13.3
Impact Factor
Theranostics 2018; 8(5):1449-1457. doi:10.7150/thno.22540 This issue Cite
Research Paper
D-alanyl-D-alanine-Modified Gold Nanoparticles Form a Broad-Spectrum Sensor for Bacteria
1. CAS Center for Excellence in Nanoscience, CAS Key Lab for Biological Effects of Nanomaterials and Nanosafety, Beijing Engineering Research Center for BioNanotechnology, National Center for NanoScience and Technology, ZhongGuanCun BeiYiTiao, Beijing, 100190, China
2. Natural Products Research Center, Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu, Sichuan, 610041, China
3. University of Chinese Academy of Science, Beijing, 100049, China
4. Department of Clinical Laboratory, Beijing You'an Hospital Affiliated to Capital Medical University, No. 8 You'an Men Wai Xi TouTiao, Fengtai District, Beijing, 100069, China
5. Present address: School of Chemistry and Chemical Engineering, University of Jinan, Jinan 250022, China
Received 2017-8-25; Accepted 2017-12-23; Published 2018-2-4
Abstract
Rationale: Rapid and facile detection of pathogenic bacteria is challenging due to the requirement of large-scale instruments and equipment in conventional methods. We utilize D-amino acid as molecules to selectively target bacteria because bacteria can incorporate DADA in its cell wall while mammalian cells or fungi cannot.
Methods: We show a broad-spectrum bacterial detection system based on D-amino acid-capped gold nanoparticles (AuNPs). AuNPs serve as the signal output that we can monitor without relying on any complex instruments.
Results: In the presence of bacteria, the AuNPs aggregate and the color of AuNPs changes from red to blue. This convenient color change can distinguish between Staphylococcus aureus (S. aureus) and methicillin-resistant Staphylococcus aureus (MRSA). This system can be applied for detection of ascites samples from patients.
Conclusion: These D-amino acid-modified AuNPs serve as a promising platform for rapid visual identification of pathogens in the clinic.
Keywords: gold nanoparticles, D-amino acid, bacteria, spontaneous bacterial peritonitis, colorimetric biosensor
Introduction
Spontaneous bacterial peritonitis (SBP) caused by ascites is a common cause of death in patients suffering from cirrhosis.[1, 2] The fundamental challenge in mitigating SBP is the early detection of these bacteria in ascites. The current standards for detecting bacteria are microbial culture or gene analysis, which need complex equipment and professional technicians to operate in specific environments. Recent studies show that colorimetric biosensors, fluorescence imaging or microelectronics can be useful for Gram-positive and Gram-negative bacteria detection.[3-12]
AuNPs are widely applicable as colorimetric biosensors, drug delivery carriers, antibacterial agents, and photothermal therapy material.[13-28] Our group has developed a series of small molecule-modified AuNPs that can kill multidrug-resistant bacteria.[29-31] We also report a number of colorimetric sensors based on AuNPs since their use can avoid the necessity of bulky instruments by obtaining signals with the naked eye.[3, 32-36] A number of investigators have reported bacterial biosensors based on AuNPs. Scientists have developed cetyltrimethylammonium bromide (CTAB)-coated AuNPs to detect bacteria.[37] CTAB is positively charged such that it can target the negatively charged bacterial surface. Researchers also use antibodies or aptamers to decorate AuNPs for bacterial targeting.[38] Unfortunately, these strategies are based on charge interactions and easily interfered by surroundings such as pH values, metal ion or proteins. A stable, easily available and navigable platform for the detection of pathogens is urgently needed.
We hypothesize that D-amino acids can play a role in AuNPs-based detection of microorganisms because they mainly exist in bacteria. In these microorganisms, D-amino acids are part of peptidoglycan, the principal component of the bacterial cell wall, which surrounds bacteria.[39, 40] Given the critical role of peptidoglycan during the synthesis of cell wall and bacterial reproduction, researchers have developed a number of antibiotics that can obstruct peptidoglycan synthesis to inhibit bacterial growth.[41] Recently, researchers have found that a number of bacteria can easily incorporate exogenous D-amino acids from the surrounding medium into their peptidoglycan via displacing the terminal D-alanine of the oligopeptide strand, due to the enzymatic promiscuity of bacterial D,D-transpeptidases.[42, 43] They used D-amino acids linked with fluorescent molecules to specifically label the peptidoglycan network.[44-46] However, these imaging methods are inconvenient and they mostly rely on complex instruments to label and detect bacteria. We thus wonder if we can modify the surface of AuNPs with D-amino acids to diagnose bacterial infection.
Here, we illustrate a novel strategy for bacterial detection by combining D-amino acids (D-alanyl-D-alanine, DADA) with AuNPs. AuNPs serve as the signal output that we can monitor without relying on any complex instruments. We utilize DADA as molecules to selectively target bacteria because bacteria can incorporate DADA in its cell wall while mammalian cells or fungi cannot.[47] When bacteria consume DADA capped on AuNPs, DADA unbinds from the AuNP surface, and the AuNPs become unstable and aggregate because DADA no longer protects the surface of the AuNPs. We prepared DADA-modified AuNPs (Au_DADA) in a one-pot reaction without any other complex modifications (Scheme 1). We employed the L-amino acid, L-Alanyl-L-alanine (LALA), as comparison to confirm the hypothesized mechanism of interaction between bacteria and Au_DADA. We chose laboratory antibiotic-sensitive bacteria and clinical multidrug-resistant (MDR) bacteria to evaluate the detection activity of our AuNPs. Interestingly, Au_DADA could distinguish between S. aureus and MRSA. To investigate the potential of Au_DADA in clinical application, we used it to diagnose bacterial infection in liver ascites from patients. Our study paves a new way to colorimetric biosensing of pathogens.
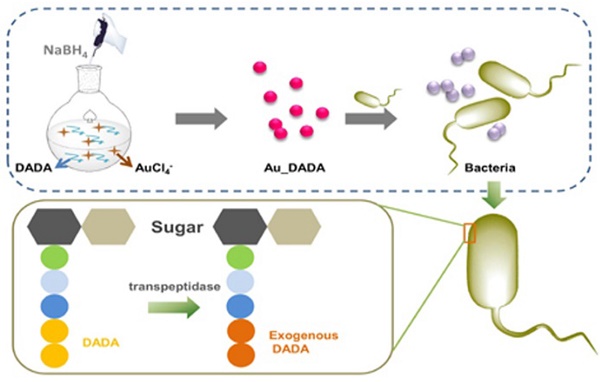
Schematic illustration of the synthesis of D-amino acid-modified AuNPs and the structural change of peptidoglycan after incubation with D-amino acid-modified AuNPs.
Materials and Methods
Materials
Gold (III) chloride hydrate (HAuCl4.3H2O), D-Alanyl-D-alanine (DADA), and sodium borohydride are from Aladdin Industrial Corporation (China). L-Alanyl-L-alanine (LALA) is from Sigma. We used the procured chemicals without further purification. We used a Milli-Q purification system to obtain Deionized (DI) water (18.2 MΩ·cm). We obtained zeta potential values of AuNPs with Zetasizer Nano ZS (Malvern Instruments). We obtained the ascites samples from Beijing You'an Hospital with patients' consent and approval by the local Ethics Committee.
Preparation of AuNPs
We stirred the mixture of D-amino acids (DADA or LALA) (16 mg, 0.1 mmol, dissolved in 6 mL of DI water, 50 μL of absolute acetic acid and 20 mg of Tween 80) and HAuCl4.3H2O (0.1 mmol, dissolved in 0.5 mL DI water) for 10 min and add NaBH4 (0.3 mmol freshly dissolved in 2.5 mL DI water) dropwise with vigorous stirring. The color of the mixture changed to dark red immediately. We kept stirring the mixture for 1 h at 0 °C. We dialyzed (14 kDa MW cutoff, Millipore) it for 48 h with DI water, sterilized it through a 0.22 μm filter (Millipore), and stored it at 4 °C for use. We synthesized different ratios of Au_DADA using the same method.
Characterization of AuNPs
We investigated the morphologies of AuNPs via transmission electron microscopy (TEM) (Tecnai G2 20 ST TEM) from the American FEI company. A microplate reader (Tecan infinite M200) gave us ultraviolet-visible (UV-vis) spectra. We prepared TEM samples by dropping 5 μL of the samples on formvar/carbon coated copper grids and dried them overnight. We measured zeta potential using Malvern Zetasizer.
Bacteria culture
We cultured bacteria in Luria-Bertani (LB) broth medium (5 g/L NaCl, 10 g/L tryptone powder, and 5 g/L beef extract powder, pH=7.2) at 37 °C on a shaker bed at 200 rpm for 4 h. Then, we diluted bacteria with LB broth to a concentration of 1.0 × 108 CFU/mL, which corresponded to an optical density of 0.1 at 600 nm measured by UV-vis spectroscopy.
Identification of bacteria
We pre-cultured Staphylococcus aureus (S. aureus, ATCC 6538P), MRSA, E. coli (ATCC 11775), MDR E. coli, Klebsiella pneumoniae (K. pneumonia, ATCC 13883), MDR K. pneumoniae, Bacillus subtilis (B. subtilis, ATCC 66633), and Listeria monocytogenes (L. monocytogenes, ATCC 19115) in LB broth to a concentration of 1.0 × 108 CFU/mL. We diluted them to a concentration of 1.0 × 106 CFU/mL. We performed the assay for identification of bacterial strains in 96-well microplates (Constar, 3599). We added 90 µL AuNPs diluted with LB medium to the plate. Then we added 10 µL bacteria prepared in the AuNPs medium (final concentration: 1.0 × 105 CFU/mL). We used AuNPs as a control group. Each group has 3 replicates. In order to keep bacteria in good form, we incubated them at 37 °C in an incubator.
The cytotoxicity of Au_DADA
We employed Cell Counting Kit-8 (CCK-8) to measure the cytotoxicity of Au_DADA. We used Dulbecco's modified Eagle's medium (DMEM; Gibco) containing fetal bovine serum (FBS, 10%; Gibco), penicillin and streptomycin (1%), and glutamine (1%) to culture human umbilical vein endothelial cells (HUVECs) and human cervical cancer (HeLa) cells and incubated in CO2 (5%) at 37 °C. HUVEC cells were grown overnight on 96-well culture plates (~10, 000 cells per well) and then we added varying concentrations of Au_DADA to the 96-well plate. After 24 h incubation, we used culture medium to wash cells and stained them with 10 μL of CCK per well. We measured the optical density of the cells at 450 nm by a microplate reader (Tecan infinite M200) after incubating for 2 h.
Stability of Au_DADA
We utilized various pH values ranging from pH=1 to pH=14 to study the stability of Au_DADA. We used 3 M HCl and 2 M NaOH to obtain the pH solutions. We incubated Au_DADA in these solutions for 4 h.
Results and Discussion
Synthesis and characterization of Au_DADA
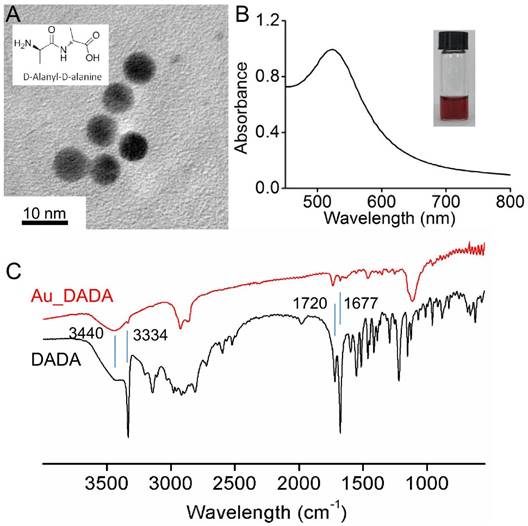
We prepared Au_DADA through a one pot process via reduction of tetrachloroauric acid by sodium borohydride in the presence of DADA molecules in deionized (DI) water. The role of DADA in this reaction is to stabilize and disperse the NPs. Our synthetic strategy is straightforward and convenient without other chemical modifications. Diameters of spherical Au_DADA are around 6 nm as indicated by TEM images and statistical analysis (Figure 1A). UV-vis spectra reveal the plasmon resonance of NPs at ~520 nm (Figure 1B). We tested zeta potential values of the NPs in water at 25 °C. The results show that Au_DADA are negatively charged (-21.9 mV), which is the same as the charges of bacterial surfaces (Table S1). We used Fourier Transform Infrared Spectroscopy (FT-IR) to further characterize the AuNPs (Figure 1C). The asymmetric stretching mode of N-H has a band with a maximum at 3334 cm-1 both in pure DADA and DADA-capped AuNPs, which confirms the presence of DADA on AuNPs. The bending mode of N-H at 1720 cm-1 and 1677 cm-1 can further verify the results.
Characterization of Au_DADA. (A) TEM image of nanoparticles. Inserted structure is DADA. (B) Absorption spectrum of Au_DADA. Inserted photo is Au_DADA in water. (C) FT-IR of Au_DADA and DADA.
Colorimetric detection of bacteria via Au_DADA
In order to obtain AuNPs for sensitive bacterial detection, we investigated the effect of Au_DADA concentration on the colorimetric response. We incubated S. aureus with various concentrations of Au_DADA (0.5, 2.5, 5, 10, 15, 20, and 25 μM). We found that the groups of 0.5, 2.5, and 5 μM are more sensitive than other groups (Figure S1). Because the color change at 5 μM is the most significant and can be read out easily, we chose it for subsequent experiments.
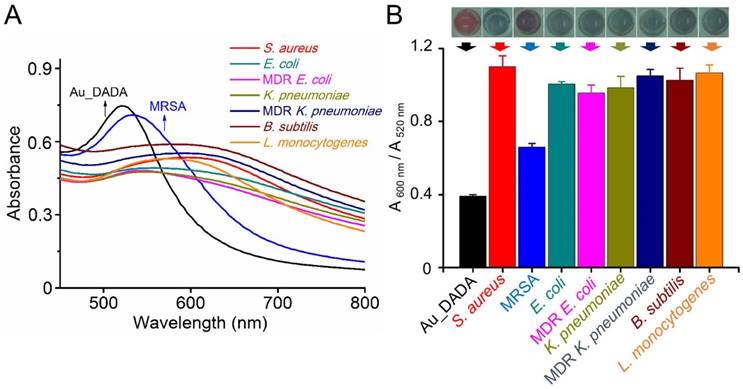
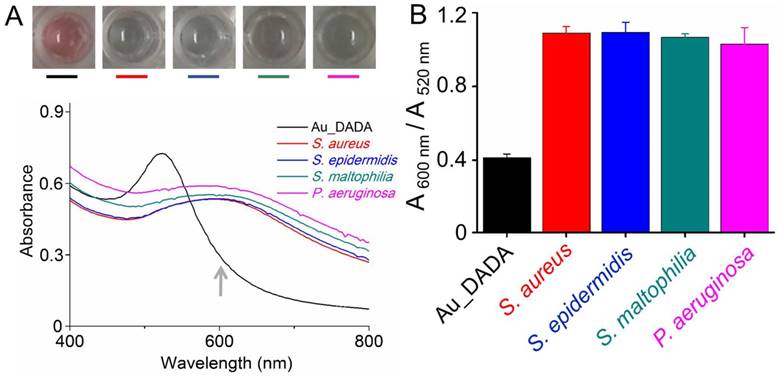
We tested the colorimetric response of Au_DADA by incubating with various types of bacteria, including laboratory antibiotic-sensitive strains (S. aureus, E. coli, K. pneumoniae, B. subtilis, L. monocytogenes) and clinical MDR isolates (MRSA, MDR E. coli, MDR K. pneumoniae). These Gram-positive (S. aureus, B. subtilis, L. monocytogenes, MRSA) and Gram-negative (E. coli, K. pneumonia, MDR E. coli, MDR K. pneumoniae) bacteria are common in clinical ascites samples. We added Au_DADA to microplate wells and mixed with LB broth (as control) and different types of bacteria with the same concentration. The color of the media from cells that contain bacteria changed to blue or purple, whereas the color of the control group remained red (Figure 2B). We can visually distinguish samples that have bacteria from those that do not. We found that the well containing MRSA turned purple instead of blue like the other species. This difference may be caused by the different activity of penicillin binding proteins (PBPs), which influences transpeptidation and synthesis of peptidoglycan.[48] This observation can potentially distinguish between S. aureus and MRSA.
To quantify the colorimetric response of AuNPs, we tested the absorption spectra of each AuNPs solution in the presence of bacteria (Figure 2A). All samples show a red-shift in their absorption spectra after addition of bacteria. The observations from the spectra are consistent with the visual observations, where the AuNPs in all wells underwent a color change from red to blue. The absorbance at 520 nm decreased and the absorbance at 600 nm increased. We used the ratio between the absorbance values of AuNPs at 600 nm and 520 nm (A 600 nm/A 520 nm) to evaluate the degree of AuNPs aggregation (Figure 2B). A low A 600 nm/A 520 nm indicates that AuNPs disperse well in the solution, and a high A 600 nm/A 520 nm indicates aggregation of AuNPs. All samples with bacteria are different from the control solution without bacteria. The only outstanding sample is MRSA, whose A 600 nm/A 520 nm is lower than the A 600 nm/A 520 nm of S. aureus and other bacterial solutions. This observation correlates well with the purple color of MRSA. Despite this difference, A 600 nm/A 520 nm of MRSA is still significantly higher than A 600 nm/A 520 nm of pure AuNPs. These facts confirm that Au_DADA can indeed detect both Gram-positive and Gram-negative bacteria.
Response of Au_DADA to different types of bacteria. (A) Absorption spectra of Au_DADA incubated with bacteria. (B) Change in color of Au_DADA caused by varying types of bacteria and the plot of A 600 nm/A 520 nm of AuNPs versus different types of bacteria. Mean values and standard deviations were obtained from five independent experiments.
The sensitivity of Au_DADA for detection of S. aureus. (A) A 600 nm/A 520 nm value of Au_DADA. (B) The linear relationship between the A 600 nm/A 520 nm value and the concentration of S. aureus. Mean values and standard deviations were obtained from five independent experiments.
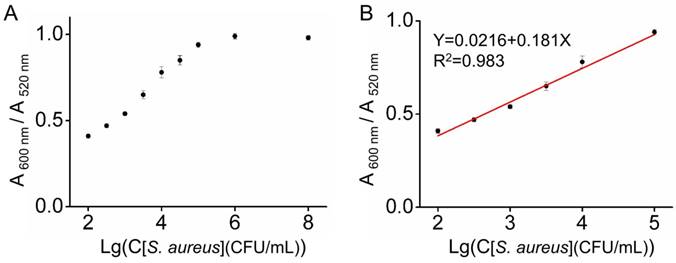
We employed S. aureus as an example to study the sensitivity of our system. We exposed Au_DADA to the S. aureus solutions with different concentrations (1×102-1×108 CFU/mL). When the concentration of bacteria is high, the value of A 600 nm/A 520 nm is high, indicating the high sensitivity of Au_DADA to high concentrations of bacteria (Figure 3). The limit of detection (LOD) was evaluated using the formula: LOD = 3S/M, where S represents the value of the standard deviation of blank samples and M is the slope of the standard curve within the concentration range. According to the formula, the LOD of Au_DADA is 3.2×102 CFU/mL. The limit of quantitation (LOQ, LOQ = 10S/M) is 1.1×103 CFU/mL. These results show that Au_DADA has relatively high sensitivity.
TEM characterization of interactions between bacteria and Au_DADA
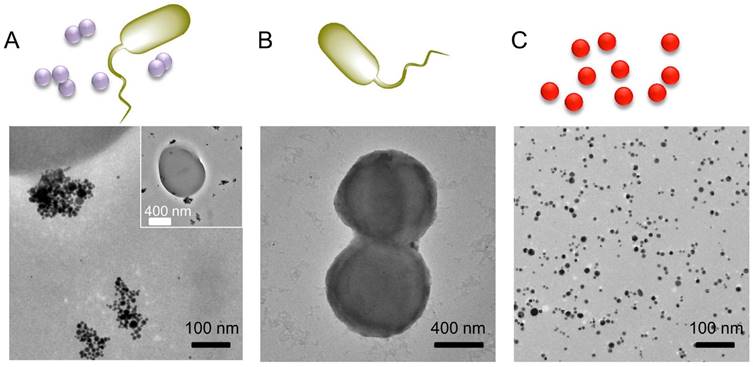
We employed TEM to characterize Au_DADA aggregation on bacteria. We used S. aureus as an example to perform this assay (Figure 4 and S2). Au_DADA aggregated around bacterial surfaces. In comparison, the TEM of the control groups show that there were no AuNPs around bacteria and Au_DADA itself did not aggregate without bacteria (Figure 4B and 4C).
The mechanism of aggregation
It is evident from other studies that bacteria can incorporate exogenous D-amino acids into their peptidoglycan. Thus, we speculate that the D-amino acid is critical for the aggregation of AuNPs in our study. We prepared L-amino acid modified-AuNPs (Au_LALA) to confirm this hypothesis. The synthetic method for Au_LALA is the same as for Au_DADA preparation. TEM and UV-vis spectra of Au_LALA demonstrate that they are similar to Au_DADA (Figure S3). We used S. aureus and E. coli to test the ability of bacterial detection about Au_LALA (Figure S4A and S4B). Au_LALA had no response to S. aureus and E. coli even when we cultured them for 16 h. This observation agrees with literature reports showing that bacteria can only incorporate exogenous D-amino acids.
To further investigate the relationship between bacterial peptidoglycan and D-amino acids, we selected C. albicans, a kind of fungi, whose cell wall biosynthesis does not involve peptidoglycan and thus cannot incorporate D-amino acid (Figure S4C and S4D).[49, 50] We cultured C. albicans with Au_DADA in potato dextrose agar (PDA) broth in 96-well plates at 25 °C. We recorded the optical density of these mixtures at different time periods via a microplate reader (Tecan infinite M200). The color of Au_DADA did not change when incubating with C. albicans for even 36 h. This observation further demonstrates that the aggregation of Au_DADA is due to incorporation of D-amino acid.
The zeta potential of Au_DADA is -21.9 mV. However, bacterial surfaces are also negative charged. Therefore, it is reasonable to deduce that there is negligible electrostatic interaction between positively charged Au_DADA and negatively charged bacterial surface.
We employed FT-IR to further characterize the aggregated Au_DADA after incubating with bacteria (Figure S5). The characteristic peaks of the asymmetric stretching mode of N-H (3334 cm-1) and the bending mode of N-H at 1720 cm-1 and 1677 cm-1 disappeared compared with DADA and the original Au_DADA, which confirms the reduction of surface density of DADA on AuNPs.
These observations lead us to deem that D-amino acid-modified AuNPs became unstable and aggregated after bacteria consumed the D-amino acid capped on AuNPs.
Cytotoxicity of Au_DADA
As biological sensors, the low cytotoxicity of materials is important. Thus, we set out to evaluate the toxicity of Au_DADA. We incubated HUVECs and HeLa cells in the presence of different concentrations of Au_DADA for 24 h and analyzed them by CCK-8. We observed no obvious loss of viability of both HUVECs and HeLa cells, which highlights the outstanding biocompatibility of Au_DADA (Figure S6).
Stability of Au_DADA
We tested the stability of Au_DADA at different pH values (pH=1-14) (Figure S7). We put Au_DADA in aqueous solutions of different pH for 4 h. The colors of Au_DADA in aqueous solutions of different pH are essentially the same (Figure S7A). The UV-vis spectra of Au_DADA at various pH levels are also essentially the same (Figure S7B).
TEM images of S. aureus and Au_DADA. (A) S. aureus incubated with Au_DADA. (B) S. aureus by itself. (C) Morphology of Au_DADA. The inserted image is the full view of the bacterium.
Bacterial detection of ascites samples. (A) Absorption spectra and photos of Au_DADA incubated with bacteria. (B) The plot of A 600 nm/A 520 nm of AuNPs versus different types of bacteria in ascites samples. Mean values and standard deviations were obtained from five independent experiments.
To further test the selectivity of our assay, we added FeCl3, CaCl2, CuCl2, BaCl2, Hg(ClO4)2, AgNO3, NaCl, KCl into Au_DADA solution at a final concentration of 500 μM and stored at 37 °C for 8 h. Au_DADA did not aggregate according to the UV-vis spectra of Au_DADA. This result demonstrates the high selectivity of Au_DADA (Figure S8).
We evaluated the stability of Au_DADA stored for almost 15 months at 4 °C and for 2 months at 37 °C and found them to be well dispersed (Figure S9). These results demonstrate that Au_DADA has outstanding stability even in highly acidic or alkaline environments.
Applicability of the detection system in a real clinical setting
To evaluate the applicability of this detection system, we applied the method to detect bacteria within ascites samples from patients (Figure 5). We incubated Au_DADA with the clinical samples of ascites that have pathogens such as S. aureus, staphylococcus epidermidis (S. epidermidis), Stenotrophomonas maltophilia (S. maltophilia), or Pseudomonas aeruginosa (P. aeruginosa). Because these bacteria are clinically important, we explored the generality of our approach by expanding beyond the types of bacteria we used in the model experiments. The color of Au_DADA changed after incubating with different kinds of pathogens compared with the control group (Au_DADA media). When pathogens existed in the samples, the color of Au_DADA changed from red to blue. The absorption spectra of the samples have obvious changes. The absorbance band at 520 nm decreased, along with peak broadening and distinctive red-shift, and a broad absorption at 600 nm emerged (Figure 5A). We employed the ratio between the absorbance values of AuNPs at 600 nm and 520 nm (A 600 nm/A 520 nm) to test the degree of AuNP aggregation. The ratio of A 600 nm/A 520 nm is also a function of the presence of bacteria in clinical samples (Figure 5B). We also tested if the matrix of ascites affects the detecting system. The ascitic fluid had no effect to our assay, which shows the highly stability of Au_DADA (Figure S8). These results further validate the potential applications in bacterial monitoring.
Conclusions
We report a versatile and stable platform based on D-amino acid-coated AuNPs for colorimetric detection of bacteria that can also distinguish between S. aureus and MRSA. This is the first report that combines D-amino acid with AuNPs to detect bacteria. Although the sensitivity of this system is not as high as reported methods and it is a broad-spectrum sensor for bacteria, its stability is superior. We believe that Au_DADA is a promising biosensor for simple detection of bacterial contaminants for clinical diagnostics. We also open the way to bacterial theranostics using D-amino acid and nanomaterials.
Abbreviations
AuNPs: gold nanoparticles; S. aureus: Staphylococcus aureus; MRSA: methicillin-resistant Staphylococcus aureus; SBP: spontaneous bacterial peritonitis; CTAB: cetyltrimethylammonium bromide; DADA: D-alanyl-D-alanine; LALA: L-Alanyl-L-alanine; MDR: multidrug-resistant; DI: Deionized; TEM: transmission electron microscopy; UV-vis: ultraviolet-visible; LB: Luria-Bertani; K. pneumonia: Klebsiella pneumoniae; B. subtilis: Bacillus subtilis; L. monocytogenes: Listeria monocytogenes; CCK-8: Cell Counting Kit-8; DMEM: Dulbecco's modified Eagle's medium; FBS: fetal bovine serum; HUVECs: human umbilical vein endothelial cells; HeLa: human cervical cancer; LOD: the limit of detection; LOQ: the limit of quantitation; PDA: potato dextrose agar; FT-IR: Fourier Transform Infrared Spectroscopy.
Supplementary Material
Supplementary figures and table.
Acknowledgements
We thank the Ministry of Science and Technology of China (2013YQ190467), Chinese Academy of Sciences (XDA09030305) and the National Science Foundation of China (81361140345, 51373043, 21535001) for financial support.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Dang Y, Lou J, Yan Y, Yu Y, Chen M, Sun G. et al. The role of the neutrophil Fc gamma receptor I (CD64) index in diagnosing spontaneous bacterial peritonitis in cirrhotic patients. Int J Infect Dis. 2016;49:154-160
2. Wiest R, Krag A, Gerbes A. Spontaneous bacterial peritonitis: Recent guidelines and beyond. Gut. 2012;61:297-310
3. Chen Y, Xianyu Y, Jiang X. Surface modification of gold nanoparticles with small molecules for biochemical analysis. Accounts Chem Res. 2017;50:310-319
4. Yang X, Wang N, Zhang L, Dai L, Shao H, Jiang X. Organic nanostructure-based probes for two-photon imaging of mitochondria and microbes with emission between 430 nm and 640 nm. Nanoscale. 2017;9:4770-4776
5. Chen W, Li Q, Zheng W, Hu F, Zhang G, Wang Z. et al. Identification of bacteria in water by a fluorescent array. Angew Chem Int Ed. 2014;53:13734-13739
6. Carrillo-Carrion C, Simonet BM, Valcarcel M. Colistin-functionalised CdSe/ZnS quantum dots as fluorescent probe for the rapid detection of Escherichia coli. Biosens Bioelectron. 2011;26:4368-4374
7. Zhu C, Yang Q, Liu L, Wang S. Rapid, simple, and high-throughput antimicrobial susceptibility testing and antibiotics screening. Angew Chem Int Ed. 2011;50:9607-9610
8. Oh S, Jadhav M, Lim J, Reddy V, Kim C. An organic substrate based magnetoresistive sensor for rapid bacteria detection. Biosens Bioelectron. 2013;41:758-763
9. Zhu C, Yang Q, Liu L, Lv F, Li S, Yang G. et al. Multifunctional cationic poly(p-phenylene vinylene) polyelectrolytes for selective recognition, imaging, and killing of bacteria over mammalian cells. Adv Mater. 2011;23:4805-4810
10. Liu H, Rong P, Jia H, Yang J, Dong B, Dong Q. et al. A wash-free homogeneous colorimetric immunoassay method. Theranostics. 2016;6:54-64
11. Yu F, Li Y, Li M, Tang L, He J. DNAzyme-integrated plasmonic nanosensor for bacterial sample-to-answer detection. Biosens Bioelectron. 2017;89:880-885
12. Gao W, Li B, Yao R, Li Z, Wang X, Dong X. et al. Intuitive label-free SERS detection of bacteria using aptamer-based in situ silver nanoparticles synthesis. Anal Chem. 2017;89:9836-9842
13. Liao J, Li W, Peng J, Yang Q, Li H, Wei Y. et al. Combined cancer photothermal-chemotherapy based on doxorubicin/gold nanorod-loaded polymersomes. Theranostics. 2015;5:345-356
14. Cheng Y, Samia AC, Meyers JD, Panagopoulos I, Fei B. et al. Highly efficient drug delivery with gold nanoparticle vectors for in vivo photodynamic therapy of cancer. J Am Chem Soc. 2008;130:10643-10647
15. Huang X, Jain PK, El-Sayed IH, El-Sayed MA. Plasmonic photothermal therapy (PPTT) using gold nanoparticles. Laser Med Sci. 2008;23:217-228
16. Tao Y, Ju E, Ren J, Qu X. Bifunctionalized mesoporous silica-supported gold nanoparticles: intrinsic oxidase and peroxidase catalytic activities for antibacterial applications. Adv Mater. 2015;27:1097-1104
17. Daniel MC, Astruc D. Gold nanoparticles: Assembly, supramolecular chemistry, quantum-size-related properties, and applications toward biology, catalysis, and nanotechnology. Chem Rev. 2004;104:293-346
18. Zhu Z, Guan Z, Jia S, Lei Z, Lin S, Zhang H. et al. Au@Pt nanoparticle encapsulated target-responsive hydrogel with volumetric bar-chart chip readout for quantitative point-of-care testing. Angew Chem Int Ed. 2014;53:12503-12507
19. Pacardo DB, Neupane B, Rikard SM, Lu Y, Mo R, Mishra SR. et al. A dual wavelength-activatable gold nanorod complex for synergistic cancer treatment. Nanoscale. 2015;7:12096-12103
20. Jin X, Jin X, Chen L, Jiang J, Shen G, Yu R. Piezoelectric immunosensor with gold nanoparticles enhanced competitive immunoreaction technique for quantification of aflatoxin B-1. Biosens Bioelectron. 2009;24:2580-2585
21. Liu D, Chen X. An ultrasensitive biosensing platform employing acetylcholinesterase and gold nanoparticles. Methods Mol Biol. 2017;1530:307-316
22. Lei Y, Tang L, Xie Y, Xianyu Y, Zhang L, Wang P. et al. Gold nanoclusters-assisted delivery of NGF siRNA for effective treatment of pancreatic cancer. Nat Commun. 2017;8:15130-15144
23. Yang X, Yang J, Wang L, Ran B, Jia Y, Zhang L. et al. Pharmaceutical intermediate-modified gold nanoparticles: Against multidrug-resistant bacteria and wound-healing application via an electrospun scaffold. ACS Nano. 2017;11:5737-5745
24. Li M, Guan Y, Zhao A, Ren J, Qu X. Using multifunctional peptide conjugated Au nanorods for monitoring beta-amyloid aggregation and chemo-photothermal treatment of Alzheimer's disease. Theranostics. 2017;7:2996-3006
25. Yang Z, Song J, Dai Y, Chen J, Wang F, Lin L. et al. Self-assembly of semiconducting-plasmonic gold nanoparticles with enhanced optical property for photoacoustic imaging and photothermal therapy. Theranostics. 2017;7:2177-2185
26. Yu R, Ma W, Liu X, Jin H, Han H, Wang H. et al. Metal-linked immunosorbent assay (MeLISA): the enzyme-free alternative to ELISA for biomarker detection in serum. Theranostics. 2016;6:1732-1739
27. Peng M, Ma W, Long Y. Alcohol dehydrogenase-catalyzed gold nanoparticle seed-mediated growth allows reliable detection of disease biomarkers with the naked eye. Anal Chem. 2015;87:5891-5896
28. Soh JH, Lin Y, Rana S, Ying J, Stevens MM. Colorimetric detection of small molecules in complex matrixes via target-mediated growth of aptamer-functionalized gold nanoparticles. Anal Chem. 2015;87:7644-7652
29. Zhao Y, Tian Y, Cui Y, Liu W, Ma W, Jiang X. Small molecule-capped gold nanoparticles as potent antibacterial agents that target gram-negative bacteria. J Am Chem Soc. 2010;132:12349-12356
30. Zhao Y, Chen Z, Chen Y, Xu J, Li J, Jiang X. Synergy of non-antibiotic drugs and pyrimidinethiol on gold nanoparticles against superbugs. J Am Chem Soc. 2013;135:12940-12943
31. Feng Y, Chen W, Jia Y, Tian Y, Zhao Y, Long F. et al. N-Heterocyclic molecule-capped gold nanoparticles as effective antibiotics against multi-drug resistant bacteria. Nanoscale. 2016;8:13223-13227
32. Qu W, Liu Y, Liu D, Wang Z, Jiang X. Copper-mediated amplification allows readout of immunoassays by the naked eye. Angew Chem Int Ed. 2011;50:3442-3445
33. Liu D, Wang Z, Jiang X. Gold nanoparticles for the colorimetric and fluorescent detection of ions and small organic molecules. Nanoscale. 2011;3:1421-1433
34. Sun J, Xianyu Y, Jiang X. Point-of-care biochemical assays using gold nanoparticle-implemented microfluidics. Chem Soc Rev. 2014;43:6239-6253
35. Xianyu Y, Xie Y, Wang N, Wang Z, Jiang X. A dispersion-dominated chromogenic strategy for colorimetric sensing of glutathione at the nanomolar level using gold nanoparticles. Small. 2015;11:5510-5514
36. Xianyu Y, Sun J, Li Y, Tian Y, Wang Z, Jiang X. An ultrasensitive, non-enzymatic glucose assay via gold nanorod-assisted generation of silver nanoparticles. Nanoscale. 2013;5:6303-6306
37. Berry V, Gole A, Kundu S, Murphy CJ, Saraf RF. Deposition of CTAB-terminated nanorods on bacteria to form highly conducting hybrid systems. J Am Chem Soc. 2005;127:17600-17601
38. Ray PC, Khan SA, Singh AK, Senapati D, Fan Z. Nanomaterials for targeted detection and photothermal killing of bacteria. Chem Soc Rev. 2012;41:3193-3209
39. Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123-136
40. Liechti GW, Kuru E, Hall E, Kalinda A, Brun YV, VanNieuwenhze M. et al. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature. 2014;506:507-510
41. Wiedemann I, Breukink E, van Kraaij C, Kuipers OP, Bierbaum G, de Kruijff B. et al. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J Biol Chem. 2001;276:1772-1779
42. Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA. et al. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;325:1552-1555
43. Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. Fems Microbiol Rev. 2008;32:234-258
44. Tiyanont K, Doan T, Lazarus M, Fang X, Rudner DZ, Walker S. Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. P Natl Acad Sci USA. 2006;103:11033-11038
45. Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F. et al. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed. 2012;51:12519-12523
46. Lupoli TJ, Tsukamoto H, Doud EH, Wang TSA, Walker S, Kahne D. Transpeptidase-mediated incorporation of D-amino acids into bacterial peptidoglycan. J Am Chem Soc. 2011;133:10748-10751
47. Bowman SM, Free SJ. The structure and synthesis of the fungal cell wall. Bioessays. 2006;28:799-808
48. Enright MC. The evolution of a resistant pathogen - the case of MRSA. Curr Opin Pharmacol. 2003;3:474-479
49. Masuoka J. Surface glycans of Candida albicans and other pathogenic fungi: Physiological roles, clinical uses, and experimental challenges. Clin Microbiol Rev. 2004;17:281-310
50. Delgado-Silva Y, Vaz C, Carvalho-Pereira J, Carneiro C, Nogueira E, Correia A. et al. Participation of Candida albicans transcription factor RLM1 in cell wall biogenesis and virulence. Plos One. 2014;9:e86270-e86282
Author contact
![]() Corresponding author: Xingyu Jiang, Email: xingyujiangcn, Tel.: +86-10-82545620; Huawu Shao, Email: shaohwac.cn, Tel.: +86-028-82890818
Corresponding author: Xingyu Jiang, Email: xingyujiangcn, Tel.: +86-10-82545620; Huawu Shao, Email: shaohwac.cn, Tel.: +86-028-82890818
 Global reach, higher impact
Global reach, higher impact