13.3
Impact Factor
Theranostics 2019; 9(13):3780-3797. doi:10.7150/thno.34493 This issue Cite
Research Paper
Circulating miR-338 Cluster activities on osteoblast differentiation: Potential Diagnostic and Therapeutic Targets for Postmenopausal Osteoporosis
1. State Key Laboratory Breeding Base of Basic Science of Stomatology (Hubei-MOST) and Key Laboratory for Oral Biomedicine of Ministry of Education (KLOBM), School and Hospital of Stomatology, Wuhan University, Wuhan 430079, China.
2. Department of Dentistry, The Central Hospital of Wuhan, Tongji Medical College Huazhong University of Science and Technology, Wuhan 430014, China.
3. Instititue of Life Sciences, Hangzhou Normal University, Hangzhou 310071.
4. Department of Developmental Dentistry, University of Texas Health Science Center, San Antonio, Texas, 78229-3700, United States.
5. Department of Periodontology, School and Hospital of Stomatology, Wuhan University, Wuhan 430079, China.
Received 2019-2-28; Accepted 2019-5-6; Published 2019-5-31
Abstract
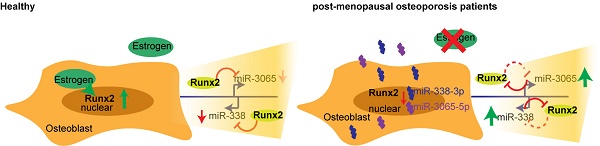
MicroRNAs (miRNAs) are the most abundant RNA species found in serum, and recently, several miRNAs have been found to be associated with osteoporosis. However, the development of such associated miRNAs into diagnostic and therapeutic targets remains unaddressed, mostly because of a lack of functional validation. Here, we identified circulating miR-338 associated with postmenopausal osteoporosis, and performed functional validation in vivo and in vitro.
Methods: We collected the serum from postmenopausal osteoporosis patients (N=15) and female volunteers of the same age but with normal bone density (N=15) and examined the enrichment of miR-338 cluster. We also confirmed such enrichment using mice subjected to ovariectomy at different stages. We employed primary bone marrow stromal cells from mice and the MC-3T3 cell line along with CRISPR, RNA-seq and ChIP-qPCR to validate the biological function of secreted miR-338 cluster on osteoblastic differentiation and their upstream regulators. Moreover, we generated miR-338 knockout mice and OVX mice injected with an inhibitor against miR-338 cluster to confirm its biological function in vivo.
Results: We observed a significant enrichment of miR-338 cluster in postmenopausal osteoporosis patients. Such enrichment was also prominent in serum from mice subjected to ovariectomy and was detected much earlier than bone density decreases revealed by micro-CT. We also confirmed the presence of an estrogen-dependent Runx2/Sox4/miR-338 positive feedback loop that modulated osteoblast differentiation, providing a possible explanation for our clinical findings. Moreover, deletion of the miR-338 cluster or direct intravenous injection of an miR-338 cluster inhibitor significantly prevented osteoporosis after ovariectomy.
Conclusion: Circulating miR-338 cluster in the serum could serve as a promising diagnostic and therapeutic target for postmenopausal osteoporosis patients.
Keywords: microRNA, miR-338 cluster, osteoporosis, Runx2
Introduction
Bone homeostasis requires a tightly controlled balance between bone-forming osteoblasts and bone-resorbing osteoclasts [1]. Postmenopausal osteoporosis is a disease in which there is a high level of bone remodeling and an imbalance between bone resorption and bone formation, resulting in decreased bone mineral density and disruption of bone microarchitecture [2-4]. However, the diagnosis of osteoporosis and osteopenia - based on measurement of bone mineral density (BMD) in the hip and spine using X-ray absorptiometry - is currently performed after the onset of the symptoms of either disease, which delays efforts at prevention or treatment.
Recently, due to advancements in high-throughput sequencing, an increasing number of novel circulating molecules such as microRNA have been found to be associated with osteoporosis and osteopenia [5-7]. Additionally, targeted deletion of Dicer, which is required for miRNA processing in mature osteoblasts, initially leads to delayed perinatal bone formation and a subsequent increase in postnatal bone acquisition [8, 9], indicating the involvement of osteoblast-related miRNAs in osteoporosis. However, there are still several challenges in developing associated miRNAs identified during fluid analysis into diagnostic or even therapeutic targets. First, most diagnostic targets have been found to be associated with the onset of the disease but have not been subject to follow-up functional study; thus, it is nearly impossible to distinguish mere association from actual biological function. During the past decade, numerous in vitro experiments have shown that several specific miRNAs are involved in modulation of osteoblast and osteoclast differentiation via direct targeting of osteoblast- or osteoclast-related genes; however, very few of them overlapped with clinically associated miRNAs. Second, virtually all diagnostic targets have been found to be the outcome of diseases, but rarely serve as candidate treatment targets. Because they were identified in patients already diagnosed with osteoporosis or osteopenia, it was not immediately clear whether their inhibition could delay or stop the progress of osteoporosis.
To address these challenges, we used an experimental pipeline to investigate the miR-338 cluster (including miR-338-3p and miR-3065-5p), of which miR-338-3p has been reported to be dramatically downregulated during osteoblast differentiation via partial arrest of the expression of FGFR2 and RUNX2 in vitro [10]. However, it has never been reported to be associated with osteoporosis, and its function during osteoblast differentiation has never been validated in vivo. Thus, we hypothesized that the in vitro culture environment resulting from the “overexpression” of miR-338-3p resembles the in vivo microenvironment in which bone marrow stromal cells (BMSCs) and/or osteoblasts are exposed to abundant levels of miR-338-3p during initiation of osteoporosis (Figure 2A). Since the microenvironment of BMSCs is tightly regulated by the circulatory system, in this study we utilized serum to serve as a biopsy sample to evaluate the levels of miR-338 cluster members. Compared with healthy controls, significant enrichment of the miR-338 cluster was observed in serum collected from postmenopausal patients diagnosed with osteoporosis and ovariectomized (OVX) mice. Given the high abundance of the miR-338 cluster in bone compared with other types of tissue, we presumed that BMSC/pre-osteoblasts were the major source of these circulating miRNAs. By using an in vitro co-culture model, we confirmed that the miR-338 cluster could inhibit osteoblast differentiation in a “paracrine” manner, which partially explained how the circulation of the miR-338 cluster promoted osteoporosis. We then identified an estrogen-dependent positive feedback loop between Runx2/Sox4 and miR-338 cluster that governs osteoblast differentiation in vitro. Direct intravenous injection of inhibitors of either or both of the miR-338 cluster members was able to greatly reduce the level of the miR-338 cluster in serum and BMSCs, and greatly reduced osteoporosis symptoms in ovariectomized mice. Finally, we confirmed that miR-338 cluster knockout mice exhibited clear increase in bone mass and pathologically decreased bone erosion. Our findings revealed the presence of an intrinsic estrogen-dependent Runx2/Sox4-miR-338 feedback loop that governs osteoblast differentiation, and revealed the circulating miR-338 cluster members to be potential diagnostic and therapeutic targets for postmenopausal osteoporosis.
Results
miR-3065-5p inhibited mouse osteoblast differentiation in vitro
We first addressed the expression pattern of the miR-338 cluster in embryonic and adult mice and found expression in bone, heart, brain, and lung we tested. We next examined the miR-338-3p and miR-3065-5p expression pattern during embryonic development in vivo. In situ hybridization using LNA-modified miRNA probes showed that miR-338-3p and miR-3065-5p were expressed in the neural crest at embryonic day 9.5 (E9.5) and were later expressed strongly in the limbs (E10.5 and E11.5) (Figure 1B). In adult tissues collected from 2-month-old mice, miR-338-3p was enriched mainly within the brain and was also highly expressed in bone tissues (normalized based on its expression level in the heart; Figure S1). However, although detectable, miR-3065-5p was not highly expressed in bone.
miR-3065-5p inhibits osteoblast differentiation in vitro. A) Screen capture of UCSC genome browser (mm9) showing the location of miR-338 and miR-3065 in Aatk locus. B) Whole mount in situ hybridization of miR-338-3p and miR-3065-5p in E9.5, E10.5 and E11.5 embryos. Blue staining indicates positive signals of miRNA of interest. C) Expression of miR-338-3p and miR-3065-5p during osteoblastic differentiation of BMSCs. Three independent groups of BMSCs were isolated from wildtype mouse. Expression levels for each miRNA were normalized to its expression level in D0 relative to U6. D-F) qRT-PCR showing the expression levels of Osx, Opn and Runx2 after overexpression of miR-3065-5p. (n=3 for each group, data represent means ± s.d, t-test, *p<0.05, ** p<0.01). G) Western blot showing RUNX2, OPN and OSX on D0, D9 after overexpression of miR-3065-5p or NC. H) Representative results of alizarin red showing mineralization changes after overexpression of miR-3065-5p or NC.
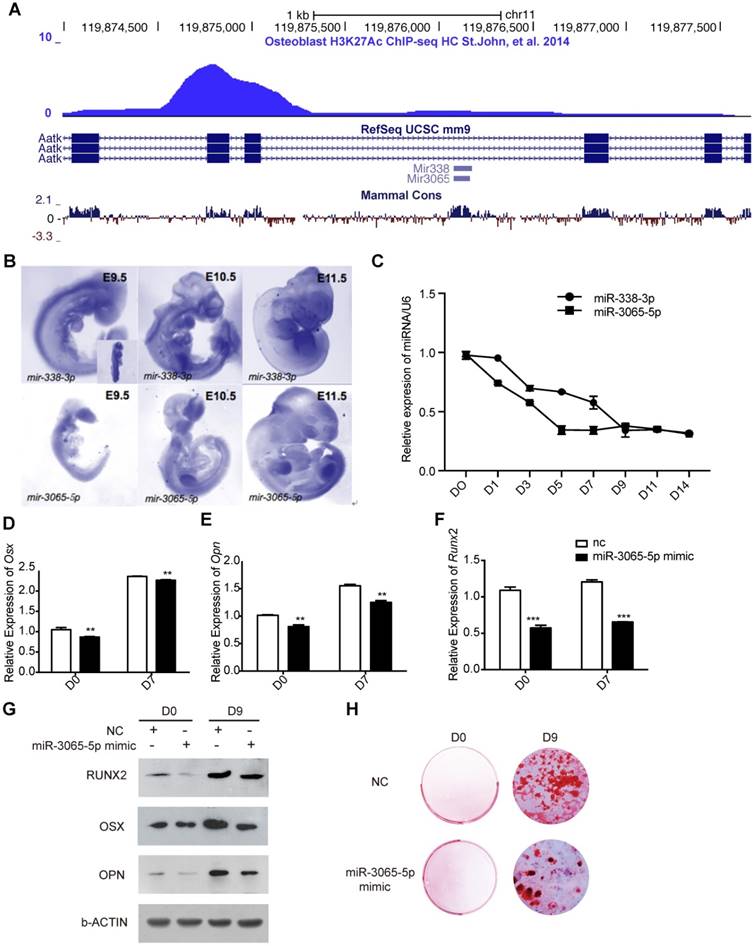
Based on data from an H3K27Ac ChIP-seq assay performed on an osteoblast cell line [11] (Figure 1A) and a similar in vivo expression pattern for both of these miRNAs in mouse embryonic limbs, we sought to determine whether miR-3065-5p exhibited expression changes during osteoblast differentiation similar to those found for miR-338-3p [10]. BMSCs were isolated from mouse femurs and subjected to osteogenic induction. The expression of miR-338-3p and miR-3065-5p at different time points after osteogenic induction was then determined using qRT-PCR. As expected, both miR-338-3p and miR-3065-5p were downregulated during in vitro osteoblast differentiation (Figure 1C).
To address whether miR-3065-5p functions similarly to miR-338-3p during osteoblast differentiation, BMSCs were transfected with either a miR-3065-5p mimic or a negative control (NC). Osteogenic induction was then initiated at 24 h post-transfection. At day 7 after induction, the key osteogenic specific genes Osx, Opn, and Runx2 were significantly downregulated due to the overexpression of miR-3065-5p (Figure 2D-F). On day 9 posttransfection, we also found that RUNX2, OSX and OPN were significantly downregulated due to the overexpression of miR-3065-5p but were still elevated compared with their levels on day 0 (Figure 1G). Additionally, alizarin red staining was decreased in miR-3065-5p-overexpressing cells (Figure 1H). These data, along with our previous findings, indicated that the two members of the miR-338 cluster downregulate osteoblast differentiation in vitro.
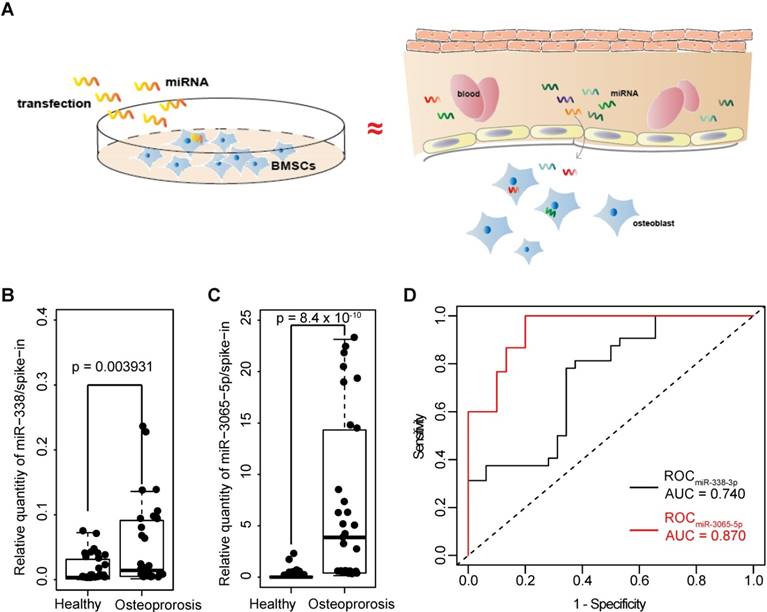
Enrichement of serum miR-338 cluster in postmenopausal osteoporosis patients. A) Hypothesis for the clinical study. In vitro culture environment resulting from the “overexpression” of miR-338-3p resembles the in vivo microenvironment in which BMSCs and/or osteoblasts are exposed to abundant miR-338-3p during initiation of osteoporosis. B) and C) Enrichment of miR-338-3p and miR-3065-5p in the serum (500uL) collected from 15 female postmenopausal osteoporosis patients (T<-2.5) and 15 postmenopausal healthy volunteers (T>-1). Expression level of each target miRNAs were normalized to the spike-in control. Kolmogorov-Smirnov test was employed to evaluate the difference between two clinical groups. D) ROC curves to show the enrichment of miR-338-3p and miR-3065-5p in determine patients with or without osteoporosis.
miR-338-3p and miR-3065-5p are significantly enriched in serum collected from female osteoporosis patients
Osteoporosis is the result of a combination of reduced osteoblast differentiation and overactivated osteoclasts [12]. Given their negative effect on osteoblast differentiation and based on our hypothesis (Figure 2A), we investigated whether both members of the miR-338 cluster were more enriched in serum (which directly contacts BMSCs and osteoblasts) that was collected from osteoporosis patients compared with serum collected from healthy controls. To examine the expression of these miRNAs in postmenopausal osteoporosis patients, we analyzed serum samples from 15 postmenopausal healthy controls (T score > -1) and 15 postmenopausal osteoporosis patients (T-score < -2.5) within a cohort of female subjects that had similar ages and Body Mass Indexes (BMIs). In these samples, the relative expression of miR-338-3p and miR-3065-5p were highly enriched in serum collected from the female osteoporosis patients (Figure 2, A and B). We employed ROC analysis to estimate the diagnostic value of miR-338-3p and miR-3065-5p. The results from the small clinical cohort showed that the dCt values (normalized to spike-in as described in Methods) of both miR-338-3p and miR-3065-5p were associated with a postmenopausal osteoporotic phenotype (AU values of 0.740 and 0.870, respectively; Figure 2C).
miR-338-3p and miR-3065-5p are gradually enriched during the progression of osteoporosis in an OVX (ovariectomy) mouse model
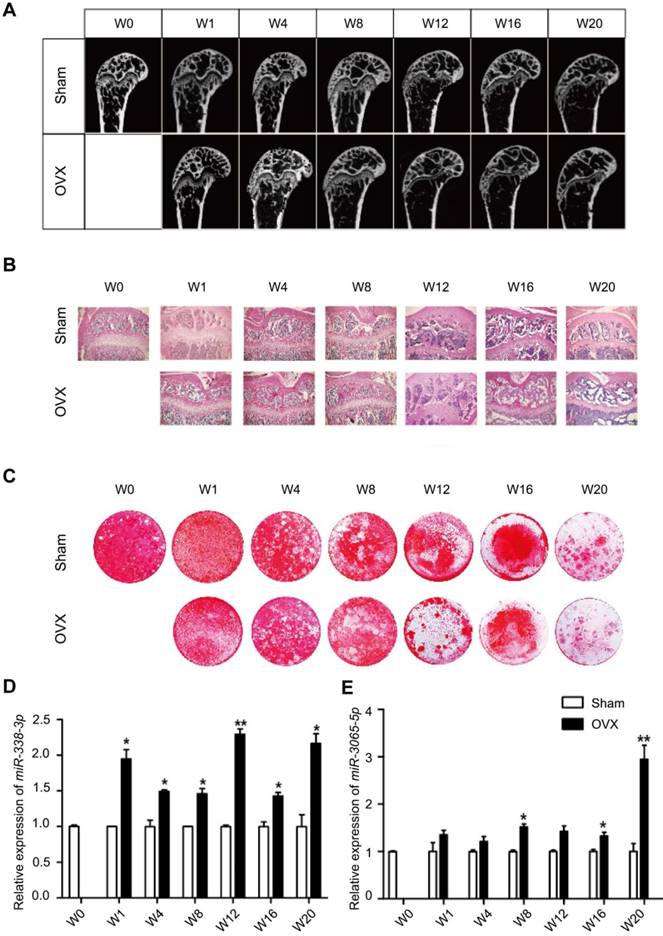
We further explored the relationship between miR-338 cluster expression and bone formation in mouse models. Eight-week-old female Balb/c mice were randomly divided into 2 groups and subjected to either a sham operation or bilateral ovariectomy. Micro-CT analysis (Figure 3A) and HE staining (Figure 3B) revealed apparent bone loss (decrease of trabecular bone volume and number) at 12 weeks post-OVX operation; the osteogenic differentiation capability was dramatically reduced as well, despite the age effect (Figure 3C). We then determined the enrichment of miR-338-3p and miR-3065-5p in serum from mice after different lengths of OVX treatment using qRT-PCR. In line with our findings in the clinical cohort, we found a bone loss-related increase in the miR-338-3p and miR-3065-5p levels (normalized to those of the sham group at the same time point) in whole serum from OVX mice. It was noted that miR-338-3p and miR-3065-5p exhibited significantly higher levels 1 week and 8 weeks post-OVX operation, much earlier than the time point when bone loss was detectable using micro-CT and HE staining (12 weeks). Collectively, the human and mouse results suggest a potential diagnostic role for miR-338-3p and miR-3065-5p in postmenopausal osteoporosis.
Secreted miR-338-3p and miR-3065-5p impair osteoblast differentiation in vitro
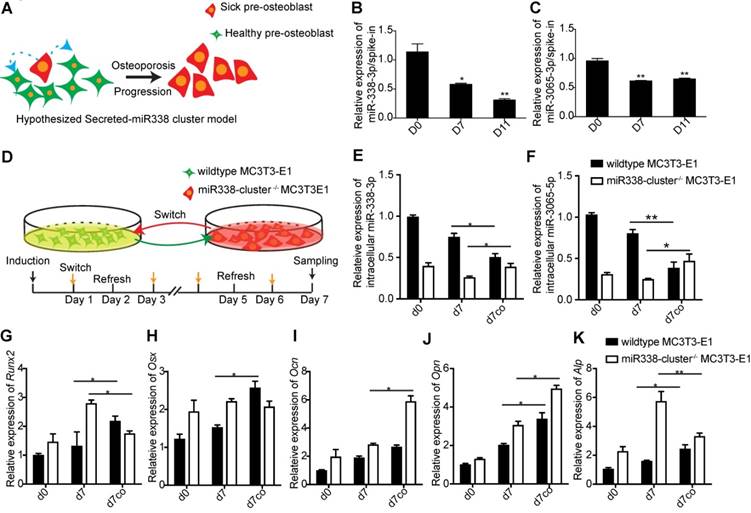
BMSCs and (pre-)osteoblasts greatly contribute to the components of serum and that bone is the second-most miR-338-cluster-enriched tissue that was examined (Figure S1). To determine the pathological role played by serum miR-338 cluster during osteoporosis, we first determined if the miR-338 cluster that was secreted by pre-osteoblasts could inhibit osteoblast differentiation. We addressed this question using a simplified “paracrine” model: in the beginning stages of osteoporosis, a small population of pre-osteoblasts secreted the miR-338 cluster into the serum around neighboring “healthy” pre-osteoblasts; consequently, the miR-338 cluster could then be transported into cells, where it inhibits osteoblast differentiation and thereby accelerates the progression of osteoporosis (Figure 4A). We first measured the relative enrichment of miR-338 in the culture medium during osteogenic induction in vitro. We found that in the supernatant used to maintain the MC3T3-E1 cell line, the levels of miR-338-3p and miR-3065-5p showed similar downregulated expression patterns during osteoblast differentiation (Figure 4, B and C). To prove that the secreted miR-338 cluster could cross into cells, we first mutated the miR-338 cluster in MC3T3-E1 cells (miR338-cluster -/-) to rule out the effects of endogenous miR-338 (Figure S2). We induced osteogenic differentiation in wildtype and miR338-cluster-/- MC3T3-E1 cells, replaced the culture medium every day, and monitored the relative expression levels of these two miRNAs during this process. We found that supernatant from wildtype MC3T3-E1 cells could rescue miR-338 cluster expression in miR338-cluster-/- MC3T3-E1 cells (Figure 4E and F). Additionally, the supernatant from miR338-cluster-/- MC3T3-E1 cells could promote osteoblast differentiation of WT MC3T3-E1 cells, as indicated by the elevation of Alp, Ocn and Opn expression (Figure 4, G to I). However, the expression of Runx2 and Osx remained unchanged (Figure 4, J and K). Overall, the results of the co-culture experiments showed that the secreted miR-338 cluster from MC3T3-E1 cells could be transported into the adjacent cells and inhibit osteoblast differentiation.
miR-338-3p and miR-3065-5p downregulated genes promoting osteoblast differentiation
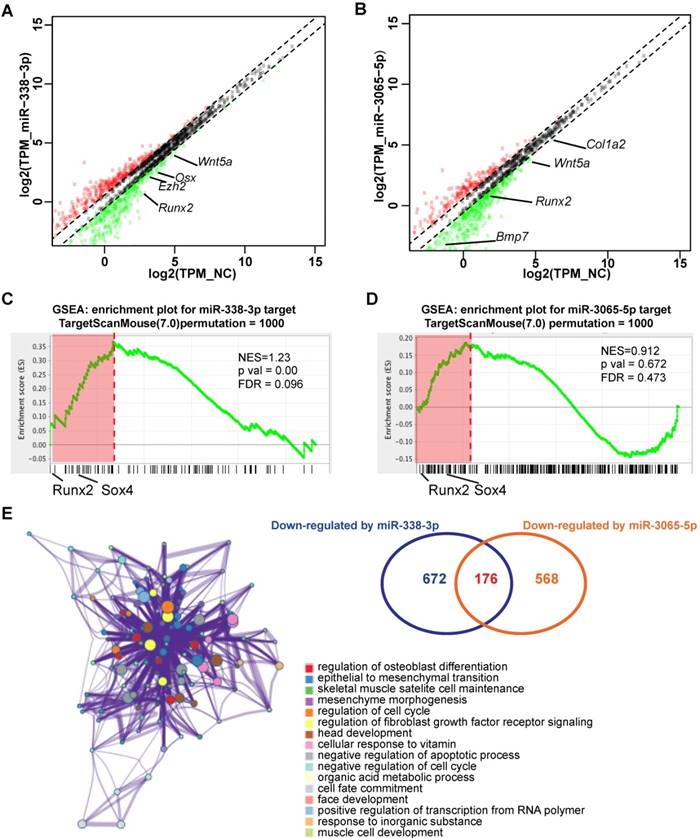
Next, to gain further insight into the molecular mechanisms by which the miR-338 cluster regulates osteoblast activity, we profiled the transcriptome in BMSCs that overexpress miR-338-3p (Figure 5A) or miR-3065-5p (Figure 5B) using RNA-seq. RNA-seq analyses revealed that both miR-338-3p and miR-3065-5p down-regulate some osteoblast-related genes such as Runx2, Bmp7, and Wnt5a. Additionally, gene set enrichment assay (GSEA) revealed that the genes downregulated by miR-338-3p were enriched with genes predicted to be directly regulated by miR-338-3p using TargetScan Mouse (v.7.0) (Figure 5C; NES = 1.23, FDR q-value = 0.096); however, no significant correlation was found for the genes downregulated by miR-3065-5p (Figure 5D; NES = 0.912, FDR q-value = 0.473). By comparing the two sets of RNA-seq results, we identified 52 genes that were significantly upregulated and 176 genes that were downregulated by both miR-338-3p and miR-3065 when compared to scrambled controls.
Enrichment of serum miR-338 cluster during osteoporosis progression in ovariectomized mice. (A) Representative micro-CT images and (B) HE staining of mouse femurs at different time points after ovariectomy. (C) Representative aliziran red staining of BMSCs induced with osteoblastic medium for 11 days harvested from femurs after ovariectomy. Relative enrichment of miR-338-3p (D) and miR-3065-5p (E), normalized to the relative expression level in control group at the same time-point. n=3, t-test, *p<0.05, ** p<0.01. OVX: ovariectomy. W: weeks after ovariectomy surgery.
A gene ontology enrichment assay (Figure 5E) revealed that the downregulated genes were significantly enriched in positive effectors of osteoblast differentiation, such as Runx2, Osx, Wnt5a and Sox4. Moreover, the miR-3065-5p/miR-338-3p-overexpression profile was negatively correlated with the Runx2-knockdown upregulated gene set in MC3T3-E1 cells that was reported previously [13] (Figure S2; NES = -1.52 and FDR q-value = 0.09 for miR-338-3p; NES = -1.12 and FDR q-value = 0.24 for miR-3065-5p), indicating that Runx2 is a major factor that affects or functions similarly to miR-338-3p and miR-3065-5p. Of all the predicted direct targets, we focused on Runx2 and Sox4 due to their association with osteoporosis [14, 15] and their location in the leading-edge subsets revealed via GESA analysis (Figure 5C and D). Dual luciferase assays were performed and showed that miR-338-3p/miR-3065-5p overexpression reduced luciferase activity in the wild-type Runx2/Sox4 3'UTR, while mutation of the putative binding sites of miR-338-3p/miR-3065-5p significantly eliminated the inhibitory effect (Figure S3). Thus, these data confirmed that overexpression of miR-338 cluster in a pre-osteoblast cell line inhibits osteoblastic differentiation via direct targeting of the related master regulators Runx2 and Sox4.
Estrogen-dependent Runx2 and Sox4 negatively regulate miR-338-3p and miR-3065-5p expression during osteoblast differentiation
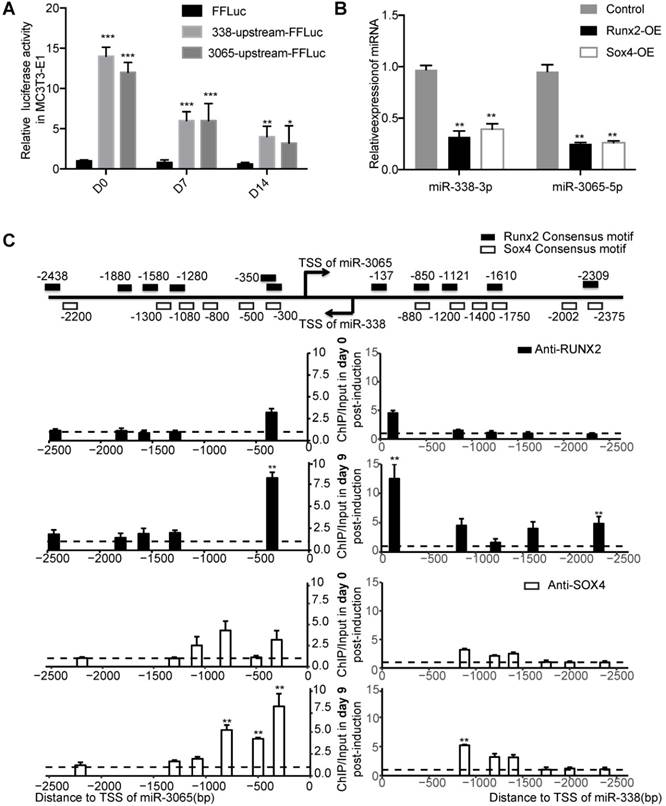
To uncover the driving force behind the upregulation and secretion of the miR-338 cluster in postmenopausal osteoporosis patients, we first determined which transcription factors directly modulated expression of these miRNAs. Although the miR-338 cluster is located within an intron in the apoptosis-associated tyrosine kinase (Aatk) gene and it has been found that miR-338-3p and AATK are co-regulated in human ß-cells [16], whether or not miR-338-3p and miR-3065-5p possess their own promoters remains unclear. We cloned the promoter regions of miR-338-3p/miR-3065-5p (approximately 2 kb upstream of the transcription start sites) and performed a luciferase activity assay. The activity of the promoter regions of miR-338-3p and miR-3065-5p exhibited a similar pattern of decrease during osteoblast differentiation in MC3T3 cells (Figure 6A), suggesting that miR-338-3p and miR-3065-5p possess their own promoters during osteoblastic differentiation.
Secreted miR-338 cluster inhibit osteoblastic differentiation. (A) Hypothesized model for secreted miR-338 cluster. Relative enrichment of miR-338-3p (B) and miR-3065-5p (C) in the culture medium during osteoblastic differentiation. N=3, t-test. (D) Co-culture model for validating hypothesis in (A). Relative expression of intracellular miR-338-3p (E) and miR-3065-5p (F) in different groups. d0: wildtype or miR-338-cluster-/- cells before osteoblastic induction, d7: gene expression in wildtype or miR-338-cluster-/- cells on day 7 post-osteoblastic induction, d7co: gene expression in wildtype or miR-338-cluster-/- cells on day 7 post-osteoblastic induction along with medium switch. Expression of Runx2 (G), Osx (H), Ocn (I), Opn (J) and Alp (K) in different groups of MC3T3-E1 cells. n=3, t-test, *p<0.05, ** p<0.01.
miR-338-3p and miR-3065-5p co-regulate osteoblast related genes. Scatter plot showing expression levels of genes responsive to miR-338-3p (A) or miR-3065-5p (B) in MC3T3-E1 cells. Values are plotted as the log2(TPM) from miR-338-3p or miR-3065-5p-overexpressing cells (vertical axis) versus NC scramble control (horizontal axis). Each point represents mean mRNA expression level from three independent biological replicates. Diagonal lines indicate the threshold for significant increase or decrease (1.5 fold) in expression. GSEA showing correlation between genes downregulated by miR-338-3p and predicted targets (C) and genes downregulated by miR-3065-5p and predicted targets (D). (E) GO enrichment network generated by metascape.org. NES: normalized enrichment score, FDR: false discovery rate.
We then scanned the promoters with JASPAR (2018) [17], which identified clusters of RUNX and SOX motifs (Figure 6C). Given their abundance in osteoblasts and the critical functions of Runx2 [18] and Sox4 [19] during bone development, we chose Runx2 and Sox4 as putative upstream factors of the miR-338 cluster in osteoblasts. Transient overexpression of Runx2 and Sox4 in MC3T3 cells significantly downregulated both miR-338-3p and miR-3065-5p (Figure 6B). Interestingly, ChIP-qPCR experiments at day 0 and day 9 after treatment of MC3T3-E1 cells with osteogenic induction medium revealed that RUNX2 and SOX4 were increasingly bound to the promoters of both miR-338-3p and miR-3065-5p (Figure 6C). Overall, the 3'UTR luciferase assay and ChIP-qPCR results revealed the presence of a negative-feedback loop between miR-338-3p/miR-3065-5p and Runx2/Sox4.
Runx2 and Sox4 are major upstream of miR-338 cluster during osteoblastic differentiation. (A) Promoter activities of the upstream of miR-338 or miR-3065 during osteoblastic differentiation of MC3T3-E1. n=3, t-test, *p<0.05, ** p<0.01. (B) Relative expression of miR-338-3p and miR-3065-5p after overexpression of either Runx2 or Sox4 in MC3T3-E1. (C) ChIP-qPCR for Runx2 and Sox4 near the TSS of miR-3065 and miR-338 during osteoblastic differentiation of mouse BMSCs. n=3, t-test, *p<0.05, ** p<0.01 (compared with the relative enrichment in D0).
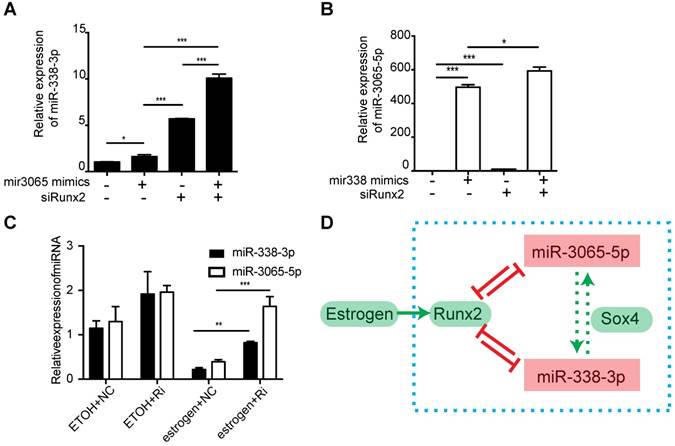
Given the similar expression patterns of miR-338-3p and miR-3065-5p during osteoblastic differentiation, we investigated whether miR-338-3p and miR-3065-5p could regulate each other via Runx2. We verified this via simple overexpression of either miR-338-3p or miR-3065 with or without perturbation of Runx2. As expected, we found that overexpression of either member of the miR-338 cluster could enhance the expression of the other member (Figure 7, A and B), and knockdown of Runx2 increased enrichment of miR-338-3p and miR-3065-5p in culture medium (Figure S4).
As we examined the abundance of the miR-338 cluster in postmenopausal women or ovariectomized mice, both of which suffer from estrogen deficiency, we hypothesized that estrogen was one of the major factors that stimulated expression of the miR-338 cluster. As Runx2 was previously reported to be enhanced by estrogen [20], we questioned whether estrogen could inhibit the level of the miR-338 cluster via stimulation of Runx2. An epistasis assay conducted in primary BMSC cultures revealed that estrogen could significantly inhibit the expression of miR-338-3p and miR-3065-5p, and siRNA targeting Runx2 could partially recover their levels (p < 0.01 according to a t-test; Figure 7C). These results collectively suggested that, in addition to serving as direct targets of the miR-338 cluster, estrogen-dependent Runx2 could directly negatively regulate transcription of the miR-338 cluster by forming a positive feedback loop during osteoblast differentiation. Additionally, the above in vitro experiments elucidated the local pathological role of miR-338 clusters during the inhibition of osteoblast differentiation in postmenopausal osteoporosis patients (Figure 7D).
Estrogen-dependent Runx2/Sox4-miR-338 cluster feedback loop in osteoblast. Expression level of miR-338-3p (A) or miR-3065-5p (B) after combinatory expression of miR-3065-5p or miR-338-3p mimics and siRunx2. (C) Expression level of miR-338-3p and miR-3065-5p after knockdown of Runx2 (Ri) with or without the presence of estrogen. (D) Schematic showing the estrogen-dependent Runx2/Sox4-miR-338 cluster in osteoblast. (n=3 per group,t-test, *p<0.05, ** p<0.01, *** p<0.001).
Inhibition of the miR-338 cluster in serum attenuates osteoporosis progression in vivo
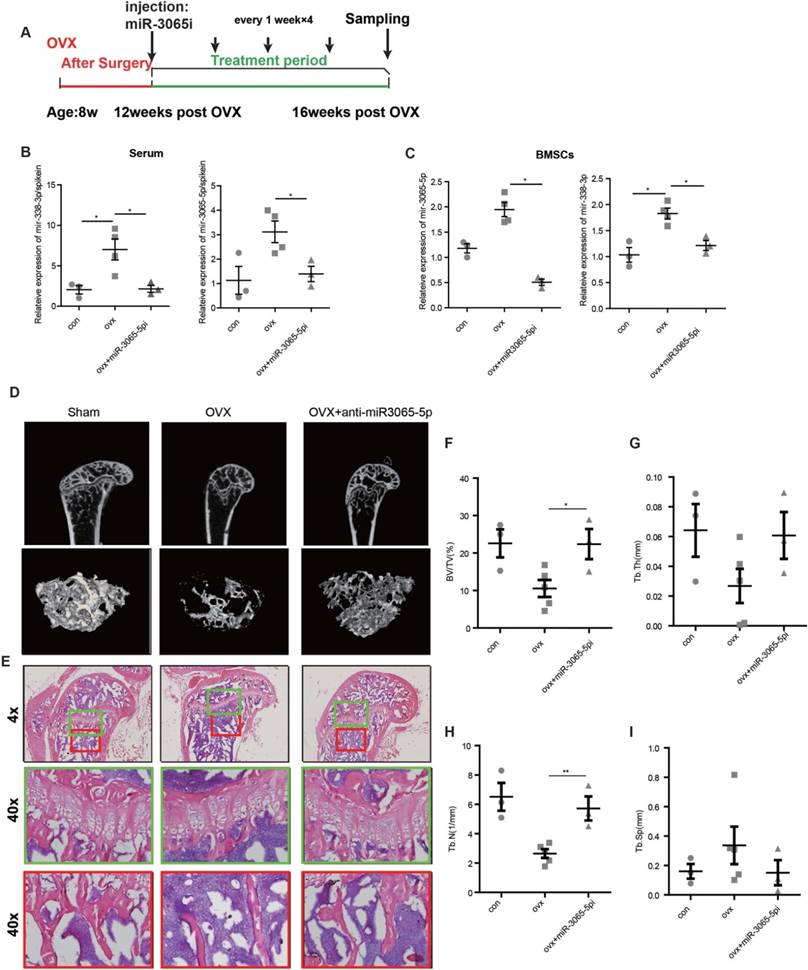
In addition to their potential roles in diagnosis, we investigated whether therapeutic inhibition of circulating miR-338-3p or miR-3065-5p could rescue or arrest osteoporosis progression in OVX mice. First, four weeks after a sham operation or OVX, mice were given either miR-338-3p or miR-3065-5p inhibitor (hereafter referred to as OVX+antimiR-338 or OVX+antimiR-3065) via a single tail vein injection to directly assess the impact of miR-338 on the regulation of osteoblast differentiation in vivo. OVX and sham mice received 4 injections of inhibitors per week to maintain the effects of the miR-338-3p (Figure S5A) or miR-3065-5p inhibitors (Figure 8A). Serum and intraosseous miR-338-3p and miR-3065-5p levels were substantially downregulated by miR-338-3p or miR-3065-5p inhibitor treatment (Figure 8 B and C; Figure S5B and C). Micro-CT images and HE staining of the distal femur showed remarkably increased bone mass in the OVX+antimiR-3065 (Figure 8D) and OVX+antimiR-338 mice (Figure S5D). Quantification analysis of the micro-CT results showed that the mice treated with inhibitors exhibited a significant increase in relative bone volume (BV/TV; Figure 8F and Figure S5F) and trabecular bone number (Tb.N; Figure 8G and Figure S5G), as well as a decrease in trabecular bone separation (Tb.Sp; Figure 8H and Figure S5H), while the thickness (Tb.Th) showed no significant differences (Figure 8I and Figure S5I) between the treated and sham mice. These results showed that intravenous injection of inhibitors against either miR-338-3p or miR-3065-5p significantly reduce the progression of osteoporosis in OVX mice via inhibition of the level of circulating and intracellular (BMSC) miR-338 cluster.
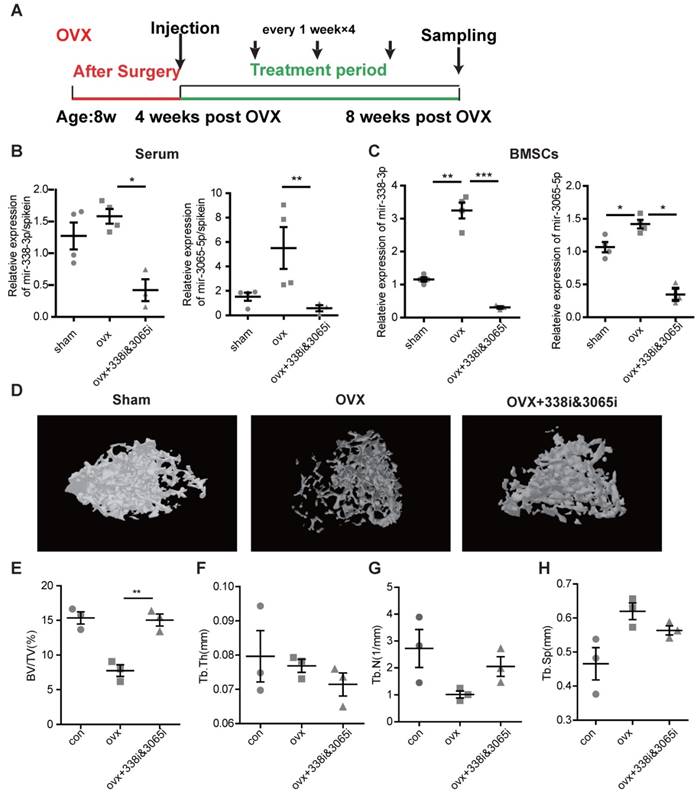
Finally, we sought to determine whether inhibition of miR-338 could prevent osteoporosis. We injected inhibitors targeting the miR-338 cluster during week 4-post OVX and collected the femurs during week 8 post-OVX (Figure 9A), earlier than the time-point when osteoporosis phenotype could be found by X-ray examination. In line with previous experiments, intravenous inhibition of miR-338 could significantly reduce its level in serum (Figure 9B) and BMSCs (Figure 9C). The micro-CT results showed that there were no significant differences in bone volume (Figure 9E) and trabecular bone number (Figure 9G) in the femurs collected from mice with early inhibition of the miR-338 cluster and the sham control mice. We also observed a significantly lower level of relative bone volume in the OVX mice compared with the mice treated with the miR-338 inhibitor (Figure 9E).
Inhibition of the miR-3065-5p in serum attenuated osteoporosis progression in vivo. (A) A schematic diagram illustrating the experimental design for the timeline of intravenous injection of miR-3065-5p inhibitor (miR-3065-5pi). Expression level of miR-338-3p and miR-3065-5p in serum (B) and BMSCs (C) isolated from different groups of mice on 16 weeks post OVX. Representative micro-CT (D) and HE staining (E) images in sham, OVX and OVX mice injected with miR-3065-5p inhibitor. Bone volume fraction (BV/TV) (F), Trabecular Thickness (Tb.Th) (G), average trabecular number (Tb.N) (H) and trabecular spacing (Tb.Sp) (I) in femurs isolated from different groups. (n=3 per group, t-test, *p<0.05, ** p<0.01, *** p<0.001).
Early inhibition of the miR-338 cluster in serum prevented osteoporosis in vivo. (A) A schematic diagram illustrating the experimental design for the timeline of intravenous injection of inhibitors targeting miR-338-3p and miR-3065-5p (338i&3065i). Expression level of miR-338-3p and miR-3065-5p in serum (B) and BMSCs (C) isolated from different groups of mice on 16 weeks post OVX. Representative micro-CT (D) images in sham, OVX and OVX mice injected with miR-3065-5p inhibitor. Bone volume fraction (BV/TV) (E), Trabecular Thickness (Tb.Th) (F), average trabecular number (Tb.N) (G) and trabecular spacing (Tb.Sp) (H) in femurs isolated from different groups. (n=3 per group, t-test, *p<0.05, ** p<0.01).
Loss of the miR-338 cluster lowers susceptibility to osteoporosis
To rule out the injection-effect or reagent effect caused by the miRNA inhibitor, we generated miR-338 cluster knockout mice (hereafter referred to as miR-338-/-) to confirm its in vivo effect on bone disease genetically. The expression levels of miR-338-3p and miR-3065-5p were significantly lower in the BMSCs collected from miR-338-/- mice compared with those from wildtype mice (Figure S6C and D). The mutant mice exhibited no different behavior or pathological phenotype compared with its heterozygous and wildtype siblings. Also, there are no significant difference in bone length between wildtype and mutant mice (Figure S6E). We then generated a model of ovariectomy in two-month-old miR-338-/- mice and their wildtype siblings. 3D reconstruction and micro-CT analysis of the trabecular bone at the same level near the growth plate of the distal femur revealed that miR-338-/- mice exhibited higher trabecular bone volume and thickness (Figure S7A-E). After ovariectomy, the bone volume in miR-338-/- mice was significantly decreased but still comparable to that in the wildtype (sham) mice (Figure S7A and B). Moreover, the osteoclast number (cells with more than 3 nuclei) was greatly decreased in miR-338-/- mice (Figure S8). We harvested BMSCs from the femurs of miR-338-/- mice and their siblings and treated them with osteogenic induction medium for 11 days. The transcription levels of Runx2, Opn, Osx, and Ocn in knockout mice were significantly higher at day 11 post-induction than in wildtype mice; however, the level of Alp was significantly lower at day 11 (Figure S7F-J). Western blotting of OSX and RUNX2 revealed the same differences (Figure S7L). Alizarin Red staining also showed that the BMSCs from miR-338-/- mice generated a greater number of mineralization nodules at day 11 than with those from wildtype mice.
Overall, our in vivo results showed direct intravenous injection of miR-338 cluster or genetic inhibition of this miRNA cluster could reduce or even prevent an osteoporosis phenotype, suggesting miR-338 cluster as a novel therapeutic target for osteoporosis in postmenopausal patients. An overall model for this study is summarized in the graphical abstract.
Discussion
This study identified the miR-338 cluster in osteoblasts as an important inhibitor of bone formation in both physiological and pathophysiological conditions. Our results suggest that therapeutic inhibition of the miR-338 cluster in osteoblasts may promote bone formation and even reverse osteoporosis. As such, anti-miR-338/miR-3065 compounds represent new therapeutics that could be used to treat and prevent postmenopausal osteoporosis. Furthermore, miR-338 levels may also be a diagnostic tool.
Multiple miRNAs are important post-transcriptional regulators of gene expression during bone formation [21, 22]. Additionally, several miRNAs are significantly enriched in osteoclasts or osteoblasts in different physical or pathological circumstances, such as aging or genetic disorders[23-26]. However, most clinical investigations have been performed using bone samples from patients, which is not feasible for noninvasive early diagnosis of osteoporosis. Additionally, most of the reported methods that have been used for in vivo bone protection via either the inhibition or overexpression of miRNAs are performed using a targeting vector [23], which increases the cost for future clinical translation. In our study, we observed high enrichment of miR-338-3p and miR-3065-5p in serum from aged postmenopausal osteoporosis patients, which was further confirmed in the ovariectomized mouse model. The ROC curves and T-scores based on the miR-338/miR-3065 levels indicate that both serum miR-338-3p and serum miR-3065-5p could be used for diagnosis. To overcome limits due to the use of a small research cohort, we employed a female mouse model of osteoporosis; we found that serum miR-338-3p and miR-3065-5p showed statistically significant increases in OVX mice at a very early stage during osteoporosis progression (1 week post-OVX for miR-338 and 8 weeks post-OVX for miR-3065). This observation provided an insight into the contribution of these two miRNAs to the pathophysiological regulation of bone formation during the progression from osteopenia to osteoporosis and indicated the potential diagnostic roles of miR-338 and miR-3065 during osteoporotic progression, which has been poorly investigated in previous osteoporosis-related studies. During this study and our previous study, we performed extensive investigations into the molecular and cellular mechanisms underlying the inhibition of osteoblast differentiation in vitro by the miR-338 cluster and generated a miR-338 knockout mouse to confirm these findings in vivo. The knockout mice exhibited normal behavior compared with their wildtype siblings, but X-ray examination revealed that they exhibited increased bone volume and were resistant to osteoporosis progression after receiving ovariectomy surgery. Similarly, when we systemically administered anti-miR-338/miR-3065 oligonucleotides that resulted in functional inhibition of the targeted miRNA, we observed markedly increased bone mass in OVX mice. This suggests that miR-338/miR-3065 is an important target in bone disorder diseases and could potentially protect patients from bone loss. Additionally, these circulating miRNAs show potential as blood-based biomarkers of skeletal disease. To validate the role of miR-338 cluster in bone development and osteoporosis progression in vivo and rule out the side effect caused by inhibitor injection, we further genetically blocked the expression of this cluster using knockout model. In consistent with our mice injected with miR inhibitor, these miR-338 mutant mice exhibited enhanced bone volume and slower progression of osteoporosis after ovariectomy.
The onset and progression of osteoporosis are mainly caused by the imbalance between osteoblast and osteoclast activity. In our miR-338 mutant mice, we found the protective effect of anti-miR-338 against osteoporosis were associated with both their negative effect on osteoblast differentiation and positive effect on osteoclast differentiation. These findings are in consistent with the previous published in vitro results that miR-338-3p could inhibit osteoblastic differentiation of BMSCs through targeting Runx2 and Fgfr2 [10], while promote osteoclast differentiation though targeting Mafb [27]. In the present study, we revealed a new mechanism whereby the miR-3065 cluster was involved in the regulation of both Runx2 and Sox4 in bone tissues. Runx2 and Sox4 activity favors bone formation by promoting osteoblast-specific gene expression, and previous studies revealed that the expression of Runx2 and Sox4 are both estrogen-dependent. Our data demonstrated that the changes in the miR-338 cluster level in physiological conditions resulted in alterations in the expression level of Runx2 and Sox4; meanwhile, Runx2 and Sox4 could greatly influence the expression of both miR-338 and miR-3065 by binding to their promoter regions. We therefore elucidated a novel positive feedback loop that utilized Runx2, Sox4, miR-338 and miR-3065 to maintain the physiological status of osteoblasts that is regulated by estrogen. The experimental evidence from in vitro and in vivo studies strongly suggested that Runx2 and Sox4 could serve as two critical functional targets of the miR-338 cluster and may mediate its regulation during bone formation. Besides our current findings on miR-338 cluster, it has been reported that miR-3065-5p can directly target NRP2 and FLT1 [28], while both of which exhibited positive effect on osteoblast differentiation thereafter increase bone mass [29, 30]. However, the regulation of this estrogen-dependent feedback loop was validated only in osteoblasts; its function and mechanism in osteoclasts remain unclear.
Given the progressive aging of the general population, postmenopausal osteoporosis is a growing public health concern. A major need during treatment of this disease is to identify safe anabolic agents that can increase bone formation on a long-term basis and to such an extent that they compensate for the increase in bone resorption caused by menopause. Given that the seed sites of miR-338-3p and miR-3065-5p are conserved in both humans and mice, and the similar degree of enrichment in serum, the intravenous injection of inhibitors that target them may be potentially applicable to patients with osteoporosis; however, a more cost-efficient method of miRNA inhibition, such as the use of plasmid-based miRNA inhibitors [31], could also be employed for further investigation. Thus, our results suggest that circulating miR-338 cluster is a potential diagnostic and therapeutic target for the treatment of postmenopausal osteoporosis.
Methods
Study approval
All animal care protocols and experiments were reviewed and approved by the Institutional Animal Care and Use Committees at the School and Hospital of Stomatology of Wuhan University (protocol No. 00266115, S07916110A). All mice were maintained in the specific pathogen-free facility of the Laboratory Animal Research Center at School and Hospital of Stomatology attached to Wuhan University. The clinical study was approved by the central hospital of Wuhan (protocol No. hospital-2015-12), and written informed consent was obtained from all participants prior to whole blood collection.
Construction of miR-338 cluster knockout Mice
We first designed a pair of CRISPR-Cas9 plasmids generating gRNAs flanking miR-338 clusters. Briefly, oligos targeting gRNA target sites GGGTCCATGCCTATGGGTG (mm10 chr11:120014710-120014731) and GGGGCTGGCCCTGTAGTGGG (mm10 chr11:120014963-120014985) were annealed and inserted into pX330-U6-Chimeric_BB-CBh-hSpCas9 (a kind gift from Feng Zhang, Addgene plasmid # 42230; http://n2t.net/addgene:42230; RRID:Addgene_42230). Pair of primers flanking miR-338 clusters were designed for genotyping. Successfully knockout of miR-338 cluster would result in a PCR product sized as 380bp while wild-type product would be 600bp.
C57BL/6 mice were used as embryo donors and were superovulated. Two CRISPR-Cas9 plasmids were linearized, purified and injected into the cytoplasm of pronuclear-stage fertilized eggs. The injected zygotes at 2-cell stage were transferred into the oviducts of pseudopregnant mice. Genomic DNA was extracted from the tail tips of the off-springs and genotyped. The heterozygotes were out-crossed with the wild-type C57BL/6 mice for three generations to reduce the off-target effect.
In-situ hybridization
In-situ hybridization probe for LNA-modified mmu-miR-338-3p and mmu-miR-3065-5p labeled with DIG on double ends were synthesized by Exiqon (Denmark). For whole-mount in situ hybridization, embryos were collected and fixed with buffer (3.7% formaldehyde, 100mM MOPS, PH 7.4 mM EGDA, 1mM MgSO4) at 4°C overnight with rotation. Following the previously reported protocol [32] all probes were used in a concentration of and hydrolyzed with hybridization buffer. After proteinase K treatment and pre-hybridization, hybridization with probes was performed at 70 °C overnight. After washing step, embryos were incubated with anti-IDIG antibody (Roche, Germany). In the end, signals were visualized with NBT/BCIP (PIERCE, USA). Samples were washed by PBS supplemented with 0.01% Triton-X and cleaned by methanol at room temperature for 2 hours. For section in-situ hybridization, embryos were fixed in 4% paraformaldehyde dissolved in PBS overnight and dehydrated with ethanol followed by embed in paraffin. Section were incubated with probes at 70 °C overnight and washed at 60 °C followed by incubation with anti-DIG antibody (Roche, Germany) at 4 °C overnight. Samples were washed and signals were visualized with NBT/BCIP (PIERCE, USA).
Mouse ovariectomy experiment and sample collection
Female mice at different age were randomly divided into two groups (at least 3 litters per group) and underwent either sham operation or bilateral ovariectomy (OVX) under general anesthesia by the dorsal approach. At different time points post-OVX operation, 1mL blood samples were drawn from orbits, after which mice were euthanized and tibias were collected followed by either BMSC collection or fixation individually in 4% paraformaldehyde (PFA) overnight at 4 °C for further analysis.
Study Population
We recruited 30 unrelated postmenopausal Chinese women, 58-68 year of age, including 15 osteoporosis patients and 15 healthy female volunteers from Tongji medical college Huazhong University of Science & Technology, the Central Hospital of Wuhan. According to the WHO criteria, subjects with the femoral neck and/or lumbar spine T score ≤ -2.5 were considered osteoporosis, T score of -1 or above were normal. Postmenopause is defined as the date of the last menses followed by at least 12 months of no menses. Patients with diabetes, malignancy, coronary heart disease or other severe diseases in the previous 5 years were excluded in our study. Also, if the subjects had received treatment with glucocorticoids, estrogens, thyroid hormone, parathyroid hormone, fluoride, bisphosphonate, calcitonin, thiazide diuretics, barbiturates, or antiseizure medication, they were also excluded. All patients' information including age, BMI, serum ALP and daily exercise frequency were recorded in Table S1.
Blood samples were collected with red-top blood handling tubes from BD (Becton Dickinson) and started RNA-extraction immediately after separation of the serum (avoid freeze-thaw cycles). To detect the level of hemolysis, we first visually inspected the color of the isolated serum before RNA extraction; we discarded any pink cloudy serum and recollected serum from the same volunteers. After reverse-transcription reaction, we determined the ratio of miR-451a to miR-23a-3p using real-time PCR, which served as indicator for hemolysis [33]. We found all the ratios for all the serum are less than 5 indicating low hemolysis rate.
Cell culture, transfection and lentivirus transduction
Following previous reported protocol [34] for mouse BMSCs isolation, bone marrow cells were flushed from femora of female mice after different treatment and maintained in a medium with Dulbecco's modified Eagle's medium (DMEM, Hyclone, Australia) and 10% fetal bovine serum (FBS, Hyclone, Australia) in a 37 °C -5% CO2 incubator for two passages to remove the blood cells and lymphocytes. Cells were split when reaching 80-85% confluence. Only the BMSCs at the third passage were subjected to induction of osteogenic differentiation, extraction of RNA and protein, and Alizarin Red staining.
Osteogenic differentiation was performed with 50 µg/mL of ascorbic acid (Sigma, St Louis, MO, USA), 10 mmol/L of sodium 393 β-glycerophosphate (Sigma) and 10 nmol/L of dexamethasone (Sigma) for 11 days. Cells were stained with 2% Alizarin Red S (Sigma-Aldrich) at pH 4.2 to evaluate the cell matrix mineralization.
Mouse pre-osteoblast cell line, MC3T3-E1 (ATCC), were maintained in DMEM supplemented with 10% FBS in 37 °C. Cells were passaged when reaching 80-85% confluence with 0.25% Trypsin-EDTA (Life Technologies, Carlsbad, CA, USA). Oligos targeting Runx2 or Sox4 were transfected using lipofectamine 2000 (Life Technologies, USA). For dual luciferase assay, MC3T3 cells were electroporated (1×106 per transfection) with Amaxa Basic Nucleofector Kit for Primary Mammalian Fibroblasts (Lonza) using Nucleofector II (Lonza, Cologne, Germany) (program: U-020). Lentivirus for miR-338 knockout was generated by GenePharma (Suzhou, China) with titration of 2x108TU/ml, and infected MC3T3. 5 colonies were picked and genotyped after drug selection, and only the colony with homozygous mutation were kept for further experiment.
To test the dependency of estrogen in Runx2/Sox4-miR-338 cluster feedback loop, MC3T3 cells were challenged by estrogen (10nM, E2758 Sigma). siRNA targeting Runx2 and Sox4 were (designed by GenePhama, Shanghai).
In vitro overexpression of miR-338-3p and miR-3065-5p was performed by transfection of mimics for miR-338-3p and miR-3065-5p (Ambion, USA).
Reporter plasmids construction and dual luciferase assay
To test the promoter activity of miR-338 and miR-3065, 2064bp and 2487bp down- and up-stream of miR-338 were cloned from mouse genomic DNA extracted from MC3T3, and inserted into pENTR/D-TOPO plasmid for Sanger sequencing validation. These DNA fragments were then shuttled into cFos-FFLuc plasmid [35] for MC3T3 electroporation with pRL-TK (Promega, USA). Cell after electroporation were recovered for two days, and subjected to osteogenic differentiation. Different time point after induction, cells were lyzed by lysis buffer provided by dual luciferase reporter assay system (Promega) and the lysates were subjected to quantification of luminescent signal using luminometer from Promega. Each transfection group were repeated for 3 times (in 3 wells), Quantification for each well was repeated for 3 times, and value for the firefly luciferase assay was normalized to the renilla luciferase value then normalized to the relative firefly value from the empty control group. For functional validation of the direct targets of miR-338-3p and miR-3065-5p, the DNA fragments of the mouse Runx2 and Sox4 3'UTR, including the predicted miRNA binding sites, were cloned and ligated into pMIR-reporter (Ambion, USA). Site-directed mutageneses were performed with overlapping PCR to remove the binding sites of miR-338-3p or miR-3065-5p. Mimics for miR-338-3p, miR-3065-5p or scramble were co-transfected with related 3'UTR reporter plasmids along with pRL-TK (Promega, USA) using Lipofectamine 2000 (Life Technologies). 48 hours after transfection, cells were subjected to dual luciferase assay as described above.
miRNA extraction from serum, tissues and cells
The total RNA from tissues and cells was extracted directly using the miRNeasy Mini Kit (QIAGEN, USA). Briefly, tissues or cells were collected in a reaction tube, lysed with 700 µl QIAzol and mixed with 140 µl chloroform. After being centrifuged at 12,000g for 15 min at 4 °C, the upper aqueous phase was transferred to an RNeasy Mini spin column in a 2-ml collection tube and mixed with 100% ethanol. After being washed with 700 µl Buffer RWT and 500 µl Buffer RPE, the total RNA was collected and quantify by NanoDrop 3000 (Life Technologies). For extraction and quantification of miR-338-3p and miR-3065-5p in the collected serum, 1 mL serum was used, and cel-miR-39-3p (hereafter as spike-in, 20 fmol spike-in/200ul serum) was used as spike-in control. Serum were mixed with 500uL of QIAzol and followed by the protocol described above. All miRNA samples were reverse-transcribed and quantified by miRNA Reverse Transcription Kit (QIAGEN) and miScript SYBR Green PCR Kit (QIAGEN). For quantification of miRNA in cells or tissues, U6 was used as control, and for miRNA in serum, spike-in was used as internal control.
Histochemistry and micro-CT analysis
Briefly, after fixation, bone samples were decalcified in 10% EDTA for 30 days and embedded in paraffin. Longitudinally oriented bone sections from three femurs harvested from 3 mice in same group were stained with H&E, TRAP (Sigma) to quantify number of osteoclasts with more than 3 nuclei in 6 independent visions.
micro-CT protocol: After fixation, bone samples were stored in PBS at 4 °C. The morphology of the bone, the trabecular bone volume/tissue volume ratio (BV/TV: %), thickness (Tb.Th: m), trabecular number (Tb.N: mm-1), and trabecular bone separation (Tb.Sp: m) were recored by micro-CT machine(70kv, 114uA, 8w, 200ms, step=10.5um, SkyScan) and analyzed by CTAn. Each batch of mouse bone samples presented in each separated Figure were scanned in one experiment day, and analyzed together to rule out the batch effect.
RNA-seq library generation and data analysis
MC3T3 cells were transfected by the mimic for miR-338-3p, miR-3065-5p or scramble. Forty-eight hours post-transfection, total RNA was isolated using miRNeasy Mini Kit (QIAGEN), and genomic DNA was digested using Turbo-DNaseI (Life Technologies). RNA-seq libraries were generated and indexed with NEBNext Ultra RNA Library Prep Kit (NEB, USA). 150-bp-paired-end sequencing was performed using HiSeq2000 sequencer (Illumina, provided by Annoroad Genomics Company (China)). Sequencing data were pseudo-aligned to mm9 genome and quantified using Kallisto (v.0.43.0) [36], followed by differential genes identification using Sleuth R package [37] with a cut-off p<0.05. Relative gene expression level for most abundant transcript of each gene was presented in TPM (transcript per Killobase Million). GO enrichment assays for the genes downregulated by miR-338-3p and / or miR-3065-5p were performed using metascape (http://metascape.org) [38].We also employed gene set enrichment assay (GSEA) (v 2.0) [39] to compare the transcriptome of miR-338-3p- or miR-3065-5p-overexpression groups with previously reported Runx2 knockdown microarray data [13].
In vivo inhibition of miR-338 cluster
The miR-338-3p and miR-3065-5p in vivo inhibitors (mirVanaTM miRNA inhibitor) were obtained from Ambion (USA). All inhibitors were dissolved in PBS into concentration and delivered by intravenous injection at 25mg/kg at the time point indicated in the result part of this study, and the control groups were just injected with same volume of PBS.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed according to the previous protocol (ref). Briefly, MC3T3 at different timepoint post osteogenic differentiation were washed and chemically cross-linked by formaldehyde at the concentration of 1% and incubated for 10 min at 37 °C followed by stop of the reaction by 125mM glycine for 5 min at 37 °C. Cells were then collected and suspended in lysis buffer, and sonicated on ice using Sonicator 3000 (Misonix, Farmingdale, NY) to generate a library of DNA sized from 200 to 1000 bp. Specific DNA was pulled down by Dynabeads (Life Technologies) pretreated by RUNX2 antibody (Santa Cruz, M-70, 5µg per 100µL Dynbeads), SOX4 antibody (Abcam, ab70598, 5µg per 100µl Dynabeads) or IgG Dynabeads (Life Technologies) as control overnight. The next day, beads were washed sequentially with different washing buffer, and cross-linked DNA bound to the beads was eluted using SDS solution, and reversed by overnight incubation with 5M NaCl. ChIPed DNA was subjected to qPCR.
Western Blot
Cultured cells were harvested and lysed with M-PER buffer (Roche, Germany) and collected through centrifuge. Total protein was measured with BCA Protein Assay Kit (Pierce, USA). Equal amounts of proteins were isolated in a 10% polyacrylamide gel and were transferred to PVDF membranes (Roche, Germany). After blocked with 5% non-fat milk, the membranes were incubated with the primary antibody for OSX (Santa Cruz, sc-22536-R), OPN (a kind gift from Dr. Shuo Chen's lab; HuaAn Biotec, 0806-6), RUNX2 (abcam,ab76956) or β-ACTIN (ABclonal, AC028) at 4 °C overnight. After wash, the membranes were then incubated with horseradish peroxide-labeled IgG for 1 hour at room temperature. ECL solution (Pierce) was used for signal visualization.
Statistical analysis
Except for RNA-seq data analysis and Kolmogorov-Smirnov test for comparing the different enrichment of miR-338 cluster between healthy and osteoporosis patients, statistical analysis was performed with a two-tailed t-test using SPSS statistics 17.0. P values < 0.05 were considered statistically significant. All experiments were independently repeated at least three times. The quantified results are represented as means± standard error.
Abbreviations
miRNA: microRNA; BMD: bone mineral density; OVX: ovariectomy; BMSC: Bone marrow stromal cell; ChIP: chromatin immunoprecipitation; ChIP-seq: ChIP-sequencing; RNA-seq: RNA-sequencing; GSEA: gene set enrichment assay.
Supplementary Material
Supplementary figures, table, and raw data for figures 1G and 8L.
Acknowledgements
We thank Dr Morgan Sturgeon from the University of Iowa and Elsevier Language Editing Services for help editing and proofreading the English in the final manuscript. The numerical calculations in this paper have been done on the supercomputing system in the Supercomputing Center of Wuhan University. We thank Prof. Yiping Chen from Tulane University for suggestions on experiment design. This study was funded by grants from National Key R&D Project of China (NO:2018YFC1105103) and National Natural Science Foundation of China (NO: 81271099; 81420108011) to Prof. Zhi Chen; Young Elite Scientist Sponsorship Program by CAST (NO. 2017QNRC001), Natural Science Foundation of Hubei Province (NO. 2017CFB515) and National Natural Science Foundation of China (NO. 81771057; 81400477) to Dr Huan Liu. The authors declare no competing conflicts of interest with respect to the authorship and/or publication of this article.
Author contributions
CJL, HL, and ZC designed the experiments. CJL, STY and HL carried out most of the experiment. XJZ, HRH, ZYZ, RZJ, and YX worked on generation and breeding of the miR-338 cluster knockout mice. MHP helped to collect the clinical samples. CJL and HL analyzed results and wrote the manuscript. SC and ZC supervised the experiment.
Data availability
The data that support the findings of this study are available from the corresponding author on request.
Competing Interests
Huan Liu, Chujiao Lin, and Zhi Chen have submitted a patent for using the inhibitors of miR-338 and miR-3065 treating osteoporosis. Patent number: 201710708188.9 (China), Application number: PTC/CN2017 (China) /100135 (United States).
References
1. Segovia-Silvestre T, Neutzsky-Wulff AV, Sorensen MG, Christiansen C, Bollerslev J, Karsdal MA. et al. Advances in osteoclast biology resulting from the study of osteopetrotic mutations. Hum Genet. 2009;124:561-77
2. Wang C, Zhang Z, Zhang H, He JW, Gu JM, Hu WW. et al. Susceptibility genes for osteoporotic fracture in postmenopausal Chinese women. J Bone Miner Res. 2012;27:2582-91
3. Liu JM, Zhang MJ, Zhao L, Cui B, Li ZB, Zhao HY. et al. Analysis of recently identified osteoporosis susceptibility genes in Han Chinese women. The Journal of clinical endocrinology and metabolism. 2010;95:E112-20
4. Xia WB, He SL, Xu L, Liu AM, Jiang Y, Li M. et al. Rapidly increasing rates of hip fracture in Beijing, China. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2012;27:125-9
5. Hackl M, Heilmeier U, Weilner S, Grillari J. Circulating microRNAs as novel biomarkers for bone diseases - Complex signatures for multifactorial diseases? Mol Cell Endocrinol. 2016;432:83-95
6. Kocijan R, Muschitz C, Geiger E, Skalicky S, Baierl A, Dormann R. et al. Circulating microRNA Signatures in Patients With Idiopathic and Postmenopausal Osteoporosis and Fragility Fractures. J Clin Endocrinol Metab. 2016;101:4125-34
7. Mandourah AY, Ranganath L, Barraclough R, Vinjamuri S, Hof RV, Hamill S. et al. Circulating microRNAs as potential diagnostic biomarkers for osteoporosis. Sci Rep. 2018;8:8421
8. Mizoguchi F, Izu Y, Hayata T, Hemmi H, Nakashima K, Nakamura T. et al. Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption. Journal of cellular biochemistry. 2009;109:866-75
9. Gaur T, Hussain S, Mudhasani R, Parulkar I, Colby JL, Frederick D. et al. Dicer Inactivation in Osteoprogenitor Cells Compromises Fetal Survival and Bone Formation, While Excision in Differentiated Osteoblasts Increases Bone Mass in the Adult Mouse. Developmental biology. 2010;340:10-21
10. Liu H, Sun Q, Wan C, Li L, Zhang L, Chen Z. MicroRNA-338-3p regulates osteogenic differentiation of mouse bone marrow stromal stem cells by targeting Runx2 and Fgfr2. J Cell Physiol. 2014;229:1494-502
11. St John HC, Bishop KA, Meyer MB, Benkusky NA, Leng N, Kendziorski C. et al. The osteoblast to osteocyte transition: epigenetic changes and response to the vitamin D3 hormone. Mol Endocrinol. 2014;28:1150-65
12. Neve A, Corrado A, Cantatore FP. Osteoblast physiology in normal and pathological conditions. Cell Tissue Res. 2011;343:289-302
13. Wu H, Whitfield TW, Gordon JA, Dobson JR, Tai PW, van Wijnen AJ. et al. Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis. Genome Biol. 2014;15:R52
14. Duncan EL, Danoy P, Kemp JP, Leo PJ, McCloskey E, Nicholson GC. et al. Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genet. 2011;7:e1001372
15. Hsu YH, Zillikens MC, Wilson SG, Farber CR, Demissie S, Soranzo N. et al. An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility Loci for osteoporosis-related traits. PLoS Genet. 2010;6:e1000977
16. Jacovetti C, Jimenez V, Ayuso E, Laybutt R, Peyot ML, Prentki M. et al. Contribution of Intronic miR-338-3p and Its Hosting Gene AATK to Compensatory beta-Cell Mass Expansion. Mol Endocrinol. 2015;29:693-702
17. Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R. et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46:D1284
18. Ducy P, Starbuck M, Priemel M, Shen J, Pinero G, Geoffroy V. et al. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 1999;13:1025-36
19. Nissen-Meyer LS, Jemtland R, Gautvik VT, Pedersen ME, Paro R, Fortunati D. et al. Osteopenia, decreased bone formation and impaired osteoblast development in Sox4 heterozygous mice. J Cell Sci. 2007;120:2785-95
20. McCarthy TL, Chang WZ, Liu Y, Centrella M. Runx2 integrates estrogen activity in osteoblasts. J Biol Chem. 2003;278:43121-9
21. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
22. Ambros V. The functions of animal microRNAs. Nature. 2004;431:350-5
23. Li CJ, Cheng P, Liang MK, Chen YS, Lu Q, Wang JY. et al. MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J Clin Invest. 2015;125:1509-22
24. Li D, Liu J, Guo B, Liang C, Dang L, Lu C. et al. Osteoclast-derived exosomal miR-214-3p inhibits osteoblastic bone formation. Nat Commun. 2016;7:10872
25. Wang X, Guo B, Li Q, Peng J, Yang Z, Wang A. et al. miR-214 targets ATF4 to inhibit bone formation. Nat Med. 2013;19:93-100
26. Inoue K, Deng Z, Chen Y, Giannopoulou E, Xu R, Gong S. et al. Bone protection by inhibition of microRNA-182. Nat Commun. 2018;9:4108
27. Sun Q, Zhang B, Zhu W, Wei W, Ma J, Tay FR. A potential therapeutic target for regulating osteoporosis via suppression of osteoclast differentiation. J Dent. 2019;82:91-7
28. Muller S, Nowak K. Exploring the miRNA-mRNA regulatory network in clear cell renal cell carcinomas by next-generation sequencing expression profiles. Biomed Res Int. 2014;2014:948408
29. Liu Y, Berendsen AD, Jia S, Lotinun S, Baron R, Ferrara N. et al. Intracellular VEGF regulates the balance between osteoblast and adipocyte differentiation. J Clin Invest. 2012;122:3101-13
30. Verlinden L, Kriebitzsch C, Beullens I, Tan BK, Carmeliet G, Verstuyf A. Nrp2 deficiency leads to trabecular bone loss and is accompanied by enhanced osteoclast and reduced osteoblast numbers. Bone. 2013;55:465-75
31. Cao H, Yu W, Li X, Wang J, Gao S, Holton NE. et al. A new plasmid-based microRNA inhibitor system that inhibits microRNA families in transgenic mice and cells: a potential new therapeutic reagent. Gene Ther. 2016;23:634
32. Hosoya A, Kim JY, Cho SW, Jung HS. BMP4 signaling regulates formation of Hertwig's epithelial root sheath during tooth root development. Cell Tissue Res. 2008;333:503-9
33. Shah JS, Soon PS, Marsh DJ. Comparison of Methodologies to Detect Low Levels of Hemolysis in Serum for Accurate Assessment of Serum microRNAs. PLoS One. 2016;11:e0153200
34. Soleimani M, Nadri S. A protocol for isolation and culture of mesenchymal stem cells from mouse bone marrow. Nat Protoc. 2009;4:102-6
35. Liu H, Leslie EJ, Carlson JC, Beaty TH, Marazita ML, Lidral AC. et al. Identification of common non-coding variants at 1p22 that are functional for non-syndromic orofacial clefting. Nat Commun. 2017;8:14759
36. Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525-7
37. Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods. 2017;14:687-90
38. Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA. et al. Meta- and Orthogonal Integration of Influenza "OMICs" Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe. 2015;18:723-35
39. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545-50
Author contact
![]() Corresponding authors: Huan Liu and Zhi Chen, State Key Laboratory Breeding Base of Basic Science of Stomatology (Hubei-MOST) and Key Laboratory for Oral Biomedicine of Ministry of Education (KLOBM), School and Hospital of Stomatology, Wuhan University, Wuhan 430079, China. E-mail: zhichenedu.cn (Z.Chen); liu.huanedu.cn (H. Liu). Tel: 13971037354. Fax: 86-27-87686198
Corresponding authors: Huan Liu and Zhi Chen, State Key Laboratory Breeding Base of Basic Science of Stomatology (Hubei-MOST) and Key Laboratory for Oral Biomedicine of Ministry of Education (KLOBM), School and Hospital of Stomatology, Wuhan University, Wuhan 430079, China. E-mail: zhichenedu.cn (Z.Chen); liu.huanedu.cn (H. Liu). Tel: 13971037354. Fax: 86-27-87686198
 Global reach, higher impact
Global reach, higher impact