13.3
Impact Factor
Theranostics 2023; 13(8):2439-2454. doi:10.7150/thno.82582 This issue Cite
Review
Technologies and applications of single-cell DNA methylation sequencing
1. Anhui Province Key Laboratory of Medical Physics and Technology, Institute of Health and Medical Technology, Hefei Institutes of Physical Science, Chinese Academy of Sciences, Hefei, 230031, China.
2. University of Science and Technology of China, Hefei, 230026, China.
3. Hefei Cancer Hospital, Chinese Academy of Sciences, Hefei, 230031, China.
4. Zhejiang ShengTing Biotech. Ltd, Hangzhou, 310000, China.
5. Department of Hematology, the First Hospital of China Medical University, Shenyang, 110001, China.
Abstract
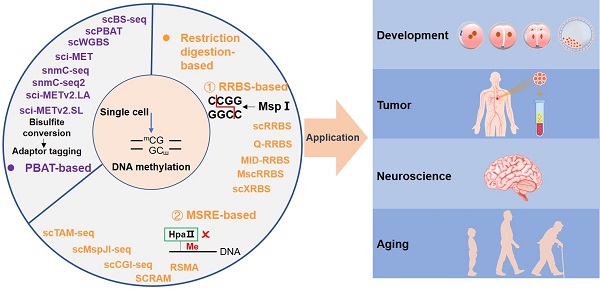
DNA methylation is the most stable epigenetic modification. In mammals, it usually occurs at the cytosine of CpG dinucleotides. DNA methylation is essential for many physiological and pathological processes. Aberrant DNA methylation has been observed in human diseases, particularly cancer. Notably, conventional DNA methylation profiling technologies require a large amount of DNA, often from a heterogeneous cell population, and provide an average methylation level of many cells. It is often not realistic to collect sufficient numbers of cells, such as rare cells and circulating tumor cells in peripheral blood, for bulk sequencing assays. It is therefore essential to develop sequencing technologies that can accurately profile DNA methylation using small numbers of cells or even single cells. Excitingly, many single-cell DNA methylation sequencing and single-cell omics sequencing technologies have been developed, and applications of these methods have greatly expanded our understanding of the molecular mechanism of DNA methylation. Here, we summaries single-cell DNA methylation and multi-omics sequencing methods, delineate their applications in biomedical sciences, discuss technical challenges, and present our perspective on future research directions.
Keywords: DNA methylation, single-cell sequencing, single-cell multi-omics sequencing
 Global reach, higher impact
Global reach, higher impact