13.3
Impact Factor
Theranostics 2012; 2(10):967-975. doi:10.7150/thno.5032 This issue Cite
Research Paper
A Novel SNPs Detection Method Based on Gold Magnetic Nanoparticles Array and Single Base Extension
1. Hunan Key Laboratory of Green Packaging and Application of Biological Nanotechnology, Hunan University of Technology, Zhuzhou 412007, P. R. China
2. State Key Laboratory of Bioelectronics, Department of Biological Science and Medical Engineering. Southeast University, Nanjing 210096, P. R. China
Received 2012-8-13; Accepted 2012-9-20; Published 2012-10-9
Abstract
To fulfill the increasing need for large-scale genetic research, a high-throughput and automated SNPs genotyping method based on gold magnetic nanoparticles (GMNPs) array and dual-color single base extension has been designed. After amplification of DNA templates, biotinylated extension primers were captured by streptavidin coated gold magnetic nanoparticle (SA-GMNPs). Next a solid-phase, dual-color single base extension (SBE) reaction with the specific biotinylated primer was performed directly on the surface of the GMNPs. Finally, a “bead array” was fabricated by spotting GMNPs with fluorophore on a clean glass slide, and the genotype of each sample was discriminated by scanning the “bead array”. MTHFR gene C677T polymorphism of 320 individual samples were interrogated using this method, the signal/noise ratio for homozygous samples were over 12.33, while the signal/noise ratio for heterozygous samples was near 1. Compared with other dual-color hybridization based genotyping methods, the method described here gives a higher signal/noise ratio and SNP loci can be identified with a high level of confidence. This assay has the advantage of eliminating the need for background subtraction and direct analysis of the fluorescence values of the GMNPs to determine their genotypes without the necessary procedures for purification and complex reduction of PCR products. The application of this strategy to large-scale SNP studies simplifies the process, and reduces the labor required to produce highly sensitive results while improving the potential for automation.
Keywords: Magnetic beads, Dual-color SBE, Single nucleotide polymorphisms, Gold magnetic nanoparticles, Microarray
Introduction
The completion of the Human Genome Project has set the stage for screening genetic mutations to identify disease genes on a genome-wide scale. Since single nucleotide polymorphisms (SNPs) are the most abundant source of variation in the human genome, genotyping large numbers of SNPs in pathological samples should provide insight into complex genetic traits associated with common human diseases [1-3]. Clinical investigators have analyzed SNPs alleles in population-based studies to identify loci that are statistically associated with certain diseases and phenotypes [4, 5]. In these studies, the number of samples available for analysis dramatically impacts data reliability. To obtain a result with a high degree of confidence, investigators must analyze multiple SNP loci from thousands of samples. To fulfill this need, the development of a new SNPs detection platform that is quick, simple, high-throughput, automated, and cost-effective is warranted. A number of techniques and commercial platforms have been developed for high-throughput SNPs genotyping. These include detection methods based on allelic specific hybridization such as Affymetrix whole genome SNP array [6], Idaho Hi-Res Melting curve analysis system [7], dynamic allele-specific hybridization (DASH) [8], and Illumina Golden Gate SNP Genotyping Arrays [9]; methods based on fluorescence resonance energy transfer (FRET) such as TaqMan [10], and molecular beacons assays [11]; and those based on extension such as Illumina Infinium bead array [12], Beckman GenomeLab SNPstream system [13], and Sequenom MassARRAY SNP system [14]. These methods have been successfully used for high-throughput SNPs genotyping. However, for large-scale SNP detection, most of these methods are not suitable for automation, and would require handling thousands of samples manually. In addition, these methods are still too expensive to be cost-effective when analyzing multiplex loci for a large number of samples.
Single base extension (SBE) involves extension of a primer located adjacent to the position of a SNP. Using DNA polymerase in the presence of fluorescently labeled dideoxynucleotide triphosphates (ddNTPs) ensures that the polymerase will incorporate only a single nucleotide for the SNP site of interest. The SBE technique provides highly accurate and effective genotyping, and has been widely used in various genotyping procedures.
Magnetic nanoparticles have found many applications in biomedical scopes [15-26]. Recent advances in microarray and magnetic nanoparticles (MNPs) have allowed high-throughput DNA analysis of multiple samples [27, 28]. As one of the most promising tools for nucleic acid detection, microarrays produce a “read-out” that is especially suitable for high-throughput detection. To take advantage of the capacity of this platform, a molecular carrier suitable for multiplexed sample preparation and automation is required.
Due to unique dispersion properties in aqueous solution, and high separation efficiency in magnetic fields, MNPs have great potential in automated and high-throughput nucleic acid analysis- especially when combined with commercial automation [29-31]. Previously, we have developed some novel genotyping methods based on MNPs and dual-color hybridization. These experiments successfully demonstrated that SNPs from a large number of samples could be efficiently and automatically genotyped using MNPs as DNA carriers. Though these methods are efficient and can be automated, selectivity and quantification are dependent upon the dissociation properties of the target DNA hybridized to the allele specific probes [32-33]. The difference in binding affinity between a perfectly matched probe and one-base mismatch is small. Thus to achieve SNP discrimination, hybridization temperatures must be optimized, and stringent wash steps must be included. In addition, a denaturation step must be performed prior to determining the fluorescence to remove high background making the assay procedure complex.
In this study, a high-throughput and automated SNP genotyping method that uses a magnetic nanoparticle (MNP) array and dual-color, single base extension was developed to address the previously identified shortcomings of dual-color hybridization and hybridization-based allele-specific discrimination (Fig. 1). The solid-phase extension primers were prepared by immobilizing biotinylated oligonucleotides onto gold magnetic nanoparticles coated with streptavidin (SA-GMNPs). In the presence of DNA polymerase, allele-specific extension using dual-color ddNTPs occurred along the stretch of hybridized target DNA, incorporating a fluorophore into the primer bound to the surface of the magnetic beads. The magnetic beads extended with fluorophores were spotted on a glass slide to fabricate a “bead-array” for SNP discrimination. By integrating 320 samples, we demonstrated that magnetic bead array based dual-color SBE is a flexible and reliable technology that can be adapted to high-throughput SNP genotyping. This new method will hopefully have wide applications in clinical diagnostic settings.
Materials and Methods
Oligonucleotides and Reagents
In this study, the methylenetetrahydrofolate reductase (MTHFR) gene C677T polymorphism was selected as target. The oligonucleotides used in this study were synthesized and purified by Shanghai Sangon Biologic Engineering Technology and Service Co. Ltd (Shanghai, China) and are summarized in Table 1. TAMRA-ddGTP, Cy5-ddATP, and unlabled ddNTPs (A, G, C, T) were purchased from Perkin-Elmer Life Sciences Inc. (Shanghai, China). Shrimp alkaline phosphatase (SAP) and Thermo Sequenase were obtained from GE Healthcare. 11-mercaptoundecanoic acid (MUA) was purchased at Sigma-Aldrich. EDC (1-Ethyl-3-(3-dimethyllaminopropyl) carbodiimide hydrochloride), MES (2-N-morpholino ethanesulfonic acid) and streptavidin were also obtained from Shanghai Sangon Biologic Engineering Technology and Service Co. Ltd (Shanghai, China). Hybridization chambers were obtained from Thermo Fisher Scientific Inc. All solutions were prepared in sterilized ddH2O. Other chemicals were of a commercially extra-pure grade.
Extension Template Preparation
Whole blood samples were obtained from Changsha Central Hospital with informed consent. Genomic DNA was extracted from 300 μL blood using the Wizard Genomic DNA Purification Kit (Promega, USA). PCR reaction of SNP polymorphism was carried out using primers shown in Table 1. The PCR product for dual-color hybridization was amplified using a biotinylated reverse primer. All PCR products containing different SNP loci were amplified according to a previously published procedure [34]. The PCR products for dual-color SBE were incubated with 1.5 U exonuclease I and 1.5 U shrimp alkaline phosphatase at 37 ºC for 90 min to inactivate excess PCR primers and dNTPs, respectively, followed by enzyme deactivation at 94 ºC for 15 min.
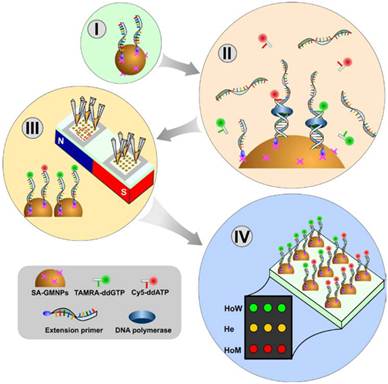
Schematic diagram showing the SNPs genotyping method based on a GMNP array and dual-color SBE. (I) Biotinyalted primers are captured by the SA-GMNPs. (II) ssPCR products are hybridized to primers and dual-color, single base extension is performed. (III) Fluorophore-GMNP complexes for each sample are immobilized on the glass slide. (IV) The genotype of each sample is determined by measuring the fluorescent intensity of the fluorophore-GMNP complexes on the array.
The oligonucleotides used in this study.
| SNPs | Sequence Name | Sequence 5'-3' |
|---|---|---|
| rs1801133 | Forward Primer | TGAAGGAGAAGGTGTCTGCGGGA |
| Reverse Primer | Biotin-(N)15- AGGACGGTGCGGTGAGAGTG | |
| Wild probe | Cy3-CGGGAGCCGATTT | |
| Mutant probe | Cy5-CGGGAGTCGATTT | |
| Extension Primer | Biotin-(N)15-AAGCTGCGTGATGATGAAATCG | |
| rs48412845 | Forward Primer | ACTGCAGAGGGAGGAGGACT |
| Reverse Primer | Biotin-(T)15- CCACCTCAAGCTCTGGTGAT | |
| Wild probe | Cy3-CTGGCGCAACAGA | |
| Mutant probe | Cy5-CTGGGGCAACAGA | |
| Extension Primer | Biotin-(T)15- CTCGTCACCCACTCTGTTGC |
Preparation of SA-GMNPs
γ-Fe2O3 magnetic nanoparticles (MNPs) were prepared by baking Fe3O4 at 380 ºC for 2 hours. The resulting MNPs were treated with 3-mercaptopropyltriethoxysilane (MPTS) to produce thiol-modified MNPs. A layer of colloid gold was formed around the thiol-modified MNPs by reduction of Au3+ in the presence of citrate to form an Au-S bond [35]. The resulting GMNPs were functionalized with carboxylic acid groups by reacting 20 mg of GMNPs with 200 mM 11-mercaptoundecanoic acid (MUA) in ethanol at room temperature.
Next the GMNPs were activated with EDC for 30 minutes at 4 ºC using slow tilt rotation. After removing the supernatant using magnetic separation, the activated GMNPs were washed once with 500 μL cold, deionized water and a second time with 500 μL 25 mM MES (pH 6) as quickly as possible to avoid hydrolysis of the activated carboxylic acid groups. Streptavidin was covalently immobilized on the GMNPs by linking the streptavidin amine groups to the carboxylic acid groups on the GMNPs. The streptavidin-coated GMNPs (SA-GMNPs) were suspended in PBS buffer at a concentration of 4 mg/mL and stored at 4 ºC. The SA-GMNPs were found to be stable for at least 6 months when stored at 4 °C.
Solid-phase Dual-color SBE Reaction
GMNP-bound extension primers were prepared by covalently linking 24 pmol of each of the biotinylated oligos with 80 μg of SA-GMNPs. Amplified product was mixed with the GMNP-bound probes and hybridized for 30 minutes. Solid-phase SBE reaction was performed in a 20 μL reaction mixture containing 20 μM relevant wild TARMA-ddGTP and mutant Cy5-ddATP, 1×PCR buffer, 60 μM of unlabeled ddNTPs, 2 U of Thermo Sequenase, and 80 μg MNPs-bound extension primer. The solid-phase SBE reactions for C677T were carried out at 95 ºC for 5 min, followed by 35 cycles of primer extension at 95 ºC for 30 s, 58 ºC for 30 s, and 72 ºC for 30 s. After SBE reaction was completed, the SBE-GMNP complexes were separated using a neodymium-boron (Nd-B) magnet and washed for 5 min using sterilized ddH2O. The wash was repeated twice to remove excess ddNTPs and PCR product. The remaining fluorophore-GMNP particles were suspended in 10 μL of sterilized ddH2O and stored at 4 ºC until arraying.
Bead array Fabrication and Signal Detection
Standard glass slides (75 mm × 25 mm × 1 mm) were used as the substrate for the GMNP array. Nd-B magnet was placed on the opposite side of the slide and each fluorophore-GMNP reaction was printed in quadruplicate directly onto the slide to fabricate a “bead-array”. After printing, the “bead-array” was snap-dried for 2 sec on a hot plate set at 100 ºC, and directly scanned with a 4100 A Microarray Analysis System (Axon). The images acquired by the scanner were analyzed using Genepix Pro 6.0 software. The average pixel intensity within each spot and the local background surrounding each spot were used to compute the mean pixel intensity for each spot. The net signal was determined by subtracting the local background from the mean pixel intensity for each spot.
SNP Detection Based on Dual-color Hybridization
SNP detection based on dual-color hybridization was used as a reference for comparing the novel dual-color SBE results. In this method, biotinylated PCR products were captured using 50 μg of SA-GMNPs. After denaturation, the ssDNA-GMNPs complexes were printed directly onto the slide to fabricate the magnetic bead array. The bead array was framed using a 65 μL hybridization chamber, 65 μL of dual-color hybridization mixture containing 22 μL of hybridization solution, and 20 pmol each of Cy3 wild probes and Cy5 mutant probes were placed in the hybridization chamber, and sealed with a cover slide. After incubating at 38 ºC for 30 min, the GMNP array was washed using 2× saline-sodium citrate (SSC) containing 1.0 g/L sodium dodecyl sulfate (SDS), 0.1×SSC containing 1.0 g/L SDS, and deionized water, respectively. Finally, the microarrays were snap-dried for 2 sec on a hot plate, and scanned with a 4100 A Microarray Analysis System (Axon).
Results and Discussion
Assay Design
The proof of concept scheme described in this paper uses solid-phase, dual-color SBE followed by a GMNP “bead-array” fabrication for SNP detection of a T→C transition. Biotinylated extension primers were captured by SA-GMNPs, and following PCR amplification of DNA templates and hybridization of the product, a solid-phase, dual-color SBE reaction was performed directly on the surface of the GMNPs (Fig 1. I - III). DNA polymerase was used for allele-specific extension with dual-color ddNTPs (TAMRA-ddGTP and Cy5-ddATP) being directly incorporated into the GMP bound primer along the stretch of target DNA. The genotype of each sample was determined simultaneously by scanning the “bead-array” containing the fluorophore-GMNP complexes.
As expected, the fluorophore-GMNP complexes containing the homozygous wild-type SNP (HoW, 677CC) produced intense green fluorescent signal, indicating the relevant TAMRA-ddGTP could elongate with template DNA from these samples. The homozygous mutant samples (HoM, 677TT) yielded strong, red fluorescent signal, indicating that the Cy5-ddATP could elongate with the template DNA. Heterozygote samples (He, 677CT) displayed strong, yellow fluorescence due to overlapping signal from both kinds of ddNTP, TAMRA-ddGTP (green) and Cy5-ddATP (red).
Genotyping Analysis
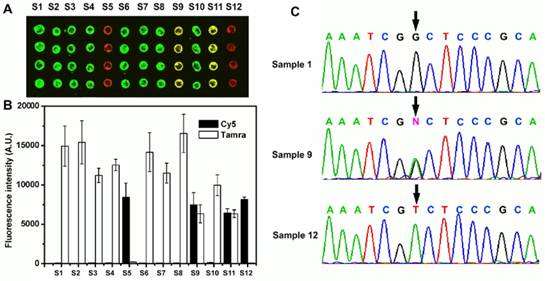
Twelve samples were analyzed for the C677T locus using this method. The results are depicted in Fig. 2. Eight homozygous wild-type samples (1-4, 6, 7, 8, 10) produced strong, green fluorescence from elongation with TAMRA-ddGTP. While the two homozygous mutant samples (5 and 12) displayed a red fluorescence from elongation with Cy5-ddATP only. The heterozygous samples (9 and 11) yielded strong yellow fluorescence, implicating incorporation of both TAMRA-ddGTP and Cy5-ddATP.
The relative fluorescence intensities were quantified and are depicted in Fig. 2B. The green fluorescent spots from Sample 1 produced an average fluorescence score of 14936.5 fluorescent units. Very low red fluorescent signal (191 fluorescent units) near background levels indicates that Sample 1 was homozygous wild type. Sample 5 was determined to be a homozygous mutant SNP where GMNPs elongated with Cy5-ddATP produced fluorescence scores of 8414, and those elongated with TAMRA-ddGTP gave fluorescence scores near background at 239 units. The wild/mutant (Tamara / Cy5) ratio for this sample was 35.2. Sample 11 yielded strong yellow fluorescence with fluorescence scores at 6439 for Tamara, and 6346 for Cy5, and a wild/mutant ratio of 1.01, indicating a heterozygote SNP. For homozygous DNA samples, the wild/mutant ratio ranged from 30.9 to 78.2, while the Cy5 to Tamara signal ratios of heterozygote samples ranged from 0.90 to 1.01. The results were as expected and good discrimination between the two alleles was obtained. The SNP results obtained with the novel method were verified by sequencing (Fig. 2C).
(A) Microarray results from 12 different samples assayed for the C677T locus of the MTHFR gene. Each column represents one sample spotted in quadruplicate. (B) Fluorescence intensities for the 12 samples are depicted in a bar graph with the standard deviation of replicates indicated by an error bar. (C) Sequencing result of the PCR products. The arrows indicate the tested SNP loci.
Comparison of Dual-color SBE with Dual-color Hybridization
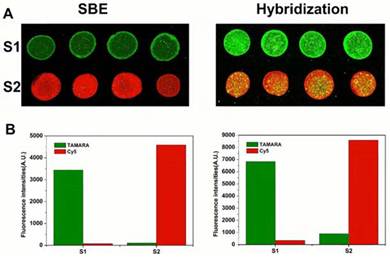
In order to evaluate if SBE increases the quality of SNP genotyping results obtained by dual-color magnetic bead arrays, we compared the dual-color SBE and hybridization methods by genotyping two different SNP loci, rs1801133 (MTHFR C677T) and rs48412845. Homozygous wild type and mutant DNA samples that were verified by sequencing were used to amplify the targets. Fig. 3A shows the genotyping results obtained from wild type and mutant samples analyzed for the C677T locus using both methods. The homozygous wild type samples produced strong green fluorescence using both methods, while mutant samples produced a strong red fluorescence. The genotype of the two samples can be easily discriminated using both methods, but the dual-color SBE method produced an S/N ratio of 44.15, whereas the hybridization method produced a wild/mutant ratio of 12.28.
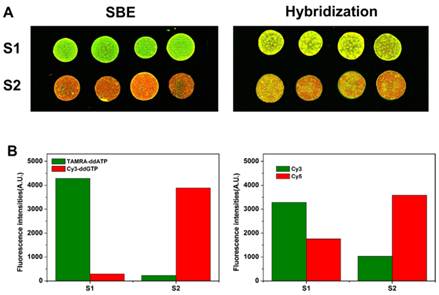
A bar graph (Fig. 4B) depicts the signal intensity of the genotyping results obtained from wild type and mutant samples analyzed for rs48412845 locus with both methods. Using dual-color SBE (Fig. 4A, left), the wild type and mutant samples can be easily discriminated by the strong green and red fluorescence, respectively. However, using dual-color hybridization method (Fig. 4A, right), the wild type and mutant samples cannot be easily discriminated. Using hybridization method, the wild/mutant ratios of the two homozygous samples are only 1.87 and 0.288 respectively. Taken together, these results suggest that the SBE method provides better SNP discrimination than hybridization. When we design the allele detection probes in order to get better discrimination, the SNP loci need to be located in the middle of detection probe. However, the rs48412845 SNP locus is too close to another SNP locus, another SNP locus in the detection probes causes inefficient hybridization and results in poor discrimination. To perform large-scale genetic studies efficiently, robust methods for SNP genotyping are required. Hybridization based techniques are generally high-throughput, but always hold the risk for cross-hybridization and unspecific bindings. Compared with techniques based on hybridization, single base extension is compatible with multiplexed analysis, and provides highly accurate genotyping results because primer extension with DNA polymerase can accurately distinguish single nucleotide differences, even for difficult SNP loci.
High-throughput SNP Genotyping of 320 Samples
To test this method for high-throughput genotyping capacity, we investigated MTHFR gene C677T locus detection using 320 diverse DNA samples. Fig. 5A shows the microarray results of the high-throughput analysis. Good discrimination between the two alleles was obtained for C677T locus. The complete array, spotted in replicates of four, produced a fluorescence pattern that was highly reproducible between replicates. The three allelic states for the locus (C/C, green; C/T, yellow; T/T, red) were easily discriminated.
Results of SNP detection for MTHFR C677T using dual-color SBE and dual-color hybridization, respectively. (A) Fluorescence images of microarrays assayed for MTHFR C677T locus (B) Relative fluorescence intensities for the 2 samples.
Results of SNP detection for rs48412845 locus using dual-color SBE and dual-color hybridization, respectively. (A) The fluorescence images of microarray assayed for rs48412845 locus. (B) Relative fluorescence intensities for the 2 samples.
Results of SNP detection of 320 samples based on SA-GMNPs assayed for MTHFR C677T. (a) The fluorescence images of microarray assayed for MTHFR C677T locus. (b) Allelic-fraction scatter plot.
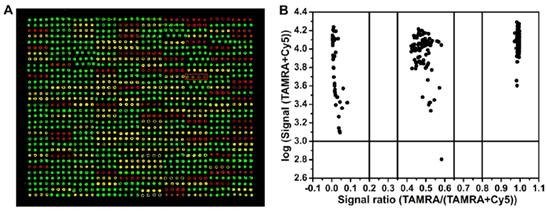
Fluorescence signals from all representative samples were quantified (Fig. 5B). Allelic-fractions were calculated as fluorescence intensity of TAMRA/ (TAMRA + Cy5) to represent the hybridization signals from matched and mismatched probes. Using this formula, we determined that heterozygous samples have allelic-fractions between 0.35 and 0.65. Homozygous wild type allelic-fractions were greater than 0.8, and mutants were less than 0.2. Samples that fell outside of these boundaries were classified as ambiguous.
Separating samples into allelic-fractions allowed unequivocal genotype assignment for the three loci. Fig. 5B illustrates the clear ratio differences between the three clusters. From left to right on the graph, allelic-fractions ranging from 0.03 to 0.075 were classified as homozygous mutant samples; allelic-fractions ranging from 0.96 to 0.98 can be classified as homozygous mutant samples. Allelic-fraction values for homozygous wild and mutant samples should be as close as possible to 1.0 and 0.0, respectively. All homozygous samples produced a match/mismatch ratio between 12.33 and 49. The middle cluster had allelic-fraction values that ranged from 0.43 to 0.58, all of which were classified as heterozygotes. One sample (Fig. 5A) gave a very low fluorescence signal due to substandard PCR product quality. Still, the genotyping results were not influenced by this problem.
Conclusion
To achieve high-throughput genotyping, the challenge lies in pairing the right assay chemistry with the right carriers to maximize efficiency with respect to accuracy, sensitivity, speed and cost. Herein, we successfully demonstrated a high-throughput SNP detection using dual-color SBE. Using SA-GMNPs as a primer carrier, and performing specific primer dual-color ddNTP incorporation at elevated temperatures avoids the low discrimination between SNPs typical of hybridization. Moreover, because of magnetic separation, SA-GMNPs were employed as useful carriers for removing excess fluorescently labeled ddNTPs - enabling a rapid change between SBE and array fabrication that resulted in robust SNP genotyping with high S/N ratios. A new technology platform was also introduced. The “bead array” utilizes two facets of SNP detection required for automation: the direct detection of beads with fluorophores, and manipulation with an external magnetic field. The results can be directly “readout” from the labels themselves, which tremendously advances the study of gene expression profiles and genomic compositions. In summary, we developed a novel high-throughput SNP detection approach based on GMNP array and dual-color SBE which offers affordable cost, rapid speed, and high flexibility with capability for genotyping of a large number of samples that are increasingly needed. The application of this strategy also simplifies the process, and reduces the labor required to produce highly sensitive results while improving the potential for automation. It is ideal for applications such as diagnostic detection of disease genes from clinical samples and screening of characteristic marker genes from many biological systems.
Acknowledgements
This research was financially supported by the National Key Program for Developing Basic Research of China (2010CB933903), NSFC (60927001, 60801007, 21205013, 61271056 and 60971045), Hunan Science and Technology Projects (2012SK3105 and 2010sk2003), Scientific Research Fund of Hunan Provincial Education Department (11A030), China Postdoctoral Science Foundation (20100471362 and 2012M511660).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sachidanandam R, Weissman D, Schmidt SC. et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928-33
2. Collins FS, Guyer MS, Charkravarti A. Variations on a theme: cataloging human DNA sequence variation. Science. 1997;278:1580-1
3. Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516-7
4. Gresham D, Ruderfer DM, Pratt SC. et al. Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science. 2006;311:1932-6
5. Ohashi J, Yamamoto S, Tsuchiya N. et al. Comparison of statistical power between 2*2 allele frequency and allele positivity tables in case-control studies of complex disease genes. Ann Hum Genet. 2001;65:197-206
6. Komura D, Shen F, Ishikawa S. et al. Genome-wide detection of human copy number variations using high-density DNA oligonucleotide arrays. Genome Res. 2006;16:1575-84
7. Graham R, Liew M, Meadows C. et al. Distinguishing different DNA heterozygotes by high-resolution melting. Clin Chem. 2005;51:1295-8
8. Prince JA, Feuk L, Howell WM. et al. Robust and accurate single nucleotide polymorphism genotyping by dynamic allele-specific hybridization (DASH): design criteria and assay validation. Genome Res. 2001;11:152-62
9. Gunderson KL, Steemers FJ, Lee G. et al. A genome-wide scalable SNP genotyping assay using microarray technology. Nat Genet. 2005;37:549-54
10. Holloway JW, Beghé B, Turner S. et al. Comparison of three methods for single nucleotide polymorphism typing for DNA bank studies: sequence-specific oligonucleotide probe hybridisation, TaqMan liquid phase hybridisation, and microplate array diagonal gel electrophoresis (MADGE). Hum Mutat. 1999;14:340-7
11. Barreiro LB, Henriques R, Mhlanga MM. High-throughput SNP genotyping: combining tag SNPs and molecular beacons. Methods Mol Biol. 2009;578:255-76
12. Oliphant A, Barker DL, Stuelpnagel JR. et al. BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques. 2002(Suppl):56-8 60-1
13. Bell PA, Chaturvedi S, Gelfand CA. et al. SNPstream UHT: ultra-high throughput SNP genotyping for pharmacogenomics and drug discovery. Biotechniques. 2002(Suppl):70-2 74, 76-7
14. Hayes BJ, Nilsen K, Berg PR. et al. SNP detection exploiting multiple sources of redundancy in large EST collections improves validation rates. Bioinformatics. 2007;23:1692-3
15. Ruan J, Song H, Li C, Bao C, Fu H, Wang K, Ni J, Cui D. DiR-labeled Embryonic Stem Cells for Targeted Imaging of in vivo Gastric Cancer Cells. Theranostics. 2012;2:618-28
16. Singh R, Nalwa HS. Medical Applications of Nanoparticles in Biological Imaging, Cell Labeling, Antimicrobial Agents, and Anticancer Nanodrugs. J Biomed Nanotechnol. 2011;7:489-503
17. Niu G, Chen X. Molecular Imaging with Activatable Reporter Systems. Theranostics. 2012;2:413-23
18. Gao X, Kan B, Gou M. et al. Preparation of Anti-CD40 Antibody Modified Magnetic PCL-PEG-PCL Microspheres. J Biomed Nanotechnol. 2011;7:285-91
19. Chen X. Integrin Targeted Imaging and Therapy. Theranostics. 2011;1:28-9
20. Naresh M, Sharma M, Mittal A. Intracellular Magneto-Spatial Organization of Magnetic Organelles inside Intact Bacterial Cells. J Biomed Nanotechnol. 2011;7:572-7
21. Ren HX, Liu L, Liu C. et al. Physiological Investigation of Magnetic Iron Oxide Nanoparticles Towards Chinese Mung Bean. J Biomed Nanotechnol. 2011;7:677-84
22. Zhang F, Zhu L, Liu G. et al. Multimodality Imaging of Tumor Response to Doxil. Theranostics. 2011;1:302-9
23. Singh SP. Multifunctional Magnetic Quantum Dots for Cancer Theranostics. J Biomed Nanotechnol. 2011;7:95-7
24. Zhen Z, Xie J. Development of Manganese-Based Nanoparticles as Contrast Probes for Magnetic Resonance Imaging. Theranostics. 2012;2:45-54
25. Llandro J, Palfreyman JJ, Ionescu A. et al. Magnetic biosensor technologies for medical applications: a review. Med Biol Eng Comput. 2010;48:977-98
26. Wang F, Ma C, Zeng X. et al. Chemiluminescence Molecular Detection of Sequence-Specific HBV-DNA Using Magnetic Nanoparticles. J Biomed Nanotechnol. 2012;8:786-90
27. Smith JR, Wang L, Tan W. Bioconjugated silica-coated nanoparticles for bioseparation and bioanalysis. Trends Analyt Chem. 2006;25:848-55
28. Fan JB, Chee MS, Gunderson KL. Highly parallel genomic assays. Nat Rev Genet. 2006;7:632-44
29. Matsunaga T, Maruyama K, Takeyama H. et al. High-throughput SNP detection using nano-scale engineered biomagnetite. Biosens Bioelectron. 2007;22:2315-21
30. Xie J, Jon S. Magnetic nanoparticle-based theranostics. Theranostics. 2012;2:122-4
31. Luo X, Hsing IM. Immobilization-free multiplex electrochemical DNA and SNP detection. Biosens Bioelectron. 2009;25:803-8
32. Liu H, Li S, Ji M. et al. Fabrication and application of single nucleotide polymorphisms library on magnetic nanoparticles using adaptor PCR. J Nanosci Nanotechnol. 2008;8:405-9
33. Li S, Liu H, Wang Z. et al. Magnetic-particles-based high-throughput genotyping method with dual-color fluorescence hybridization. Anal Biochem. 2006;359:277-9
34. Frosst P, Blom HJ, Milos R. et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111-3
35. Liu L, Liu H, Li S. et al. Fabrication and Characterization of Streptavidin γ-Fe2O3@Au Nanocomposites for Biological Application. Acta Chimica Sinica. 2010;68:2041-6
Author contact
![]() Corresponding author: Nongyue He PhD, Sipailou No.2, Southeast University, Nanjing, 210096, China. Tel: +86-25-83790885, Fax: +86-25-83790885. email: nyhe1958com.
Corresponding author: Nongyue He PhD, Sipailou No.2, Southeast University, Nanjing, 210096, China. Tel: +86-25-83790885, Fax: +86-25-83790885. email: nyhe1958com.
 Global reach, higher impact
Global reach, higher impact