13.3
Impact Factor
Theranostics 2022; 12(10):4703-4717. doi:10.7150/thno.71676 This issue Cite
Research Paper
Genome instability is associated with ethnic differences between Asians and Europeans in hepatocellular carcinoma
1. Genome Institute of Singapore, Agency for Science, Technology and Research, Singapore, 138672, Singapore.
2. Key Laboratory of Zoological Systematics and Evolution, Institute of Zoology, Chinese Academy of Sciences, Beijing, 100101, China.
3. School of Biological Sciences, Nanyang Technological University, Singapore, 637551, Singapore.
4. Centre for Quantitative Medicine, Program in Health Services and Systems Research, Duke-NUS Medical School, Singapore, 169857, Singapore.
5. Institute of Pathology, Faculty of Medicine and University Hospital Cologne, University of Cologne, Cologne, 50937, Germany.
6. Division of Medical Oncology, National Cancer Centre Singapore, Singapore, 169610, Singapore.
7. Singapore General Hospital, Singapore, 169608, Singapore.
8. Institute of Molecular and Cell Biology, Agency for Science, Technology and Research (A*STAR), Singapore, 138673, Singapore.
9. CAS Key Laboratory of Quantitative Engineering Biology, Shenzhen Institute of Synthetic Biology, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, 518055, China.
10. Guangzhou Laboratory, Guangzhou International Bio Island, Guangzhou 510005, Guangdong Province, China.
11. Hepatopancreatobiliary and Transplant Surgery, Singapore General Hospital, Singapore, 169608, Singapore.
12. Division of Surgery and Surgical Oncology, National Cancer Centre, Singapore, 169610, Singapore.
13. Office of Clinical Sciences, Duke-NUS Graduate Medical School, Singapore, 169857, Singapore.
14. Center for Excellence in Animal Evolution and Genetics, Chinese Academy of Sciences, Kunming, 650223, China.
15. School of Life Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei 230027, China.
Abstract
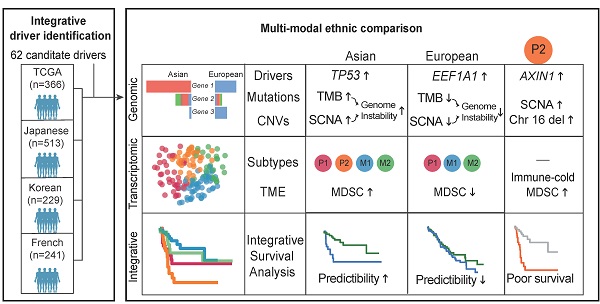
Hepatocellular carcinoma (HCC) is one of the deadliest cancer types with diverse etiological factors across the world. Although large scale genomic studies have been conducted in different countries, integrative analysis of HCC genomes and ethnic comparison across cohorts are lacking.
Methods: We first integrated genomes of 1,349 HCC patients from five large cohorts across the world and applied multiple statistical methods in identifying driver genes. Subsequently, we systematically compared HCC genomes and transcriptomes between Asians and Europeans using the TCGA cohort.
Results: We identified 29 novel candidate driver genes, many of which are infrequent tumor suppressors driving late-stage tumor progression. When we systematically compared ethnic differences in the genomic landscape between Asian and European HCCs using the TCGA cohort (n = 348), we found little differences in driver frequencies. Through multi-modal integrative analysis, we found higher genomic instability in Asians together with a collection of molecular events ranging from tumor mutation burden (TMB), copy number alterations as well as transcriptomic subtypes segregating distinctively between two ethnic backgrounds. Strikingly, we identified an Asian specific transcriptomic subtype with multiple ethnically enriched genomic alterations, in particular chromosome 16 deletion, leading to a clinically aggressive RNA subgroup unique to Asians. Integrating multi-modal information, we found that survival models predict patient prognosis much better in Asians than in Europeans, demonstrating a higher potential for precision medicine applications in Asia.
Conclusion: For the first time, we have uncovered an unprecedented amount of genomic differences segregating distinctively across ethnicities in HCC and highlighted the importance of differential disease biology and management in HCC across ethnic backgrounds.
 Global reach, higher impact
Global reach, higher impact