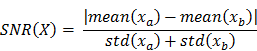13.3
Impact Factor
Theranostics 2013; 3(9):719-728. doi:10.7150/thno.5010 This issue Cite
Research Paper
Bioengineering Silicon Quantum Dot Theranostics using a Network Analysis of Metabolomic and Proteomic Data in Cardiac Ischemia
1. San Jose State University, California.
2. University at Buffalo, New York.
3. Lanzatech, Auckland.
4. North Shore Hospital, Auckland.
5. Princess Alexandria hospital, Brisbane.
6. Green Lane Cardiovascular Service, Auckland.
7. CCRep, Middlemore hospital, Auckland.
8. School of Biological Science, Auckland.
9. CAMO Software, New Jersey.
10. KEDRI, Auckland University of Technology.
11. Lerner Research Institute, Cleveland.
Received 2012-8-9; Accepted 2013-7-5; Published 2013-9-4
Abstract
Metabolomic profiling is ideally suited for the analysis of cardiac metabolism in healthy and diseased states. Here, we show that systematic discovery of biomarkers of ischemic preconditioning using metabolomics can be translated to potential nanotheranostics. Thirty-three patients underwent percutaneous coronary intervention (PCI) after myocardial infarction. Blood was sampled from catheters in the coronary sinus, aorta and femoral vein before coronary occlusion and 20 minutes after one minute of coronary occlusion. Plasma was analysed using GC-MS metabolomics and iTRAQ LC-MS/MS proteomics. Proteins and metabolites were mapped into the Metacore network database (GeneGo, MI, USA) to establish functional relevance. Expression of 13 proteins was significantly different (p<0.05) as a result of PCI. Included amongst these was CD44, a cell surface marker of reperfusion injury. Thirty-eight metabolites were identified using a targeted approach. Using PCA, 42% of their variance was accounted for by 21 metabolites. Multiple metabolic pathways and potential biomarkers of cardiac ischemia, reperfusion and preconditioning were identified. CD44, a marker of reperfusion injury, and myristic acid, a potential preconditioning agent, were incorporated into a nanotheranostic that may be useful for cardiovascular applications. Integrating biomarker discovery techniques into rationally designed nanoconstructs may lead to improvements in disease-specific diagnosis and treatment.
Keywords: metabolomics, silicon quantum dots, theranostics, cardiac ischemia, myocardial infarction.
Introduction
The heart is highly metabolic and consumes more energy than any other organ.[1] Unsurprisingly, cardiac energetics and metabolism is an area of intense study. Metabolomic profiling takes a high-throughput, global approach and screens thousands of metabolites using either targeted or untargeted methods. Due to its agnostic approach, and breadth of observation, its strength is in identifying novel pathways, not previously associated with disease.[2-4]
Metabolomics and proteomics have been applied to biomarker discovery for diseases such as myocardial infarction.[5] Although integrating data from both of these methods has been performed, published research in this area is rare.[6] Whilst biomarker discovery is one use for this method, the ideal goal is the elucidation of pathways for drug targeting or compound discovery for therapeutic applications.
Integrating drugs and imaging agents into a single platform to create theranostics has become a highly active area of research because it may enable earlier detection and treatment of diseases and may allow the assessment of patient response to therapy. To this end, nanoplatforms are actively being investigated as delivery vehicles for theranostics because they can readily incorporate multiple active agents (drugs, dyes, quantum dots, etc) in their core or on their surface. In addition to incorporation of multiple functionalities, nanotechnology platforms have shown the potential to reduce the side effects of free drug molecules by enhancing delivery to a specific targeted site, increasing the circulation time, and improving the toxicity profile of the drug. Rational design of nanocarriers allows for their functionalization with targeting agents that promote localization of the nanoplatforms at a disease site. Combining the strengths of proteomics and metabolomics with those of nanoplatforms enables the acquisition of rich information directly from patient samples to be translated into theranostics that can then be evaluated in suitable candidates. This approach may have real implications for personalized medicine when extended to single patients.
Recent developments in optical imaging provide quantitative information that offers interesting possibilities for in vivo diagnosis and monitoring of treatment efficacy in cancer and other various diseases.[7] The advantages of optical imaging are that is uses non-ionizing radiation and has high spatial resolution. The strategy of creating multifunctional optical systems for diagnosis, treatment and monitoring of cancer has been explored by several groups. Optical imaging has been separately combined with chemotherapy, photodynamic therapy, photothermal therapy, and gene therapy. It has also been combined with MRI agents and therapeutics to allow for multimodal imaging and therapy. The investigations of optical based theranostic platforms show promising potential for cancer treatment and diagnosis. The benefit of multifunctional nanomedicines in cancer is also relevant to cardiovascular diseases (CVDs) as there are many commonalities to the two disease entities. McCarthy et al reported the use of a light-activated theranostic nanoagent for targeted macrophage ablation in inflammatory atherosclerosis.[8] Their approach showed promise for detection and treatment of atherosclerotic lesions. Creating CVD theranostics is highly desirable; however, limited information on the active agents and targeting sites is presently available.
Quantum dots are light emitting semiconductor spheres 1-10nm in diameter and have been investigated as the optical component of theranostics, but investigations have been limited by toxicity concerns. The toxicity concerns are associated with the presence of heavy metals in first generation of quantum dots (e.g. CdSe-based QDs). This toxicity concern can be largely eliminated by using silicon quantum dots (SiQDs). SiQDs have been shown to be a promising non-toxic candidate for imaging and multimodal applications.[9] SiQDs have been investigated as imaging agents because they have bright emission, tunable luminescence color, and low toxicity. Significant demonstrations with SiQDs have been made in biological applications such as tumor targeting, sentinel lymph node mapping, and multicolor imaging. In this paper, we demonstrate the potential of integrating metabolomic and proteomic data in a human model of cardiac ischemia and preconditioning. We then translated this into a potential therapeutic diagnostic (theranostic), using a multimodal non-toxic nanoimaging agent (i.e. SiQDs, FDA approved F127 and DSPE-PEG-Amine) combined with compounds identified in the metabolomics analysis. Using data directly obtained from human models for the immediate fabrication of a theranostic nanoconstruct is a novel, highly multidisciplinary approach that has a strong potential to be adopted on the pathway to personalized medicine.
Materials and Methods
This study was approved by the Northern Regional Ethics Committee for New Zealand. 33 patients gave informed consent to be enrolled into the trial. Patients with a first non ST elevation myocardial infarction and serum troponin T >0.1 mmol/l undergoing coronary angiography between 24 hours and six days following were screened.
Exclusion criteria included: acute myocardial infarction within the preceding 24 hours, haemodynamic instability (including cardiogenic shock, systolic blood pressure <100 mmHg, uncontrolled heart failure or significant LV impairment (EF <35 percent)), significant (moderate-severe) valvular disease, renal impairment (creatinine >0.16 mmol/l), occluded vessel or extensive angiographic thrombus on diagnostic angiography.
All non-study medications including use of heparins, direct thrombin inhibitors, clopidogrel and glycoprotein IIb/IIIa inhibitors were administered at the discretion of the leading physician.
Study procedure
Baseline angiography was performed in the usual manner. Baseline blood samples were simultaneously taken from the great cardiac vein and ascending aorta. Angioplasty was then performed with a mandated initial predilatation of 60 seconds, unless otherwise indicated on clinical grounds. 20 minutes after the first balloon inflation a final blood sample was taken from the great cardiac vein (CS), ascending aorta (AO) and femoral vein.
Laboratory Procedures
Metabolomics GC-MS analysis
Metabolites were extracted according to an optimised protocol developed by the Villas-Bôas laboratory. A methyl chloroformate (MCF) derivatization was performed according to Villas-Bôas et al.[10] GC-MS analysis was undertaken according to the parameters established by Villas-Bôas & Bruheim.[11]
Metabolite identification
The approach for metabolite identification was developed by the Villas-Bôas laboratory[12] and is summarised as follows; metabolites in the samples were identified using our in-house MCF MS library of derivatized metabolites. The library contains MS spectra obtained from ultra-pure standards that have been derivatized, with the mass spectrum saved in the AMDIS 2.65 software (www.amdis.net). The relative level of the metabolites was based on the base peak height. Values were normalised by the base peak height of the internal standard (d4-alanine). A metabolite profile for each sample was thus built up and subjected to further analyses as described below.
Proteomics substudy (n=8)
Blood was collected into 5 ml EDTA (ethylenediaminetetraacetic acid) vacutainer tubes (Becton, Dickinson and Company) and plasma extracted by centrifugation within 2 minutes of collection.
Depletion of plasma samples
To allow for detection of moderately abundant plasma proteins, plasma samples were depleted of the 12 most abundant proteins using the ProteomeLab IgY-12 High Capacity SC Spin Column Kit (Beckman Coulter), according to the instructions provided by the manufacturer.
Sample preparation and liquid chromatography-tandem mass spectrometry (LC-MS/MS)
Depleted samples were prepared for iTRAQ labeling according to our previously devised procedure [13] with some modification. Protein concentrations were determined using the Bradford method (BioRad). Digested plasma protein from each preparation was labeled with 4-plex iTRAQ reagent, according to the manufacturer's description. The combined pools of iTRAQ-labeled samples were then fractioned by on-line cation exchange using 15 salt steps and the resulting LC effluent was directed into the ion spray source of a QSTAR XL hybrid mass spectrometer (Applied Biosystems, Foster City, CA) set to scan from 300 to 1600 m/z and with the top three most abundant multiply charged peptides selected for MS/MS analysis (80-1600 m/z). Protein Pilot 1.0 software (Applied Biosystems, Foster City, CA) was then used with the “rapid” search effort to search the output data against the human IPI v3.27 database with carbamidomethyl cysteine as fixed modification and trypsin as the enzyme specificity.
Statistics and Bioinformatics
Proteomic data analysis
The significance of the protein expression changes were determined using a two-tailed Student's t-test assuming unequal variance, p<0.05 was considered significant and p<0.10 was considered trending. Further to this first pass analysis, proteins found to change with p<0.10 were subjected to a second test in order to predict the relevance of these findings across patients, i.e. their potential usefulness as biomarkers. For this, protein log ratios were obtained for each post-PCI versus the non-corresponding pre-PCI value analyzed within the same LC-MS/MS run. Significance was then determined using an unpaired two-tailed Student's t-test assuming unequal variance, p<0.05 was considered significant and p<0.10 was considered trending.
Metabolomic data analysis
The metabolomic data was initially transformed using Principal Component Analysis (PCA) before bioinformatic methods were applied. A signal to noise (SNR) method was used to rank data from most important to least important based on SNR.
(Where xa are the values of variable X belonging to class A and xb are the variables of belonging to class B).
The data was pre-processed (missing values filled in, normalization) and then split several times (three to six folds cross validation) into a training part (e.g. 70 percent) and test part (e.g. 30 percent). Features were extracted and ranked from the training part and then these features and the training data were used to build a personalized model (PM) for every sample from the test data.
Functional analysis
The proteins and metabolites that featured commonly between the first principal component and the bioinformatic methods, used above, were mapped into the Metacore network database (GeneGo, MI, USA). Genes known to interact with or affected by the key metabolites from comparative toxicogenomics studies were also included in the analysis.
Preparation of Theranostic Nanoconstructs
Synthesis of silicon nanoparticles
Luminescent silicon nanoparticles were synthesized as detailed in previous reports [14]. Non luminescent powders were synthesized by laser pyrolysis, etched with a 10:1 hydrofluoric acid:nitric acid mixture to obtain luminescence, and functionalized with ethyl undecylenate by UV initiated hydrosilylation. The samples were then stored for further use.
Synthesis of theranostic nanoconstructs
The silicon quantum dots, myristic acid (Aldrich), Pluronic® F127 (BASF) and DSPE PEG amine (Avanti Polar Lipids) were suspended in chloroform (1:0.3:19:1, w/w ratio) and sonicated. The chloroform was evaporated using a Labconco rotary evaporator, leaving a lipidic film. The film was sonicated in HPLC grade water to form a micellar suspension. In subsequent experiments, the nanoconstructs were conjugated to hyaluronic acid (Lifecore Biomedical LLC) via EDC chemistry to form an amide bond between carboxyl groups on the hyaluronic acid and the amine groups of the DSPE-PEG amine. The particles were washed three times and stored for further use.
Results
Proteomics
The initial analysis of changes within patients identified 31 proteins as significantly different between pre- and post-PCI samples (p<0.05). A second analysis identified 27 proteins as significantly different (p<0.05). Combining the information from the two analyses revealed 13 proteins, displayed in Table 1, which exhibited absolute consistency and which changed with p<0.10 in both analyses. Of these, four decreased by PCI had calculated ratios (post-PCI Vs pre-PCI, paired and crossed) consistently lower than <0.6, and a further three proteins were increased with all ratios > 1.4.
Protein expression changes due to percutaneous coronary intervention (PCI).
| Protein Name | IPI Accession | average pre-PCI Vs post-PCI (paired) | Range of ratios | average pre-PCI Vs post-PCI (crossed) | Range of ratios | ||
|---|---|---|---|---|---|---|---|
| Min | Max | Min | Max | ||||
| Thyroxine-binding globulin | IPI00292946.1 | 0.26 | 0.11 | 0.52 | 0.25 | 0.15 | 0.46 |
| Alpha-1-acid glycoprotein 2 | IPI00020091.1 | 0.37 | 0.19 | 0.94 | 0.37 | 0.15 | 0.90 |
| Complement C1r-like protein | IPI00009793.3 | 2.47 | 1.54 | 5.85 | 2.47 | 1.40 | 4.92 |
| Isoform 1 of Complement factor B | IPI00019591.1 | 1.98 | 1.24 | 3.19 | 1.98 | 1.16 | 6.98 |
| CD44 antigen | IPI00828192.1 | 5.10 | 1.79 | 14.41 | 5.10 | 1.70 | 15.18 |
| Isoform 1 of Alpha-1-antichymotrypsin | IPI00550991.3 | 0.08 | 0.03 | 0.19 | 0.08 | 0.02 | 0.23 |
| Alpha-2-antiplasmin | IPI00029863.4 | 0.24 | 0.07 | 0.92 | 0.24 | 0.05 | 0.91 |
| Kallistatin | IPI00328609.3 | 0.17 | 0.09 | 0.47 | 0.13 | 0.02 | 0.82 |
| Heparin cofactor 2 | IPI00292950.4 | 0.12 | 0.07 | 0.23 | 0.12 | 0.04 | 0.35 |
| Isoform HMW of Kininogen-1 | IPI00032328.2 | 0.06 | 0.03 | 0.13 | 0.06 | 0.02 | 0.10 |
| Apolipoprotein(a) | IPI00739995.2 | 3.70 | 2.19 | 5.52 | 3.70 | 1.77 | 5.32 |
| Apolipoprotein B-100 | IPI00022229.1 | 0.42 | 0.22 | 0.75 | 0.42 | 0.12 | 1.00 |
| Alpha-1B-glycoprotein precursor | IPI00745089.2 | 2.14 | 1.22 | 4.20 | 2.14 | 1.32 | 2.86 |
Min and max ratios (paired and crossed comparisons) for the 13 proteins that exhibited absolute consistency and changed with p<0.10 in both analyses. In bold are four decreased proteins where the highest individual post-PCI vs. pre-PCI ratio (paired or crossed) was below 0.6, and three increased proteins where the lowest post-PCI Vs pre-PCI ratio (paired or crossed) was at least 1.40.
Western blotting
Obtaining reliable antibodies for most of the proteins of interest was difficult and therefore restricted confirmation of the proteomic results by western blotting to ceruloplasmin (CP). The behavior of CP levels in non-depleted plasma was largely consistent with the proteomic findings in that seven of the eight patients showed a marked decrease in CP levels post-PCI.
Metabolomics
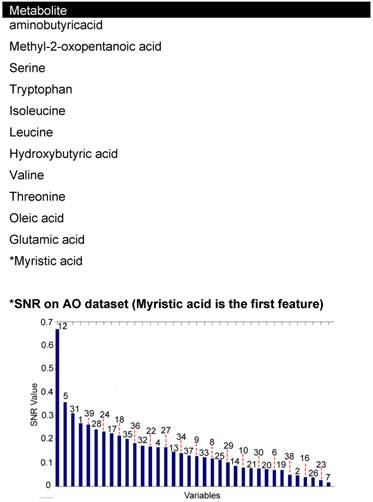
Principal components analysis showed that 31 metabolites accounted for 42 percent of the variance in the results. 21 of these metabolites, in the first PCA (Table 2, Figure 1), were cross-referenced with the metabolites identified by the highest signal to noise ratio in the coronary sinus plasma dataset. 12 common metabolites (Table 3) were used for the functional analysis.
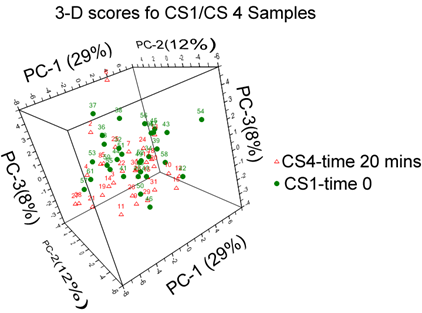
The scores plot of the first three PCs, describe 48 percent of the variance in the data, and the separation of samples into groups can be seen clearly.
3D score plot for coronary sinus samples at baseline and post procedure (an enlarged image is included in supporting information.)
Metabolites identified in first Principal Component (PCA1).
| Metabolite |
|---|
| aminobutyricacid |
| Methyl-2-oxopentanoic acid |
| Serine |
| Tryptophan |
| Glycine |
| Lysine |
| Isoleucine |
| Leucine |
| Hydroxybutyric acid |
| Phenylalanine |
| Valine |
| Creatinine |
| Threonine |
| Oleic acid |
| Aspartic acid |
| Glutamic acid |
| Pyroglutamic acid |
| Alanine |
| Myristic acid |
| Cysteine |
| Lactic acid |
Common 12 features seen between PCA1 and SNR ranking of coronary sinus plasma.
Biomarker analysis
Metabolomics
Discriminatory analysis using bioinformatics was applied to systemic blood taken from the aorta. Three methods were used, including wKNN classification model, a SVM classification method and an evolving spiking neural network classification model (eSNN). After 1,000 iterations of the algorithm a discriminatory accuracy of 81 percent was made. 11 features were identified using these models, five were common with the metabolites identified by PCA1 and SNR from the coronary sinus dataset. The potential biomarkers identified by bioinformatics analysis of aortic blood and cross-referenced with coronary sinus were methyl-2-oxopentanoicacid, serine, tryptophan, oleic acid and *myristic acid. Structures of all molecules are listed in Supplementary Material: table S1.
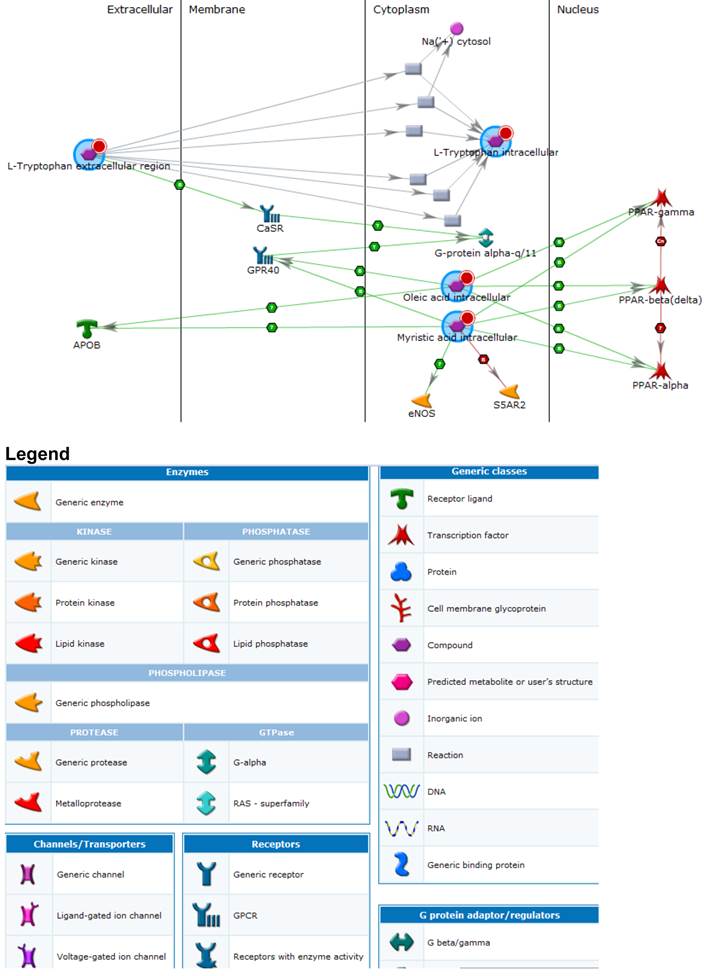
Functional network analysis
Cross-referencing proteins, metabolites and putative genes identified a number of pathways of interest (Figure 2 and Legend). These included; immune response, clotting cascade, steroid, lipid, cholesterol and sterol metabolic processes as well as vLDL particle assembly. General biomarkers fell into the categories of glucose metabolism disorders, hyperinsulinism, nutritional and metabolic disorders. The top five diseases associated with the key metabolites and interacting genes included; myocardial ischemia, glucose metabolism, metabolic disorders, vascular diseases and eye disease. Statistically significant pathways and process networks showed glutathione metabolism, and EDG3 and EDG1 signaling pathways to be involved as well as networks related to hypoxia and oxidative stress, signal transduction ESR2, inflammation and complement system, immune response - phagocytosis, cell adhesion (platelet-endothelium-leukocyte interactions), translational selenium pathway, cell cycle G1-5 growth factor regulation, cell adhesion glycoconjugates, signal transduction WNT signaling and cytoskeleton regulation and rearrangement.
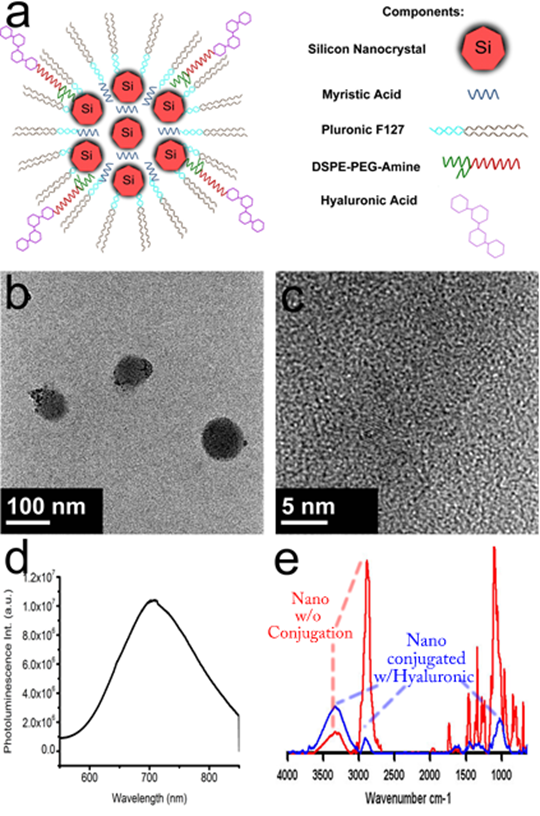
To fabricate a theranostic based on the information from the metabolomics study, the myristic acid and silicon quantum dots were encapsulated in the core of a micelle composed of a mixture of Pluronic ® F127 and DSPE-PEG-NH2. Hyaluronic acid, capable of targeting CD44, was bioconjugated to the surface of the nanoparticle by creating peptide bonds between the carboxyl group and the amine groups of the DSPE-PEG-NH2. Figure 3a depicts a schematic showing the components of the potential theranostic. Transmission electron microscopy images in Figure 3b show that the theranostic is near-spherical and about 100 nm or less in diameter. Upon close observation in figure 3c, crystalline silicon is observed (Supplementary Material: Figure S2 has a larger image to allow lattice fringes to be observed clearly). The silicon nanoparticles are dispersed throughout the micelle, not densely packed. This is attributed to the presence of myristic acid. The emission of the silicon is at 700 nm as seen in Figure 3d. This is a particularly good emission wavelength for biological imaging because biological samples have the highest transparency between 700 nm and 900 nm. FTIR confirms the attachment of hyaluronic acid to the surface of the micelles, as shown in figure 3e. The red line shows the spectra of silicon nanoconstructs before bionconjugation. It has characteristic peaks around for amines around 3300 cm-1 and CH bending around 2800 cm-1. The blue line shows spectra after bioconjugation and suggests that hyaluronic acid is bound to the surface of the micelle, because the OH peak of hyaluronic acid becomes more prominent around 3400 cm-1 and 1000 cm-1, and the CH peaks associated with DSPE-PEG decrease. Preliminary release analysis by HPLC indicates that 3.7% of the myristic acid was released from the micelle in 3 hours. The targeting ability of the particles to CD44 was evaluated by targeting them to pancreatic cancer cells with CD44 receptors [15]. The signal from the silicon quantum dots was readily seen.
Discussion
This exploratory experiment of coronary angioplasty identified altered expression in a number of proteins and metabolites. Furthermore merging the metabolite and proteomic data in a network analysis identified pathways and disease mechanisms consistent with the perturbation being examined, that being cardiac ischemia. A number of the changes seen were probably the result of heparin administration (e.g. antithrombin III, beta-2-glycoprotein 1, hydroxybutyrate) [16] however the key features of CD44 and various fatty acids were notable.
CD44 appeared as an interesting potential diagnostic biomarker for ischemia/reinfarction in this study. An independent proteomics study by Addona et al has also showed upregulation of CD44 in patients undergoing alcohol induced myocardial infarction [17]. CD44 antigen has been shown to be up-regulated in response to ischemic injury of the kidney as well as myocardial infarct and it may mediate infarct healing through the regulation of inflammatory and fibrotic responses [18]. CD44 is expressed on a number of cell types including not only solid organs but also neutrophils, macrophages and bone marrow derived mesenchymal stem cells (MESCs).[19] The multifunctional CD44 antigen is a hyaluronan receptor primarily involved in cell-cell and cell-matrix interactions. Hyaluronic acid, the ligand for CD44, appears critical in the localisation of MESCs after ischemic injury. It has been suggested that CD44 represents an interesting drug target and disruption of CD44 appears to protect against renal reperfusion injury in rats.[20] Hence we considered it worthwhile to develop a theranostic nanocontruct, using CD44 as the target molecule.[21]
Processes identified amongst key metabolites and proteins. An enlarged version of this image is available in the supporting information.
Composite diagram containing formulation schematic, TEM, photoluminescent emission, and FTIR spectra. (a) schematic of theranostic nanoconstruct (further details of self-assembly of lipid micelles are shown in Supplementary Material: Figure S1); (b) low-resolution TEM of nanoconstructs; (c) high resolution TEM showing crystalline Si nanocrystals within the constructs (larger image is shown in Supplementary Material: Figure S2); (d) photoluminescence emission spectrum of nanoconstructs; and (e) FTIR spectra of nanoconstructs before (red) and after (blue) conjugation with hyaluronic acid.
The metabolomic network analysis revealed a number of interesting findings with converging functional relevance. Both myristic and oleic acid demonstrated a functional connectivity with mechanisms related to peroxisome proliferator-activated receptor gene (alpha, beta, gamma) function (PPARs). This mechanism has similarly been shown to be important using metabolomic analysis of serum in patients following myocardial infarction.[22] Myristic acid itself has previously been shown to act as a protective agent against reperfusion injury in vitro[23], as a result of preconditioning. Preconditioning is caused by a endogenous humoral factor, probably a small molecule, and can be transferred between animals and species, using dialysate from treated animals.[24-26] Myristic acid has been shown to have active biological properties when administered ex vivo. Myristic, pentadecanoic, palmitic, and palmitoleic acid cause downregulation of TNF-alpha, via PPAR agonism, in inflammatory diseases such as falciparum malaria infection.[27] We hypothesized that myristic acid may be physiologically active in the context of ischemia and therefore developed a theranostic micelle using hyaluronic acid as the targeting ligand and incorporating myristic acid for therapeutic delivery or as an excipient. This targeted construct could also be a carrier for other reperfusion injury therapeutics, such as dexamethasone, which if delivered systemically has significant adverse consequences.[28]
Combining this information to create a nanoconstruct is a logical approach and the demonstrations here indicate that the fabrication of such a rationally designed nanoconstruct is possible. Toxicity is a concern for optical imaging agents based on quantum dots because many demonstrations of the potential of QD imaging used heavy metal-based QDs. Investigations into properly using heavy metal based quantum dots (generation 1 QDs) are still ongoing and are not yet conclusive as to the nature of toxicity in humans. SiQDs have been suggested as a suitable replacement for first-generation heavy metal-based quantum dots because they reduce the toxicity concerns and maintain their promise as optical imaging agents.
SiQDs have been used for cancer application such as tumor targeting. Here, we use knowledge of that design to show that the information from metabolomics and proteomics can be used to create a potential theranostic. The rational design includes the use of amine terminated DSPE-PEG that allows for further functionalization of the nano-construct. FTIR verifies that hyaluronic acid is present on the surface of the nanoparticle. We have previously demonstrated that CD44 can be targeted with hyaluronic acid.[29] The myristic acid in the core of the nanoconstruct is released at a slow rate however this may be desirable for delayed release kinetics. The optical signal from the SiQDs is at 700nm, which is a desirable wavelength for biological applications because it can provide a better signal to noise ratio than emission at visible wavelengths where tissue is less transparent. Implementation of this potential nanoconstruct will require testing and modification of the nanoconstruct to evaluate its effectiveness, which will be reserved for future studies in cells and animals. There is potential to expand the theranostic functionality of the nanoparticle by including other desirable modalities. Silicon quantum dots have already been combined with dyes, iron oxide, gold shells, siRNA and drugs in nanoformulations.
The overall vision is to be able to obtain rich patient information with proteomic and metabolomics and rationally design platforms that can be evaluated and yield diagnosis and treatment information in a timely manner.
Limitations
This experiment was performed on a small sample size (n=33) with the potential for variability due to uncontrollable factors. The inability to perform multiple parallel MS experiments on the same samples limits the reproducibility of results. An additional limitation was the fact that the effects of pharmacological agents (heparin and glyceryl trinitrate) administered in conjunction with PCI cannot be easily distinguished from that of ischemia induced by balloon inflation during PCI. Optical imaging can suffer from poor tissue penetration and biological autofluorescence which would limits its use in imaging the heart, however we use particle that emit at 700nm, a wavelength that has the advantage of reduced autofluorescence, reduced tissue scattering and greater depth of penetration.[30] It is also quite plausible that the nanoconstruct will identify peripheral circulating blood cells which could be used for diagnostic imaging through skin or blood assays.
Conclusion
This mechanistic study has demonstrated a method for metabolomic profiling of plasma and integrating this with proteomic data. This method has value in identifying novel biomarkers of disease and elucidating disease mechanisms through a holistic network analysis and in guiding development of novel therapeutic targets and compounds. These results are consistent with reports from independent laboratories. We have demonstrated the feasibility of using this data to construct a nanotheranostic that can be used for targeted imaging and therapy purposes.
Supplementary Material
Table S1-S2, Fig.S1-S2.
Acknowledgements
The authors acknowledge the ford foundation.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Neubauer S. The failing heart--an engine out of fuel. N Engl J Med. 2007;356:1140-51
2. Huffman KM, Shah SH, Stevens RD, Bain JR, Muehlbauer M, Slentz CA. et al. Relationships between circulating metabolic intermediates and insulin action in overweight to obese, inactive men and women. Diabetes Care. 2009;32:1678-83
3. Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E. et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448-53
4. Huang Y, Zhou M, Sun H, Wang Y. Branched-chain amino acid metabolism in heart disease: an epiphenomenon or a real culprit? Cardiovasc Res. 2011;90:220-3
5. Lewis GD, Wei R, Liu E, Yang E, Shi X, Martinovic M. et al. Metabolite profiling of blood from individuals undergoing planned myocardial infarction reveals early markers of myocardial injury. The Journal of clinical investigation. 2008;118:3503-12
6. Lin D, Hollander Z, Meredith A, Stadnick E, Sasaki M, Cohen Freue G. et al. Molecular signatures of end-stage heart failure. Journal of cardiac failure. 2011;17:867-74
7. Mura S, Couvreur P. Nanotheranostics for personalized medicine. Advanced drug delivery reviews. 2012;64:1394-416
8. McCarthy JR, Korngold E, Weissleder R, Jaffer FA. A light-activated theranostic nanoagent for targeted macrophage ablation in inflammatory atherosclerosis. Small. 2010;6:2041-9
9. Liu L, Law WC, Yong KT, Roy I, Ding H, Erogbogbo F. et al. Multimodal imaging probes based on Gd-DOTA conjugated quantum dot nanomicelles. The Analyst. 2011;136:1881-6
10. Villas-Boas SG, Delicado DG, Akesson M, Nielsen J. Simultaneous analysis of amino and nonamino organic acids as methyl chloroformate derivatives using gas chromatography-mass spectrometry. Anal Biochem. 2003;322:134-8
11. Villas-Boas SG, Bruheim P. Cold glycerol-saline: the promising quenching solution for accurate intracellular metabolite analysis of microbial cells. Anal Biochem. 2007;370:87-97
12. Smart KF, Aggio RB, Van Houtte JR, Villas-Boas SG. Analytical platform for metabolome analysis of microbial cells using methyl chloroformate derivatization followed by gas chromatography-mass spectrometry. Nat Protoc. 2010;5:1709-29
13. Jüllig M, Hickey AJ, Middleditch MJ, Crossman DJ, Lee SC, Cooper GJS. Characterization of proteomic changes in cardiac mitochondria in streptozotocin-diabetic rats using iTRAQ™ isobaric tags. Proteomics Clin Appl. 2007;1:565-76
14. Erogbogbo F, Yong K-T, Roy I, Hu R, Law W-C, Zhao W. et al. In Vivo Targeted Cancer Imaging, Sentinel Lymph Node Mapping and Multi-Channel Imaging with Biocompatible Silicon Nanocrystals. ACS Nano. 2010;5:413-23
15. Erogbogbo F, Liu X, May JL, Narain A, Gladding P, Swihart MT. et al. Plasmonic gold and luminescent silicon nanoplatforms for multimode imaging of cancer cells. Integrative Biology. 2013;5:144-50
16. Brunner MP, Shah SH, Craig DM, Stevens RD, Muehlbauer MJ, Bain JR. et al. Effect of heparin administration on metabolomic profiles in samples obtained during cardiac catheterization. Circulation Cardiovascular genetics. 2011;4:695-700
17. Addona TA, Shi X, Keshishian H, Mani DR, Burgess M, Gillette MA. et al. A pipeline that integrates the discovery and verification of plasma protein biomarkers reveals candidate markers for cardiovascular disease. Nature biotechnology. 2011;29:635-43
18. Kocak B, Orug T, Turhan N, Ozcay N, Gonenc F. CD44 expression in renal ischemia-reperfusion injury in rats. Int Urol Nephrol. 2009Dec;41(4):791-4
19. Gyongyosi M, Posa A, Pavo N, Hemetsberger R, Kvakan H, Steiner-Boker S. et al. Differential effect of ischaemic preconditioning on mobilisation and recruitment of haematopoietic and mesenchymal stem cells in porcine myocardial ischaemia-reperfusion. Thrombosis and haemostasis. 2010;104:376-84
20. Rouschop KM, Roelofs JJ, Claessen N, da Costa Martins P, Zwaginga JJ, Pals ST. et al. Protection against renal ischemia reperfusion injury by CD44 disruption. Journal of the American Society of Nephrology: JASN. 2005;16:2034-43
21. Krettek A, Sjoberg S. CD44 - a new cardiovascular drug target or merely an innocent bystander? Cardiovascular & hematological disorders drug targets. 2009;9:293-302
22. Bodi V, Sanchis J, Morales JM, Marrachelli VG, Nunez J, Forteza MJ. et al. Metabolomic profile of human myocardial ischemia by nuclear magnetic resonance spectroscopy of peripheral blood serum: a translational study based on transient coronary occlusion models. Journal of the American College of Cardiology. 2012;59:1629-41
23. Kaplan LJ, Blum H, Bellows CF, Banerjee A, Whitman GJ. Reversible injury: creatinine kinase recovery restores bioenergetics and function. J Surg Res. 1996;62:103-8
24. Shimizu M, Tropak M, Diaz RJ, Suto F, Surendra H, Kuzmin E. et al. Transient limb ischaemia remotely preconditions through a humoral mechanism acting directly on the myocardium: evidence suggesting cross-species protection. Clin Sci (Lond). 2009;117:191-200
25. Wang L, Oka N, Tropak M, Callahan J, Lee J, Wilson G. et al. Remote ischemic preconditioning elaborates a transferable blood-borne effector that protects mitochondrial structure and function and preserves myocardial performance after neonatal cardioplegic arrest. J Thorac Cardiovasc Surg. 2008;136:335-42
26. Jean-St-Michel E, Manlhiot C, Li J, Tropak M, Michelsen MM, Schmidt MR. et al. Remote Preconditioning Improves Maximal Performance in Highly-Trained Athletes. Med Sci Sports Exerc. 2011Jul;43(7):1280-6
27. Debierre-Grockiego F, Schofield L, Azzouz N, Schmidt J, Santos de Macedo C, Ferguson MA. et al. Fatty acids from Plasmodium falciparum down-regulate the toxic activity of malaria glycosylphosphatidylinositols. Infection and immunity. 2006;74:5487-96
28. Alisky JM. Dexamethasone could improve myocardial infarction outcomes and provide new therapeutic options for non-interventional patients. Medical hypotheses. 2006;67:53-6
29. Erogbogbo F, Liu X, May JL, Narain A, Gladding P, Swihart MT. et al. Plasmonic gold and luminescent silicon nanoplatforms for multimode imaging of cancer cells. Integrative biology: quantitative biosciences from nano to macro. 2012;5:144-50
30. Janib SM, Moses AS, MacKay JA. Imaging and drug delivery using theranostic nanoparticles. Advanced drug delivery reviews. 2010;62:1052-63
Author contact
![]() Corresponding author: Patrick Gladding, Address: Northshore Hospital, Waitemata DHB. Email: patrickgcom.
Corresponding author: Patrick Gladding, Address: Northshore Hospital, Waitemata DHB. Email: patrickgcom.
 Global reach, higher impact
Global reach, higher impact