this study, patients with chronic periodontitis often have poorly differentiated tumours within the oral cavity due to chronic inflammation [18]. In addition, there is also a synergy between chronic periodontitis and oral HPV infection [22]. The periodontal pocket consists of stratified squamous epithelium and is continuously undergoing epithelial proliferation, migration, rete-ridge formation and ulcerations, providing ample opportunity for HPV infection and persistence [22].
A recent study from Furquim et al., (2016) investigated the influence of salivary microbiome in fanconi anemia (FA) patients with regard to their oral health status and OSCC risk factors [23]. FA patients are more likely to develop HNSCC compared to the general population due to pancytopenia [24]. FA patients are aware of their susceptibility to cancers at a young age and therefore do not typically engage in high-risk behaviours such as excessive consumption of tobacco and alcohol [23]. The salivary microbiome of FA patients who are at a high risk of developing HNSCC showed a similar diversity pattern when compared to non-FA patients with oral leucoplakia and OSCC. Based on their findings, the authors suggested that environmental exposures (such as the local microbiome) other than the classical risk factors may play a role in promoting OSCC in FA patients alongside their inherent genomic instability.
A population-based case-control study from China collated data from esophageal squamous cell carcinoma (ESCC) patients (n=616) and normal healthy controls (n=770) to investigate the association between poor oral hygiene and ESCC [25]. Based on the data collected, poor oral hygiene has a significant association with the development of ESCC independent of other risk factors such as tobacco and alcohol consumption, signifying the role of oral microbiome in ESCC. Although poor oral hygiene is also known to be associated with tobacco and alcohol consumption, the association between poor oral hygiene and ESCC in this case was significantly stronger in non-smoking and non-drinking ESCC patients. The outcome of this case-control study further exemplifies the potential risk associated with oral microbiome changes in the development of cancers.
The role of bacteria in the pathogenesis of cancers
The role of gut microbiota in promoting gastric cancer is well established [26]. However, the influence of oral microbiota in the pathogenesis of oral cancers is not fully elucidated. We speculate that this may be due to the temporal changes of the oral microbiota due to daily oral hygiene treatment and nutrients intake [3]. The flux of oral bacteria profiles throughout the day makes it difficult to carry out functional studies due to their obligated dependencies for growth and survival [3, 27]. In addition, different types of microbiome sampling methods such as saliva collections and buccal swab may also affect the collection of oral bacteria and thereby influence downstream applications [28, 29]. However, based on the data pertaining to gut microbiome and gastrointestinal cancers from literature, we were able to discern possible mechanisms leading to the pathogenesis of oral and oropharyngeal cancers.
Provoke chronic inflammatory responses
Studies have established that chronic inflammation is responsible for 25% of human malignancies and represents the seventh hallmark in the development of cancers [30]. Chronic inflammatory mediators cause or facilitate increased cell proliferation, mutagenesis, oncogene activation, and angiogenesis that ultimately lead to the loss of normal growth control and cancer [30, 31]. Bacterial infection is one of the major causes of chronic inflammation. The strongest link established between bacterial infection and the development of cancer due to chronic inflammation to date is the association between Helicobacter pylori (H. pylori) and adenocarcinoma of the stomach, while other known associations include Salmonella typhi and gallbladder cancer, Streptococcus bovis and colon cancer, Chlamydia pneumonia and lung cancer, and Bartonella species and vascular tumour formation [26, 31]. In general, studies have shown that bacteria alone are unable to induce cancer; the process is commonly accompanied by chronic inflammation and requires independent mutations in oncogenic signalling pathways [26].
As a major transcription factor that regulates genes responsible for both the innate and adaptive immune response, nuclear factor KB (NF-KB) is a pivotal link between inflammation and cancer [32]. While the activation of NF-KB is part of the immune response in acute inflammatory processes to eliminate transformed cells; it is also highly activated in many cancer types and exhibits a variety of pro-tumourigenic functions [33]. When activated, NF-KB induces cytokines to recruit phagocytes/sentinel cells to the site of inflammation [33]. It is postulated that the increase in reactive oxygen species released from the recruited neutrophils to kill invading pathogens may also damage the host cell DNA, causing genetic mutations that ultimately trigger tumour initiation [33].
Clinical studies have shown that the activation of NF-KB is a crucial component in bacteria associated cancer formation [26]. The innate immune system is the first line of defence against invading bacteria through the deployment of sentinel cells with highly sensitive pattern recognition receptors (PRRs) that are responsible for the recognition of pathogen signature and the activation of cytokine production via NF-KB signalling pathway [34]. Amongst the PRR family members, Toll-like receptors (TLRs) bacterial pattern recognition is one of the major factors in bacteria-induced inflammation as well as the contributor to carcinogenesis [34]. Lipopolysaccharide (LPS, also known as endotoxin) is a major component of the Gram-negative bacterial outer membrane [35]. Upon infection, LPS are released from Gram-negative bacteria and detected first by surveying sentinel cells expressing TLR4 [35]. Gram-negative bacteria are known to promote carcinogenesis in the colon, liver, pancreas and skin through TLR4 signalling pathways via activation of NF-KB [34].
Direct manipulation of host cell biology
Bacteria may influence the pathogenesis of cancers directly via the secretion of bacterial effector proteins using type 3 or type 4 secretion systems (T3SS/T4SS) [26, 34]. Bacterial effector proteins are generally critical for virulence and have many different roles during disease [26, 34]. As an example, Bartonella secretes a cocktail of Bartonella effector proteins (Beps) into host cells using T4SS to modulate cellular processes to the benefit of the bacteria [36, 37]. These processes involve direct stimulation of endothelial cell proliferation, cytoskeletal rearrangements, activation of NF-KB and inhibition of cellular apoptosis [26]. Similarly, H. pylori secretes cytotoxin-associated gene A (CagA) into the host cytoplasm using T4SS to promote cell proliferation, trigger NF-KB signalling pathway, alter cell cytoskeleton and promote cell migration and invasion [26]. Researchers have shown that H. pylori strains that are CagA-positive are associated with more severe gastritis and are at a higher risk of carcinogenesis [26]. While Beps and CagA are examples of powerful bacterial effector proteins, bacteria-derived metabolites such as acetaldehyde, hydrogen sulphide and superoxide radicals also contribute to cancer formation by causing genomic instability [34]. In particular, acetaldehyde, a local carcinogen produced by the bacteria in the human digestive tract is known to increase the risk of gastric cancer among gastritis patients [34]. Furthermore, the secretion of bacterial toxins such as cytolethal distending toxin (CDT), cytotoxic necrotising factor 1 and colibactin can directly modulate carcinogenesis via triggering DNA damage response and genomic instability, leading to the cell cycle arrest during the G2/M phase [34].
Alter tissue stem-cell homeostasis
While bacterial effector proteins are able to alter the cell biology of epithelia and endothelial cells, these effector proteins have to be able to affect the cell biology of self-renewing cells that have the potential for malignant transformation to promote carcinogenesis in vivo [26]. One of the functions of tissue-specific stem-cells is to regenerate differentiated cells such as epithelial and endothelial cells [26, 38]. In order to secure a vast supply of differentiated cells, stem-cell homeostasis has to be regulated in a stringent manner to avoid uncontrolled proliferation that would lead to carcinogenesis [26, 38]. Stem-cell niche (complex milieu composed of cells, extracellular matrix and signalling molecules associated with the respective stem-cell population) regulates stem-cell homeostasis through the secretion of a repertoire of growth factors produced from supporting cells and stem-cell anchorage; both features are known to be conserved in mammalian stem-cell niches [26, 38]. Although the long term effects of external factors that increase the growth factors in stem-cell tissue as well as exogenous signals that alter stem-cell anchorage are poorly understood, these features are known to promote the acquisition of an invasive phenotype [26]. Among the aforementioned bacterial effector proteins, CagA secreted from H. pylori is able to achieve both features by activating the growth factor (epidermal and transforming) pathway in stem-cell tissue and disturb the anchorage via increasing the transcriptional activity of β-catenin, a protein responsible for cell-cell adhesion regulation and coordination [26]. The overexpression of β-catenin is often associated with many human cancers such as breast, colorectal and prostate [39-41]. In the colon, Citrobacter rodentium (C. rodentium) infection promotes β-catenin expression in the cell cytosol and in the nucleus of epithelial cells, ultimately leading to the carcinogenesis of colon cancer [26]. The signals directed to the epithelium from infecting C. rodentium are able to instigate an oncogenic pathway in the tissue. This demonstrates that infecting bacteria are also capable of interacting with stem-cell niche indirectly [26].
Since the role of gut bacterial infection in causing or promoting gastrointestinal carcinogenesis is reasonably well-understood, attention should be given to oral bacteria as the oral cavity is one of the most colonised anatomies in humans [28]. From available information on the mechanisms of bacterial infection implicated in carcinogenesis it is reasonable to ask, therefore, if shifts in the composition of oral bacteria could be the promoter or cause of oral cancers [28].
The oral microbiome profiles in oral and oropharyngeal cancer patients
The link between oral and systemic diseases has encouraged researchers to investigate the influence of oral microorganisms in the development of human cancers. It is well-documented in literature that there are indeed significant changes in the abundance of oral microbiota (e.g., Bifidobacteriales, Lactobacillaceae and Firmicutes) in oral cancer patients compared to healthy individuals [28, 42]. However, to the best of our knowledge, Schmidt et al., (2014) and Guerrero-Preston et al., (2016) were the only groups to report observable differences in the oral microbiome that could potentially serve as a biomarker for oral cancers [28, 29]. This is due to the limitations in earlier studies using small numbers of known and cultivable oral bacterial species and other molecular methods that focus on specific phyla instead of high-throughput methods such as next-generation sequencing [28].
The microbial influence in the pathogenesis of oral cavity and oropharyngeal cancers.
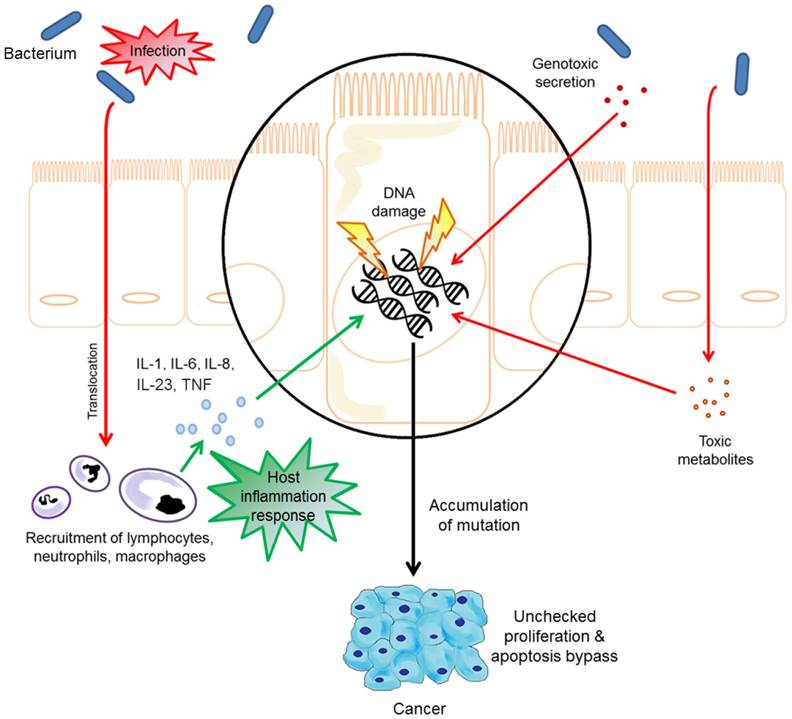
The study carried out by Schmidt et al., (2014) investigated the oral microbiome of five oral cancer patients and eight oral pre-cancer patients using 16s rRNA gene amplicon next-generation sequencing. The biospecimens were collected using swabs on the oral lesion and a contralateral normal site. This study reported a significant decrease in abundance of Firmicutes and Actinobacteria in cancer patients. A significant decrease in these phyla were also confirmed in pre-cancer patients, suggesting that oral lesion-associated shifts in oral microbiome may occur early in oral cancer development and/or herald cancer progression. The study from Guerrero-Preston et al., (2016) utilised oral rinse as biospecimens. The oral microbiome of 19 HNSCC patients and 25 normal healthy individuals were investigated using 16s rRNA gene amplicon next-generation sequencing and a decrease in microbial richness and diversity was reported in cancers. The enriched presence of Lactobacillus or the loss of Haemopilus, Neisseria, Gemellaceae or Aggregatibacter in saliva was reported as a potential biomarker for HNSCC. While HPV status did not have a significant impact on the oral microbiome, it is speculated that the small sample size may have influenced the outcomes. The findings from both studies indicated that microbial diversity and taxonomic composition of the oral microbiome may be useful biomarkers for HNSCC as well as provide a solid framework for future oral microbiome research.
Aside from saliva oral microbiome sampling, the oral microbiome in OSCC tumour samples has also been investigated. Pushalkar et al., (2012) were able to observe a substantial shift in bacterial colonisation between OSCC tumours and adjacent non-tumour mucosa [43]. The study consisted of 10 OSCC tumour samples with their respective adjacent histologically confirmed normal mucosa. The total bacterial diversity (bacteria on the surface as well as within tissue) was detected using denaturing gradient-gel electrophoresis (DGGE) assays coupled with 16S rRNA gene amplicon next-generation sequencing. The initial results from DGGE fingerprinting showed different bacterial colonies in tumour and non-tumour oral sites. Clonal libraries with clinical distinctions were then constructed and sequenced to provide a more detailed analysis. While the relative abundance of bacteria in tumour samples is significantly different compared with their adjacent normal mucosa, no phylogenetic differences were detected. Streptococcus sp. oral taxon 058, Peptosteptococcus stomatis, S. salivarius, S. gordonii, G. haemolysans, G. morbillorum, J. ignava and S. parasanguinis I, were found to be associated with tumour site while Granulicatella adiacens was more prevalent at normal mucosa. Furthermore, there was a relative shift from Gram-negative to Gram-positive microbiota (19%) in tumour samples. Albeit of modest sample size, this study was able to associate microbial dysbiosis and OSCC.
In contrast, a recent study from Wang et al., (2017) demonstrated that the microbiomes in HNSCC are fundamentally similar in terms of diversity and richness in tumour tissue compared with their respective histologically normal adjacent tissue [44]. The microbiomic differences in paired tumour and non-tumour tissue samples from HNSCC patients (n=121) were identified using Sanger sequencing. While there were no major shifts in the overall diversity (Shannon index or phylogenetic diversity) between tumour and non-tumour tissues, there was a significant decrease in the genus Actinomyces and its parent taxonomic up to the phylum level. In addition, the decrease of Actinomyces was found to be more pronounced in tumours at higher tumour-node-metastasis (TNM)-stage. Although the authors acknowledge the limitation of Sanger sequencing may have under-predicted the true phylogenetic diversity of patient samples compared with other saliva studies that utilise next-generation sequencing, the consistent depletion of Actinomyces in tumours at higher TNM-stage may be a useful biomarker for disease monitoring.
Limitations
Oral microbiome sampling
The human oral microbiome can be collected by using multiple different formats depending on the application and downstream analysis. The most commonly used method in literature is the use of oral swabs. Since microbes in the oral cavity travel around the oral region through the movement of the tongue and saliva, swab sampling may not provide an accurate representation of the oral microbiome, especially in cases where the tumours are small and located in hidden areas such as the tonsillar crypts or within the pits and crevices in the lingual tonsils of the tongue base [8, 45]. As such, saliva sampling is another common method to access the oral microbiome. However, the diurnal variations of analytes present in saliva posse a problem. Although, recent findings from Belstrom et al., (2016) demonstrated that the human microbial profiles remained unchanged within 24 h, different saliva collection methods could still potentially influence the experimental endpoints [46]. As an example, in stimulated saliva, the muscle movements from the paraffin gum chewing action may provide a larger microbial coverage compared with passive drooling; while oral rinse sampling (through the gargling action of saline solution) provides access into the microbial communities residing within the oropharyngeal area. Nevertheless, there is a need to determine how well the microbial composition in saliva mirrors the overall oral microbiome as certain microbes in the oral cavity (such as Aggregatibacter, actinomycetemcomitans and Porphyromonas gingivalis) form biofilms in cryptic regions that may not be captured by saliva [7]. The lack of a standardised protocol for oral microbiome sampling makes it a challenge to discern microbial profiles within the oral cavity when comparing published articles [47, 48].
Poor culturability rates of oral microbiota
The profiling of oral microbiome relies heavily on molecular techniques as one-third of the oral microbiota remain uncultivated [49]. This is due to the obligated dependencies of microorganisms for growth and survival. While advance next-generation sequencing such as the Human Oral Microbe Identification using Next Generation Sequencing (HOMINGS) is able to simultaneously identify ~600 oral taxa at the species-level, the data can only be interpreted as a metagenomic snapshot [27, 46, 50]. Molecular techniques such as the HOMINGS do not take into consideration if the bacteria are alive or not as well as the fact that certain dental plaque in the oral cavity develop temporally. Data from molecular techniques have to be validated in culture to accurately determine the oral microbiome profiles present within a given time.
Other host factors
Host factors are known to affect the oral microbiome such as age, gender, ethnicity, geographical region, diet, risky behaviours (smoking and drinking) and the overall health condition of an individual [51-55]. The oral microbiome abundance shift in oral and oropharyngeal cancers cases, therefore, may not be solely due to the tumorigenicity effect. However, this problem could be reduced by careful and stringent selection of the control and patient cohorts to avoid unwanted bias.
Future outlook
HNSCC patients presenting early clinical stages have a 5 year survival rate of 70-90%, whilst 50% of HNSCC patients presenting advanced clinical stages die within 2 years following the initial diagnosis [56]. With HPV on the rise, one of the major clinical challenges for managing HNSCC today is the lack of early screening/diagnostic methods. This may be due to the lack of cancer awareness in addition to the fact that some of the more obvious HNSCC symptoms only present at a later stage. Furthermore, the low patient survival is mainly due to the lack of correlation between histological grade of dysplasia and prognosis which leads to late diagnosis [57, 58]. A retrospective study was carried out in the United Kingdom to investigate the correlation between the direct cost associated with HNSCC and the point of diagnosis, with 147 case reviews from two different hospitals [59]. According to the study, early detection of HSNCC offers potential to reduce overall expenditure by 14% (half a billion on a global-scale) [59, 60]. Clinical registry data in the United States also showed that 50% of the HNSCC cases are diagnosed at an advanced clinical stage due to the lack of early diagnostic tools [59]. While the prevention of HNSCC is the most cost-saving scenario, early detection are the alternatives with marginal cost of $30, 000 to $50, 000 per quality-adjusted life-year gained [59]. Hence, there is an urgent need of early detection biomarkers for HNSCC.
In recent years, saliva diagnostics has played a central role in the discovery of biomarkers for the diagnosis of oral and systemic diseases [61]. This is attributable to the fact that most of the biomarkers present in blood and urine can also be detected in a sample of saliva [61]. However, none of these host salivary biomarkers were able to proceed with a clinical trial due to the high amount of biological variation between individuals. To minimise individual biological variation, a variety of salivary biomarker combinations were used in a panel to detect oral cancers [62]. While the diagnostic accuracy of these salivary biomarker panels has improved, the sensitivity and specificity are not adequate to be accepted for clinical tests [63]. Recent advances in salivary biomarker diagnostics have broadened the discovery of microbial pathogens associated with systemic and oral diseases [61]. The oral microbiota is dynamic and responsive to environmental and biological changes, so shifts in composition and/or function of the oral microbiota might offer new biomarkers useful for the diagnosis of oral and oropharyngeal cancers [5]. While host biomarkers are subjected to individual biological variation as mentioned, oral microbiome is relatively conserved among unrelated individuals [4]. Hence, the incorporation of oral microbiome with salivary tumour biomarkers such as DNA methylation and miRNA expression may help reduce the human biological variation that prevents the actualisation of molecular diagnosis for oral and oropharyngeal cancers [4].
Abbreviations
Beps: Bartonella effector proteins; CagA: cytotoxin-associated gene A; CDT: cytolethal distending toxin; DGGE: denaturing gradient-gel electrophoresis; ESCC: esophageal squamous cell carcinoma; FA: fanconi anemia; HNSCC: head and neck squamous cell carcinoma; HOMINGS: human oral microbe identification using next generation sequencing; LPS: lipopolysaccharide; NF-KB: nuclear factor KB; OPSCC: oropharyngeal squamous cell carcinoma; OSCC: oral squamous cell carcinoma; PRRs: pattern recognition receptors; T3SS/T4SS: type 3 or type 4 secretion systems; TLRs: Toll-like receptors; TNM: tumour-node-metastasis.
Acknowledgements
This study was supported by the Queensland Centre for Head and Neck Cancer funded by Atlantic Philanthropies, the Queensland Government, the Princess Alexandra Hospital, the Queensland University of Technology Vice Chancellor Fellowship (CP) and the QUT postgraduate research scholarship (YKL).
Author's Contributions
YKL wrote the manuscript and designed the review content. MT and MM revised the manuscript critically for content. CP advised and participated in content designing as well as revised the manuscript critically for content. All authors have read and approved the content of this manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Turnbaugh PJ, Ley RE, Hamady M. et al. The Human Microbiome Project. Nature. 2007;449:804-10
2. Dewhirst FE, Chen T, Izard J. et al. The human oral microbiome. J Bacteriol. 2010;192:5002-17
3. Kolenbrander PE, Andersen RN, Blehert DS. et al. Communication among Oral Bacteria. Microbiol Mol Biol Rev. 2002;66:486-505
4. Zaura E, Nicu EA, Krom BP. et al. Acquiring and maintaining a normal oral microbiome: current perspective. Front Cell Infect Microbiol. 2014;4:85
5. Aas JA, Paster BJ, Stokes LN. et al. Defining the Normal Bacterial Flora of the Oral Cavity. J Clin Microbiol. 2005;43:5721-32
6. Takahashi N. Microbial ecosystem in the oral cavity: Metabolic diversity in an ecological niche and its relationship with oral diseases. Int Congr Ser. 2005;1284:103-12
7. Dowd SE, Wolcott RD, Sun Y. et al. Polymicrobial Nature of Chronic Diabetic Foot Ulcer Biofilm Infections Determined Using Bacterial Tag Encoded FLX Amplicon Pyrosequencing (bTEFAP). PLoS ONE. 2008;3:e3326
8. Danser MM, Gómez SM, Van der Weijden GA. Tongue coating and tongue brushing: a literature review. Int J Dent Hyg. 2003;1:151-8
9. Wilson M. Bacteriology of humans: an ecological perspective. Blackwell Publishing LTYD. 2008
10. Hull MW, Chow AW. Indigenous microflora and innate immunity of the head and neck. Infect Dis Clin North Am. 2007;21:265-82 v
11. Taylan I, Özcan İ, Mumcuoğlu İ. et al. Comparison of the Surface and Core Bacteria in Tonsillar and Adenoid Tissue With Beta-Lactamase Production. Indian J Otolaryngol Head Neck Surg. 2011;63:223-8
12. Weidlich P, Cimões R, Pannuti CM. et al. Association between periodontal diseases and systemic diseases. Braz Oral Res. 2008;22:32-43
13. Leishman SJ, Do HL, Ford PJ. Cardiovascular disease and the role of oral bacteria. J Oral Microbiol. 2010;2:10.3402 /jom.v2i0.5781
14. Huck O, Tenenbaum H, Davideau JL. Relationship between periodontal diseases and preterm birth: recent epidemiological and biological data. J Pregnancy. 2011;2011:164654
15. Mealey BL, Oates TW. Diabetes mellitus and periodontal diseases. J Periodontol. 2006;77:1289-303
16. Gomes-Filho IS, Passos JS, Seixas da Cruz S. Respiratory disease and the role of oral bacteria. J Oral Microbiol. 2010:2
17. Wang L, Ganly I. The oral microbiome and oral cancer. Clin Lab Med. 2014;34:711-9
18. Tezal M, Grossi SG, Genco RJ. Is periodontitis associated with oral neoplasms? J Periodontol. 2005;76:406-10
19. Moraes RC, Dias FL, Figueredo CM. et al. Association between Chronic Periodontitis and Oral/Oropharyngeal Cancer. Braz Dent J. 2016;27:261-6
20. Laprise C, Shahul HP, Madathil SA. et al. Periodontal diseases and risk of oral cancer in Southern India: Results from the HeNCe Life study. Int J Cancer. 2016;139:1512-9
21. Galvão-Moreira LV, da Cruz MCFN. Oral microbiome, periodontitis and risk of head and neck cancer. Oral Oncol. 2016;53:17-9
22. Tezal M, Sullivan Nasca M, Stoler DL. et al. Chronic periodontitis-human papillomavirus synergy in base of tongue cancers. Arch Otolaryngol Head Neck Surg. 2009;135:391-6
23. Furquim CP, Soares GM, Ribeiro LL. et al. The Salivary Microbiome and Oral Cancer Risk: a Pilot Study in Fanconi Anemia. J Dent Res. 2017;96:292-9
24. Alter BP. Fanconi anemia and the development of leukemia. Best Pract Res Clin Haematol. 2014;27:214-21
25. Chen X, Yuan Z, Lu M. et al. Poor oral health is associated with an increased risk of esophageal squamous cell carcinoma - a population-based case-control study in China. Int J Cancer. 2017;140:626-35
26. Vogelmann R, Amieva MR. The role of bacterial pathogens in cancer. Curr Opin Microbiol. 2007;10:76-81
27. Jenkinson HF. Beyond the oral microbiome. Environmental Microbiology. 2011;13:3077-87
28. Schmidt BL, Kuczynski J, Bhattacharya A. et al. Changes in abundance of oral microbiota associated with oral cancer. PLoS One. 2014;9:e98741
29. Guerrero-Preston R, Godoy-Vitorino F, Jedlicka A. et al. 16S rRNA amplicon sequencing identifies microbiota associated with oral cancer, Human Papilloma Virus infection and surgical treatment. Oncotarget. 2016;32:51320-34
30. Multhoff G, Molls M, Radons J. Chronic Inflammation in Cancer Development. Front Immunol. 2011;2:98
31. Shacter E, Weitzman SA. Chronic inflammation and cancer. Oncology (Williston Park). 2002;16:217-26 29; discussion 30-2
32. Karin M. NF-κB as a Critical Link Between Inflammation and Cancer. Cold Spring Harbor Perspectives in Biology. 2009;1:a000141
33. Hoesel B, Schmid JA. The complexity of NF-kappaB signaling in inflammation and cancer. Mol Cancer. 2013;12:86
34. Agassandian M, Shurin GV. Bacterial Infections and Cancar Development. In: (ed.) Shurin MR, Thanavala Y, Ismail N. Infection and Cancaer: Bi-Directorial Interactions; 1 ed. Springer International Publishing. 2015:408
35. Zhang G, Ghosh S. Molecular mechanisms of NF-kappaB activation induced by bacterial lipopolysaccharide through Toll-like receptors. J Endotoxin Res. 2000;6:453-7
36. Siamer S, Dehio C. New insights into the role of Bartonella effector proteins in pathogenesis. Curr Opin Microbiol. 2015;23:80-5
37. Okujava R, Guye P, Lu Y-Y. et al. A Translocated Effector Required for Bartonella Dissemination from Derma to Blood Safeguards Migratory Host Cells from Damage by Co-translocated Effectors. PLoS Pathog. 2014;10:e1004187
38. Chen S, Lewallen M, Xie T. Adhesion in the stem cell niche: biological roles and regulation. Development (Cambridge, England). 2013;140:255-65
39. Lin SY, Xia W, Wang JC. et al. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci U S A. 2000;97:4262-6
40. Chen Z, He X, Jia M. et al. beta-catenin overexpression in the nucleus predicts progress disease and unfavourable survival in colorectal cancer: a meta-analysis. PLoS One. 2013;8:e63854
41. Guturi KKN, Mandal T, Chatterjee A. et al. Mechanism of β-Catenin-mediated Transcriptional Regulation of Epidermal Growth Factor Receptor Expression in Glycogen Synthase Kinase 3 β-inactivated Prostate Cancer Cells. J Biol Chem. 2012;287:18287-96
42. Ahn J. et al. Oral microbiome and risk of head and neck cancer, a nested case-control study. Cancer Res. 2014:74
43. Pushalkar S, Ji X, Li Y. et al. Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma. BMC Microbiol. 2012;12:144 -
44. Wang H, Funchain P, Bebek G. et al. Microbiomic differences in tumor and paired-normal tissue in head and neck squamous cell carcinomas. Genome Med. 2017;9:14
45. Lim Y, Wan Y, Vagenas D. et al. Salivary DNA methylation panel to diagnose HPV-positive and HPV-negative head and neck cancers. BMC Cancer. 2016;16:1-12
46. Belstrom D, Holmstrup P, Bardow A. et al. Temporal Stability of the Salivary Microbiota in Oral Health. PLoS One. 2016;11:e0147472
47. Pepe MS, Etzioni R, Feng Z. et al. Phases of Biomarker Development for Early Detection of Cancer. J Natl Cancer Inst. 2001;93:1054-61
48. Lucs AV, Saltman B, Chung CH. et al. Opportunities and challenges facing biomarker development for personalized head and neck cancer treatment. Head Neck. 2013;35:294-306
49. Wade W, Thompson H, Rybalka A. et al. Uncultured Members of the Oral Microbiome. J Calif Dent Assoc. 2016;44:447-56
50. Gomes BP, Berber VB, Kokaras AS. et al. Microbiomes of Endodontic-Periodontal Lesions before and after Chemomechanical Preparation. J Endod. 2015;41:1975-84
51. Kumar PS, Matthews CR, Joshi V. et al. Tobacco Smoking Affects Bacterial Acquisition and Colonization in Oral Biofilms. Infect Immun. 2011;79:4730-8
52. Thomas AM, Gleber-Netto FO, Fernandes GR. et al. Alcohol and tobacco consumption affects bacterial richness in oral cavity mucosa biofilms. BMC Microbiol. 2014;14:250
53. Mason MR, Nagaraja HN, Camerlengo T. et al. Deep sequencing identifies ethnicity-specific bacterial signatures in the oral microbiome. PLoS One. 2013;8:e77287
54. Li J, Quinque D, Horz HP. et al. Comparative analysis of the human saliva microbiome from different climate zones: Alaska, Germany, and Africa. BMC Microbiol. 2014;14:316
55. Oh C, Lee K, Cheong Y. et al. Comparison of the Oral Microbiomes of Canines and Their Owners Using Next-Generation Sequencing. PLoS One. 2015;10:e0131468
56. Duray A, Lacremans D, Demoulin S. et al. Prognosis of HPV-positive head and neck cancers: implication of smoking and immunosuppression. Advan Cell Mol Otolaryngol. 2014;2:1-10
57. Ovchinnikov DA, Cooper MA, Pandit P. et al. Tumor-suppressor Gene Promoter Hypermethylation in Saliva of Head and Neck Cancer Patients. Transl Oncol. 2012;5:321-6
58. Ralhan R. How will increasing our knowledge of the head and neck cancer cells' secretome benefit head and neck squamous cell carcinoma patients? Expert Rev Proteomics. 2012;9:115-8
59. Menzin J, Lines LM, Manning LN. The economics of squamous cell carcinoma of the head and neck. Curr Opin Otolaryngol Head Neck Surg. 2007;15:68-73
60. Wissinger E, Griebsch I, Lungershausen J. et al. The economic burden of head and neck cancer: a systematic literature review. Pharmacoeconomics. 2014;32:865-82
61. Malamud D. Saliva as a diagnostic fluid. Dent Clin North Am. 2011;55:159-78
62. Righini CA, de Fraipont F, Timsit JF. et al. Tumor-specific methylation in saliva: a promising biomarker for early detection of head and neck cancer recurrence. Clin Cancer Res. 2007;13:1179-85
63. Drucker E, Krapfenbauer K. Pitfalls and limitations in translation from biomarker discovery to clinical utility in predictive and personalised medicine. EPMA Journal. 2013;4:1-10
Author contact
![]() Corresponding author: Associate Professor Chamindie Punyadeera, The School of Biomedical Sciences, Institute of Health and Biomedical Innovations, Queensland University of Technology, 60 Musk Avenue, GPO Box 2434, Brisbane QLD 4059, Australia. +61 7 3138 0830
Corresponding author: Associate Professor Chamindie Punyadeera, The School of Biomedical Sciences, Institute of Health and Biomedical Innovations, Queensland University of Technology, 60 Musk Avenue, GPO Box 2434, Brisbane QLD 4059, Australia. +61 7 3138 0830
 Global reach, higher impact
Global reach, higher impact