13.3
Impact Factor
Theranostics 2017; 7(7):2015-2032. doi:10.7150/thno.19500 This issue Cite
Research Paper
Melatonin Suppresses Neuropathic Pain via MT2-Dependent and -Independent Pathways in Dorsal Root Ganglia Neurons of Mice
1. Department of Neurology, Tangdu Hospital, Fourth Military Medical University, Xi'an, Shannxi, 710004, China;
2. Department of Anatomy, Fourth Military Medical University, Xi'an, Shannxi, 710032, China;
3. Department of Neurosurgery, Tangdu Hospital, Fourth Military Medical University, Xi'an, Shannxi, 710004, China.
* Jia-Ji Lin, Ye Lin and Tian-Zhi Zhao contributed equally to this work.
Received 2017-2-5; Accepted 2017-3-15; Published 2017-5-12
Abstract
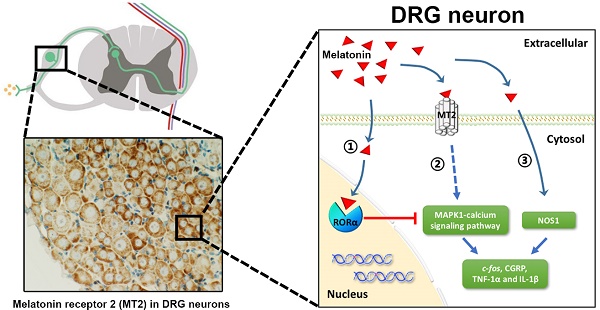
Melatonin (Mel) and its receptors (MT1 and MT2) have a well-documented efficacy in treating different pain conditions. However, the anti-nociceptive effects of Mel and Mel receptors in neuropathic pain (NP) are poorly understood. To elucidate this process, pain behaviors were measured in a dorsal root ganglia (DRG)-friendly sciatic nerve cuffing model. We detected up-regulation of MT2 expression in the DRGs of cuff-implanted mice and its activation by the agonist 8-M-PDOT (8MP). Also, Mel attenuated the mechanical and thermal allodynia induced by cuff implantation. Immunohistochemical analysis demonstrated the expression of MT2 in the DRG neurons, while MT1 was expressed in the satellite cells. In cultured primary neurons, microarray analysis and gene knockdown experiments demonstrated that MT2 activation by 8MP or Mel suppressed calcium signaling pathways via MAPK1, which were blocked by RAR-related orphan receptor alpha (RORα) activation with a high dose of Mel. Furthermore, expression of nitric oxide synthase 1 (NOS1) was down-regulated upon Mel treatment regardless of MT2 or RORα. Application of Mel or 8MP in cuff-implanted models inhibited the activation of peptidergic neurons and neuro-inflammation in the DRGs by down-regulating c-fos, calcitonin gene-related peptide [CGRP], and tumor necrosis factor-1α [TNF-1α] and interleukin-1β [IL-1β]. Addition of the MT2 antagonist luzindole blocked the effects of 8MP but not those of Mel. In conclusion, only MT2 was expressed in the DRG neurons and up-regulated upon cuff implantation. The analgesic effects of Mel in cuff-implanted mice were closely associated with both MT2-dependent (MAPK-calcium channels) and MT2-independent (NOS1) pathways in the DRG.
Keywords: melatonin, neuropathic pain, melatonin receptor, dorsal root ganglia, neuron.
Introduction
Neuropathic pain (NP) is a chronic pain that results from injury of the peripheral nervous system (PNS) or the central nervous system (CNS) [1]. The most prominent features of NP include dysesthesia, allodynia, and hyperalgesia [2, 3]. In the USA, it is estimated that 25 million (11.2%) adults suffer from chronic daily pain and up to 23 million (10%) adults have severe pain [4]. In Europe, the situation is not much different, with up to 7-8% of the European population complaining of refractory chronic pain that was reported to be severe in 5% of participants [5, 6].
Melatonin (Mel) is a circadian hormone that has been widely reported to possess anti-oxidant and anti-inflammatory properties [7-9]. It was previously demonstrated that Mel receptors in the CNS are critical in melatonin-induced anti-nociception in inflammatory and NP models [10-12]. However, the existence and biological function of Mel receptors in the PNS remains largely unknown. The dysfunction of pain transmission in the PNS, particularly the dorsal root ganglia (DRG), is likely involved in NP development. Injury to a peripheral afferent fiber results in hyper-excitability in axotomized DRG neurons, which promote ectopic firing and induce central sensitization and clinical allodynia [13-16]. Further, the neuroimmune activation of neurons in the DRG can also sensitize and lower the threshold for neuronal firing, leading to peripheral and central sensitization as well as the development of chronic NP [17, 18]. Stimulation of the DRG reduces the intensity of pain as well as stabilizes and decreases the hyper-excitability of DRG neurons [19]. It has been reported that extracellular application of Mel inhibits high-voltage activated calcium channels and reduces the intracellular free Ca2+ concentration in cultured DRG neurons based upon the whole-cell patch clamp and the fura-2 fluorescence ratio Ca2+-imaging techniques [20]. Mel can also modulate the Ca2+ entry induced by diabetes or 2.45-GHz electromagnetic radiation through the transient receptor potential melastatin 2 (TRPM2) and transient receptor potential vanilloid type 1 (TRPV1) [21, 22]. In light of these observations, it is essential to explore the regulatory mechanism of Mel in the DRG.
Although melatonin has been widely reported to possess significant analgesic effects in several pain models [23-27], the anti-nociceptive effect of Mel in neuropathic pain (NP) is controversial and largely unknown. In mice with NP induced by sciatic nerve ligation (SNL), Mel treatment reduced the thermal hyperalgesia without significantly improving mechanical allodynia [28], while it was only effective in treating mechanical allodynia in a chronic constriction injury (CCI) model [29]. These inconsistencies could be attributed to the characteristics of these NP models, which always promote significant injury on the DRG and primary afferents [30, 31]. The SNL model is easily exposed to infection and often causes a severe motor deficit [30]. The degree of allodynia produced in the CCI models is subject to variation in the snugness of ligation that leads to pronounced individual differences among the treated animals [30]. Therefore, we choose the cuff implantation model to re-assess the analgesic effects of Mel on NP in the DRG [31]. Cuff implantation promotes ipsilateral allodynia and hyperalgesia as early as on day 1 of post-surgery day without an inflammatory component and behavior injury. In this study, we explored the analgesic effects of Mel in a cuff implantation NP model as well as the biological function of Mel receptors in the DRG, and the findings will ultimately lead to advances in therapeutic techniques for NP.
Materials and Methods
Reagents
Mel, luzindole (MT1 and MT2 antagonist), ramelteon (MT1 and MT2 agonist), 8-methoxy-2-propionamidotetralin (8MP, MT2 agonist) 4-phenyl-2-propionamidotetralin (4PP, MT2 antagonist), CGP52608 (RAR-related orphan receptor alpha [RORα] receptor agonist), and ML-176 (RORα receptor inverse agonist) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). All reagents were dissolved in vehicle (1% ethanol diluted in sterile saline). The lysis buffer, protease and phosphatase inhibitors, bovine serum albumin (BSA), Tris-HCl buffer, bicinchoninic acid (BCA) kit, hematoxylin, and Tris-buffered saline with Tween-20 were purchased from Sigma-Aldrich (St. Louis, MO, USA). Sodium dodecyl sulfate (SDS) polyacrylamide gels and diaminobenzidine tetrahydrochloride (DAB) were purchased from Bio-Rad Laboratories (Hercules, CA, USA) and Doujin (Kumamoto, Japan), respectively. Trizol reagent and the PrimeScript RT reagent kit were purchased from Takara (Dalian, Liaoning, China). Mouse interleukin-1 beta (IL-1β) and tumor necrosis factor alpha (TNF-α) enzyme-linked immunosorbent assay (ELISA) kits, Advanced Dulbecco's modified Eagle medium (DMEM), lipofectamine 3000, and Opti-MEM were acquired from Thermo Fisher Scientific (Waltham, MA, USA). The antibodies and siRNA sequences used in this study are listed in Table 1.
Sciatic nerve cuffing model and experimental design
A total of 100 adult male C57BL/6J mice weighing 22-25 g were obtained from the experimental animal center of the Fourth Military Medical University. Mice were allowed a 2-week acclimatization period and housed under a 12-hour light/dark cycle with food and water available ad libitum. The experimental protocol was revised and approved by the Institutional Animal Care and Use Committee of Fourth Military Medical University. All measures were taken to minimize the animal suffering and decrease the number of mice used in this study.
The detail information of primers, antibody and siRNA.
| Primers | |
|---|---|
| Gene ID | Primer |
| MAPK1 | Forward: 5'-TGTTCCCAAATGCTGACTCC-3', Reverse: 5'-AGCCTGTTCAACTTCAATCCTC-3'; |
| Cacna1b | Forward: 5'-ACCATCCGCATCCTATTGTG-3', Reverse: 5'-GATACTGGTGTCATCATCAAGGG-3'; |
| Cacna1e | Forward: 5'-CTAGGCCTCGGCAAGAGATG-3', Reverse: 5'-GCACCACCTTTGGCGATTTT-3'; |
| Grm1 | Forward: 5'-GGGTCAGATTAAGGTCATACGG-3', Reverse: 5'-CGAGGTAACGGATAGTAATGGG-3'; |
| Grin2a | Forward: 5'-TTTGAGGACGCCAAGACAC-3', Reverse: 5'-AGACTGAAATGAGACCCGATG-3'; |
| NOS1 | Forward: 5'- CGATCGGCCCTTGGTAGA-3', Reverse: 5'-AGGCAATGCCCCTGAGAAC-3'; |
| β-actin | Forward: 5'-CTGTCGAGTCGCGTCCAC-3', Reverse: 5'-GTTTGTGTAAGGTAAGGTGTG-3'. |
| Antibodies used in Western blot | |
| rabbit anti-MT1: ab203038, 1:200, Abcam; rabbit anti-MT2: ab203346, 1:200, Abcam; rabbit anti-MAPK1: ab32081, 1:400, Abcam; rabbit anti-Cacna1b: ab97284, 1:300, Abcam; rabbit anti-Cacna1e SAB4300837 1:200 Sigma; rabbit anti-Grm1: ab82211, 1:200, Abcam; rabbit anti-Grin2a: ab124913, 1:400, Abcam; rabbit anti-NOS1: ab76067, 1:300, Abcam; rabbit anti-c-fos: ab190289 1:200, Abcam; rabbit anti-CGRP: AB5920, 1:1000, Millipore; anti-RORα: ab60134, 1:200, Abcam; rabbit anti-β-actin: ab8227, 1:400, Abcam; HRP-conjugated anti-rabbit donkey IgG: 1:5000; Zhongshan, Beijing, China. | |
| Antibodies used in immunohistochemical staining | |
| rabbit anti-MT1: ab203038, 1:100, Abcam; rabbit anti-MT2, ab203346, 1:100, Abcam; biotinylated goat anti-rabbit IgG, ab64256, 1:400, Abcam. | |
| Antibodies used in immunofluorescent staining | |
| rabbit anti-MT1: ab203038, 1:100, Abcam; rabbit anti-MT2: ab203346, 1:100, Abcam; chicken anti-NeuN: ab134014, 1:1000, Abcam; chicken anti-GFAP: ab50738, 1:100, rabbit anti-CGRP: AB5920, 1:1000, Millipore; anti-rabbit-conjugated AlexaFluor 594: ab150116, 1:1000, Abcam; anti-chicken-conjugated AlexaFluor 488: A-11039, 1:1000, Thermo Fisher Scientific, Shanghai, China. The IB4 were detected with fluorochromes with FITC (FITC-IB4: FL-1201, 1:200, Vector). | |
| The siRNA used for gene knocking down | |
| MAPK1 (sc-35336), Cacna1b (sc-42699), Cacna1e (sc-42703), Grm1 (sc-61027), Grin2a (sc-36084), RORα (sc-38863) or control siRNA (sc-37007) were purchased from Santa Cruz Biotechnology. | |
The sciatic nerve cuffing model was prepared as described previously [31]. Briefly, following anesthesia, mice were placed on a warm mat (37°C) in a left lateral position, and the surgical procedure was performed under aseptic conditions. The common branch of the right sciatic nerve was exposed, and a 2-mm split polyethylene tubing (PE-20, Harvard Apparatus, Dave Hollis, MA, USA) was placed around it. The shaved skin layer was closed using a suture. The sham-operated mice underwent the same procedures described above without the cuff implantation (Sham group).
Following recovery, all mice underwent pain behavioral testing, and only mice displaying significant pain behavior were used for the different treatment protocols: 1) Cuff+Vel group: mice received i.p. injection of 100 ml/kg vehicle (1% ethanol/normal saline) via intraperitoneal (i.p.) injection; 2) Cuff+Mel group: mice received i.p. injection of 100 mg/kg Mel [28]; 3) Cuff+8MP group: mice received i.p. injection of 100mg/kg 8MP; 4) Cuff+R+4PP group: mice received i.p. injection of 50 mg/kg ramelteon and 20 mg/kg 4PP. Additionally, biochemical test data were obtained for the following groups: 1) Cuff+Mel+L group: mice received i.p. injection of 100 mg/kg Mel and 20 mg/kg luzindole; and 2) Cuff+8MP+L group: mice received i.p. injection of 100 mg/kg 8MP and 20 mg/kg luzindole. Drug treatments were administered every day for 21 days following cuff implantation.
Behavioral testing for pain
Mice were placed in clear Plexiglas boxes (7 cm × 9 cm × 7 cm) and were allowed 30 min for acclimatization. Also, mice were allowed an additional 15 min for exploration and grooming behaviors. The mechanical allodynia was performed using the von Frey hair test as detailed previously [32]. Briefly, calibrated von Frey filaments (Stoelting, Kiel, WI, USA) were applied to the plantar surface of each hindpaw in a series of ascending forces (range, 0.4-15 g). Rapid pulling back and shaking of the ipsilateral hind limb of the cuffing model was regarded as a positive sign of withdrawal response. Each filament was tested 5 times per paw, and the mechanical threshold was defined as 3 or more withdrawals observed out of the five trials. The thermal hyperalgesia was evaluated using the Hargreaves method [33]. The infrared beam of a radiant heat source (8 V, 50 W; Ugo Basile, Comerio, VA, Italy) was applied to the plantar surface of each hindpaw. The cut-off period was set at 15 s to prevent skin damage. Three measures of the paw withdrawal latency were taken in the ipsilateral hind limb of the cuffing model and averaged for each hindpaw. Behavioral exams were performed before cuffing or sham surgery and at days 2, 5, 7, 14, and 21 post-surgery.
Culture and treatment of primary DRG neurons
The neonatal mouse DRG neurons were isolated using the Felix method as described previously [34]. Briefly, 5-9 day old neonatal mice were decapitated using surgical scissors. Next, mice were placed in a prone position, and a longitudinal incision was made to expose the cervical, thoracic, and lumbar regions of the vertebral column. Another midline incision was made through the first vertebral arch and extended to expose the spinal cord. Next, the spinal cord was carefully pulled away and the DRG was removed from the vertebral column and placed into a 35-mm Petri dish containing 2 ml Advanced DMEM. Subsequently, DRG neurons were transferred into a dissociating medium solution and incubated for 40 min at 37°C with 5% CO2. Next, cells were dissociated using a fire-polished glass Pasteur pipette and centrifuged for 2 min at 300 r/min at room temperature. The supernatant was discarded and 1 ml fresh Advanced DMEM was added. Finally, the cell suspension was seeded on coverslips coated with poly-L-lysine and incubated for at least 3 h at 37°C with 5% CO2 in 1 ml Advanced DMEM. These culture conditions enabled the maintenance of neurons under optimal conditions for up to 7 d. By the end of the fourth day, we added different drug treatments to the medium, and the cells were incubated for an additional 24 h until the final collection followed by subsequent experiments.
Western blotting
The DRG neurons and primary cultured neurons were lysed in a lysis buffer supplemented with protease and phosphatase inhibitors on ice. Protein concentrations were estimated using the BCA method. Samples were heated in boiling water for 5 min, loaded onto 10% SDS polyacrylamide gels, and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The membranes were then blocked with a 5% BSA solution for 2 h and probed with the specific primary antibodies overnight at 4°C. Subsequently, the membranes were incubated with horseradish peroxidase (HRP)-conjugated antibody for 2 h and rinsed three times with Tris-buffered saline with Tween-20. Protein blot densities were analyzed using Labworks Software (Ultra-Violet Products, UK).
Immunostaining
Paraffin sections of DRGs were prepared a detailed previously [35]. Briefly, following anesthesia, mice were intracardially perfused with 4% paraformaldehyde for 10 min. The DRG neurons were quickly removed and fixed overnight at 4°C before being directly dehydrated in a graded ethanol series and processed for embedding in paraffin wax. Sections of 3-4 μm thickness were cut according to standard procedures, mounted on gelatin-coated slides, and finally air-dried. Before immunostaining, sections were placed in a bath solution of 3% H2O2 and 60% methanol PBS (pH 7.4) for 30 min and treated with 0.01 mol/L sodium citrate buffer using a pressure cooking method for antigen retrieval [36]. Sections were blocked with 10% donkey serum. For ABC-DAB staining, sections were incubated with the primary antibody at 4°C overnight. Subsequently, sections were incubated with biotinylated IgG diluted in PBS for 4 h followed by analysis with the avidin-biotin-peroxidase complex (ABC) Elite Kit (1:100; Vector, Burlingame, CA, USA) in 0.01 M PBS (pH=7.4) for 2 h. Finally, antibody immunoreactivity was visualized in 0.05M Tris-HCl buffer (pH=7.6) containing 0.04% DAB and 0.003% H2O2 followed by hematoxylin staining of nuclei. For immunofluorescence, sections were incubated with the primary antibody overnight at 4°C. Subsequently, sections were detected by the corresponding secondary antibody with DAPI nuclear staining. The specificities of the immunohistochemical and immunofluorescent staining were examined by omitting the specific primary antibodies.
Image analysis
For immunofluorescence staining, images were captured by a Leica SP5 confocal microscope (Leica) and recorded sequentially using Leica Application Suite Software (Leica). For DAB staining, images were obtained by a DMR-X microscope coupled to a DC500 digital camera (Leica, Wetzlar, Germany) and the image analysis system Quantimet Q550 (Leica). Nine randomly selected discontinuous fields (magnification, 20×) per sample were evaluated, and the integrated optical density of MT2 for each neuron was quantified using Image-Pro Plus software (Media Cybernetics, Baltimore, MD, USA). According to the MT2 immunostaining intensity, neurons were classified as either high MT2-expressing neurons (optical density >50%) or low MT2-expressing neurons (optical density <50%). Next, the average soma diameters for the high-expressing neuron were calculated by averaging the major diameter with the minor diameter; the major and minor diameters were the longest and shortest axes, respectively, through the nucleolus.
Real-time qPCR analysis
Total RNA from DRGs and the primary cultured neurons was extracted using Trizol reagent. RNA was transcribed to cDNA with PrimeScript RT reagent kit following the standard protocol. Then, real-time qPCR was performed using the CFX96TM real-time system (Bio-Rad) and the relative gene expression was normalized according to the internal control β-actin. The primer sequences for the SYBR Green probes for the target genes are presented in Table 1.
Microarray and computational analyses
DRG neurons were cultivated for 4 days and 1 mM Mel, 100 μM 8MP, or control vehicle were added to the culture medium for an additional 24 h. For microarray analysis, the Agilent Array platform was employed. Briefly, total RNA from each sample was isolated, amplified, and transcribed into fluorescent cRNA using Agilent's Quick Amp Labeling protocol (version 5.7). The arrays were scanned and analyzed by the Agilent Scanner G2505B and Agilent Feature Extraction software (version 10.5.1.1). Quantile normalization and subsequent data processing were performed using the GeneSpring-GXv11.5 software. The microarray data discussed in this manuscript were deposited in NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSE89289.
Differentially expressed genes (DEGs) in the 100 μM 8MP group (between 100 μM 8MP and control vehicle treatment) and the 1 mM Mel group (between 1 mM Mel and control vehicle treatment) with an up- or down-regulated fold change of ≥2.0 were identified through the random variance model among the different treatment groups. The Gene Ontology was analyzed for the main functions of the DEGs, and Fisher's exact test, χ2 test, and the false discovery rate were calculated to adjust the P-value [37-39]. The Gene Ontology terms were considered significant if the P-value and false discovery rates were <0.05. Also, the DEGs in pain-related Gene Ontology terms were identified including GO0048265 (in response to pain), GO0051930 (regulation of sensory perception of pain), GO0050966 (detection of mechanical stimulus involved in sensory perception of pain), GO0048266 (the behavioral response to pain), GO0019233 (the sensory perception of pain), GO0050968 (detection of chemical stimulus involved in sensory perception), and GO0050965 (detection of temperature stimulus involved in sensory perception). The KEGG database and Cytoscape were also used to build the network of genes according to the relationship among these pain-related DEGs [40-43].
Cell treatment and siRNA interference
The DRG neurons were cultivated for 4 days, and the culture medium was refreshed. The different treatments were directly added to the medium starting from the fourth day, and the cells were incubated for an extra 24 h. Cells were plated in 6-well plates at 80% confluence before transfection. Individual siRNAs (at 5 nM), Lipofectamine 3000, and Opti-MEM were mixed and incubated at room temperature for 20 min. The siRNA-lipofectamine complexes were added to the DRG neurons for 24 h, and the medium was replaced by fresh Advanced DMEM after transfection. Experiments were performed 48 h after transfection. The knockdown efficiency of siRNA was assessed by Western blotting (Suppl Figure S1).
ELISA
Following extraction and quantification of the DRG neurons protein lysates, the IL-1β and TNF-α concentrations were determined using the commercially available ELISA kits according to the manufacturer's protocols (Sangon Biotech, Shanghai, China). Each cytokine sample was analyzed in duplicate, and the mean cytokine concentration was calculated. IL-1β and TNF-α levels were expressed as pg/mg.
Statistical analysis
Data associated with both DRG neurons were normalized according to the left side of the Sham group for each experiment. There was no statistical difference between the right/left DRGs in the Sham group and the right DRG. Statistical analyses were performed by Student's t-test, one-way analysis of variance (ANOVA), or two-way repeated-measures ANOVA followed by Bonferroni's multiple comparison tests as appropriately indicated. All data are expressed as mean ± standard error of the mean (SEM). A P value <0.05 was considered to be statistically significant.
Results
Increased MT2 activation in DRG neurons of the cuff-implanted mice suppressed mechanical allodynia and thermal hyperalgesia
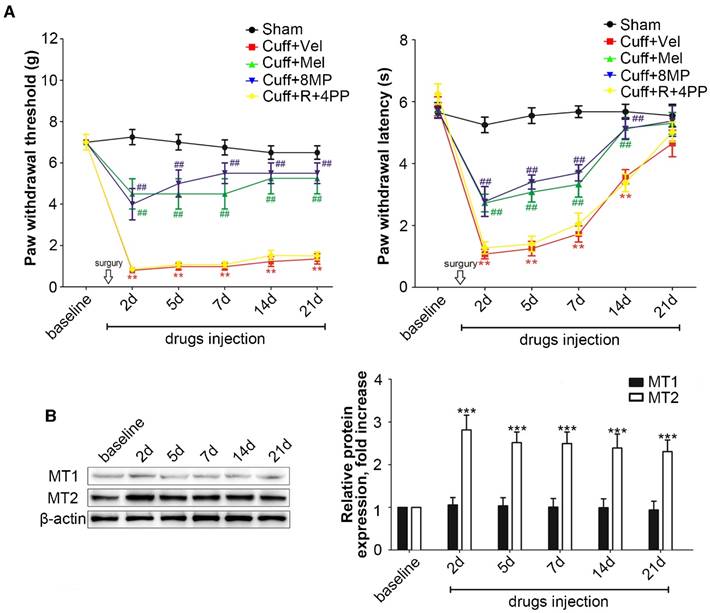
In the cuff group, unilateral cuff implantation caused a significant ipsilateral decrease in the latency for paw withdrawal at days 2, 5, 7, 14, and 21 post-surgery compared to that in mice in the Sham group (P<0.01, Fig. 1A). The exogenous i.p. administration of 100 mg/kg Mel or 50 mg/kg 8MP induced a moderate but significant attenuation of the thermal allodynia response as well as mechanical allodynia compared with that in the Sham group (P<0.01, Fig. 1A). The exogenous i.p. administration of 50 mg/kg ramelteon and 20 mg/kg 4PP did not affect the thermal or mechanical allodynia response induced by cuff implantation (Fig. 1A).
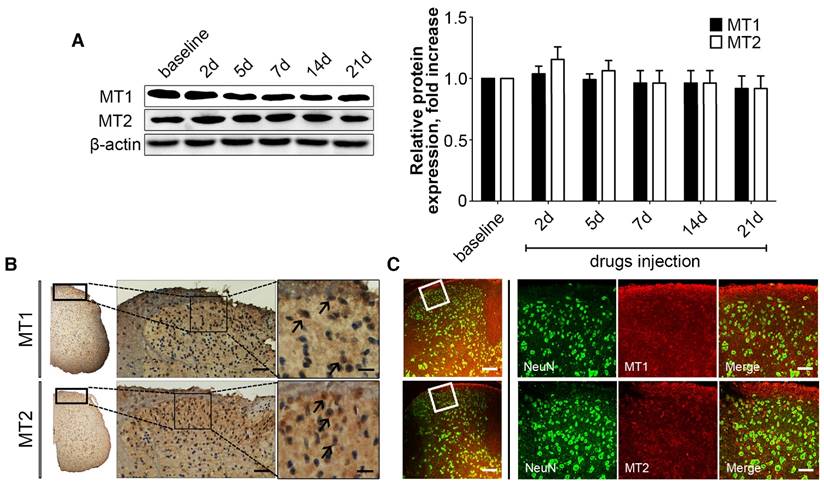
Western blotting revealed that the cuff implantation caused a significant increase in MT2 expression in the ipsilateral DRGs compared with levels in the Sham group at days 2, 5, 7, 14 and 21 post-surgery without altering the MT1 expression in ipsilateral DRGs (P<0.001, Fig. 2B).
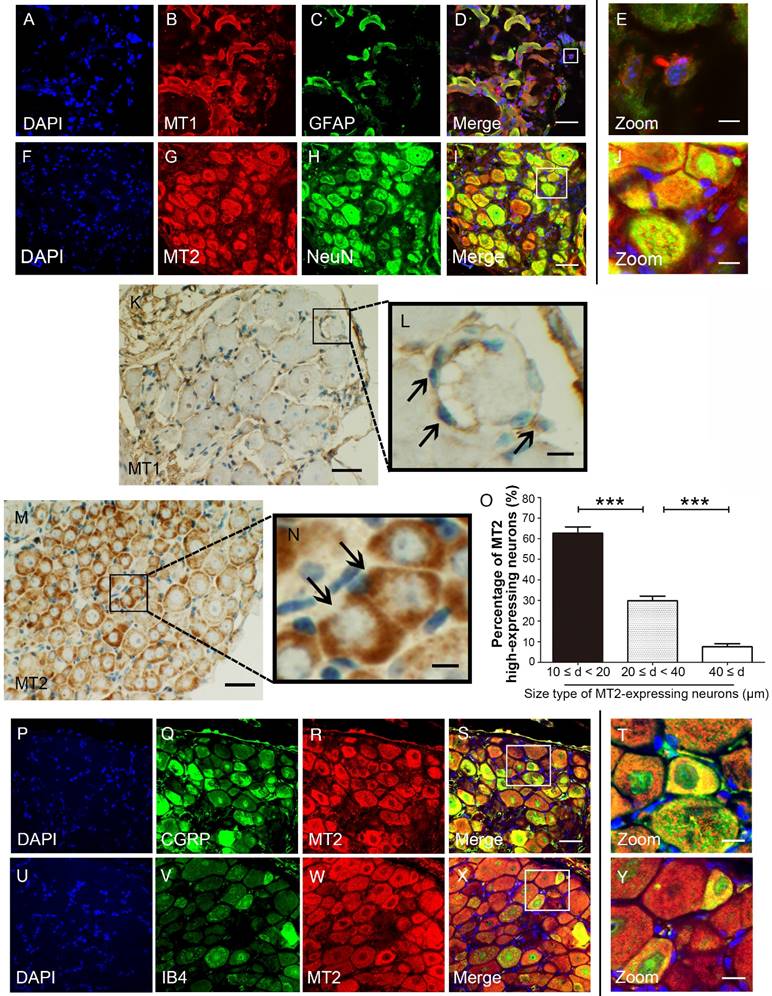
MT1 and MT2 exhibit different expression patterns in mouse DRG
We used immunohistochemical and immunofluorescence staining to examine the exact distribution of MT1 and MT2 receptors in control C57BL/6J mice. MT1 was localized in the DRG mainly with the glial population as verified by MT1 and GFAP double immunostaining (Fig. 2A-E). In contrast, MT2 was mainly expressed in neurons as verified by MT2 and NeuN double immunostaining (Fig. 2F-I). Further analysis revealed that MT1 receptor immune-reactive product was localized in the satellite cells of the DRG (Fig. 2K-L), while expression of MT2 was observed in the cytoplasm of DRG neurons (Fig. 2M). Measurement of soma diameters of the high MT2-expressing neurons demonstrated that 63.61±2.21% had a diameter of 10-20 μm, 29.14±3.11% had a diameter of 20-40 μm, and 7.25±1.25% had a diameter greater than 40 μm (Fig. 2N). Immunofluorescence staining also verified that all calcitonin gene-related peptide (CGRP)- and isolectinB4 (IB4)-positive cells expressed MT2 (Fig. 2O-X).
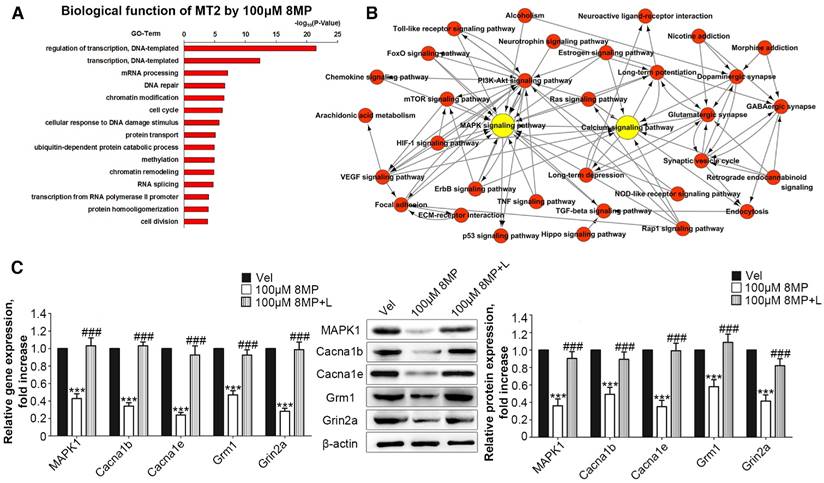
MT2 activation by 100 μM 8MP suppressed calcium channel activity via MAPK1 in DRG neurons
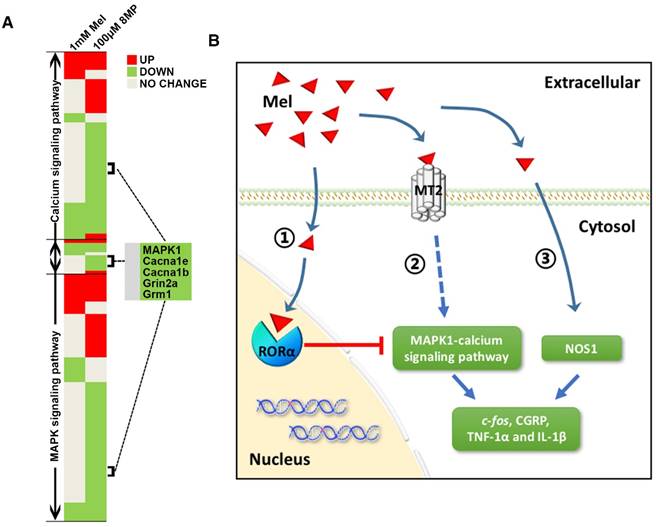
Microarray analysis was used to assess the biological functions of MT2 activation by 100 μM 8MP (Fig. 3A). Gene Ontology analysis revealed that the in the group treated with 100 μM 8MP, DEGs were mainly involved in the regulation of transcription and translation (Fig. 3A). A total of 39 pain-related DEGs were selected in the 100 μM 8MP group based on pain-related Gene Ontology terms. The pathway network analysis indicated that the MAPK and calcium signaling pathways were the chief players in the MT2-related pain signaling (Fig. 3B). Compared with the control group, DRG neurons treated with 100 μm 8MP for 24 h showed obvious down-regulation of the gene and protein expression of mitogen-activated protein kinase 1 (MAPK1), voltage-dependent N-type calcium channel subunit alpha-1B (Cacna1b), voltage-dependent R-type calcium channel subunit alpha-1E (Cacna1e), metabotropic glutamate receptor1 (Grm1), and the glutamate receptor ionotropic, NMDA2A (Grin2a; Fig. 3C). Additionally, incubation with 100 μm luzindole (MT1 and MT2 antagonist) reversed the inhibitory effects of 100 μm 8MP (P<0.001, Fig. 3C).
Activation of increased MT2 in the DRG suppressed mechanical allodynia and thermal hyperalgesia in cuff-implanted mice. (A) Sham group included the sham-operated mice without cuff implantation. The mechanical and thermal latency of different time points were measured in the Sham group and cuff-implanted mice with different treatments (the ipsilateral left paws): the Cuff+Vel group (treated with i.p. injection of vehicle), Cuff+Mel group (treated with i.p. injection of 100 mg/kg Mel), Cuff+8MP group (treated with i.p. injection of vehicle), and Cuff+R+4PP group (treated with i.p. injection of 50 mg/kg ramelteon and 20 mg/kg 4PP), **P<0.01 vs. the Sham group; ##P<0.01 vs. Cuff group, n=8 for each group; (B) Representative immunoblots of MT1/MT2 in the DRG of mice subjected to cuff implantation with β-actin as an internal standard. The expression of MT2 was up-regulated by cuff implantation (***P<0.001 vs. baseline); n=8 for all groups.
MT1 and MT2 had different expression patterns in the DRG in control C57BL/6J mice. (A-E) Immunofluorescence staining of MT1 and GFAP in the DRG in control C57BL/6J mice: Panel D is the merged image of panel A, B, and C. Panel E shows a magnified section of the panel D (bar=30 μm in A-D; bar=9 μm in E); (F-J) Immunofluorescence staining of MT2 and NeuN in the DRG in control C57BL/6J mice: Panel I is the merged image of panel F, G and H. Panel J is a magnified section of the panel I (bar=30 μm in F-I; bar=9 μm in J); (K-N) DAB staining of MT1 and MT2 in the DRG in control C57BL/6J mice: Panels L and N show magnified sections of panel K and M, respectively. The arrows in L indicate the MT1-immunoreactive satellite cells. The arrows in N represent the MT2-immunoreactive neurons. (bar=80 μm in K and M; bar=18 μm in L and N.); (O) Bar chart representing the soma diameters of the high MT2-expressing neurons (***P<0.001; n=8 for each group); (P-Y) Double-label staining of MT2 and CGRP/IB4. Panels T and Y are magnified sections of merged panels S and X, respectively (bar=30 μm in P-S and U-X; bar=9 μm in T and Y).
MT2 activation by 100 μM 8MP suppressed calcium channel signaling via MAPK1 in DRG neurons (Part I). (A) The biological process of Gene Ontology analysis of MT activation by 100 μM 8MP in primary DRG neurons; (B) The pathway network analysis of 39 pain-related DEGs in the 100 μM 8MP group; (C) Real-time PCR and Western blotting results obtained from different groups with β-actin as an internal standard. Bar chart represents the fold increases in expression of different genes and proteins isolated from primary cultured DRG neurons following 24-h incubation with different treatments: Vel group (treated with vehicle), 100 μM 8MP group (treated with 100 μM 8MP) and 100 μM 8MP+L group (treated with 100 μM 8MP and 100 µM luzindole). (***P<0.001 vs. Vel; ###P<0.001 vs. 100 μM 8MP; n=8 for each group).
We examined the interaction between the MAPK1 and calcium signaling pathways. We treated the DRG neurons with siRNA for MAPK1, Cacna1b, Cacna1e, Grm1, or Grin2a, which significantly down-regulated the expression of each target protein (Suppl Fig. S1). Suppression of MAPK1 expression inhibited the gene and protein expression of Cacna1b, Cacna1e, Grm1, and Grm1 (P<0.001, Fig. 4A). However, MAPK1 expression was not affected by knock down of any of the four genes (Fig. 4B).
MT2 activation by low-dose Mel suppressed calcium channel and MAPK1 signaling in DRG neurons
Next, we added low-dose Mel (100 μm) to activate MT2 in the primary DRG neurons. Real-time PCR and Western blotting demonstrated that 100 μm Mel suppressed the expression of MAPK1, Cacna1b, Cacna1e, Grm1, and Grin2a at the protein and RNA levels (P<0.001, Fig. 5A), and this down-regulation was blocked by 100 μm luzindole (P<0.001, Fig. 5A). However, there were no significant differences in the expression levels of any of the genes and their respective proteins upon addition of high dose (1 mM) Mel compared with the levels in the vehicle-treated neurons (Fig. 5B).
Activation of RORα receptor by Mel inhibited down-regulation of MAPK1
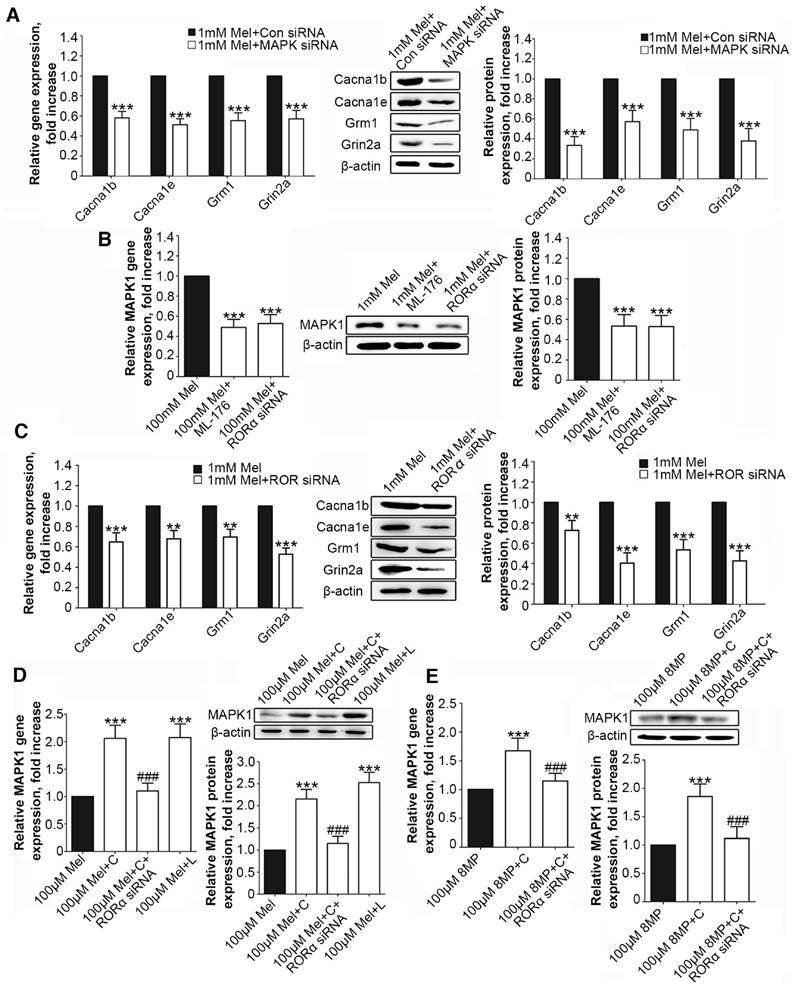
Following MAPK1 knockdown, the treatment of DRG neurons with 1 mM Mel induced significant decreases in the gene and protein expression of Cacna1b, Cacna1e, Grm1, and Grin2a (P<0.001, Fig. 6A). We assessed the effects of RORα receptor on MAPK1 expression in DRG neurons. Compared with 1 mM Mel alone, the combination of 1 mM Mel with 200 µM ML-176 (RORα receptor inverse agonist) induced decreases in MAPK1 gene and protein expression (P<0.001, Fig. 6B). The knockdown of RORα receptor induced a significant decrease in the expression of RORα receptor (Suppl Fig. S1F). Following knock down of RORα receptor, 1 mM Mel could also induce down-regulation of MAPK1 gene and protein expression (P<0.001, Fig. 6B). The combination of 1 mM Mel with RORα siRNA induced significant decreases in the gene and protein expression of Cacna1b, Cacna1e, Grm1, and Grin2a (P<0.001, Fig. 6C). However, addition of 200 µM CGP52608 (RORα receptor agonist) significantly increased the expression of MAPK1 in the primary DRG neurons treated with 100 μM Mel or 100 μM 8MP (P<0.001, Fig. 6D and E, respectively), which could be reversed by knock down of RORα receptor (P<0.001, Fig. 6D and E, respectively). The application of the MT2 antagonist luzindole (200 µM) promoted the expression of MAPK1 in 100 μM Mel-treated DRG neurons (P<0.001, Fig. 6D).
MT2 activation by 100 μM 8MP suppressed calcium channel signaling via MAPK1 in DRG neurons (Part II). Real-time PCR and Western blotting from different groups with β-actin as an internal standard: Primary cultured DRG neurons were subjected to different treatments as follows: (A) Con siRNA group (treated with the control siRNA) and MAPK1 siRNA (treated with the MAPK1 siRNA), ***P<0.001 vs. Con siRNA group; (B) Con siRNA group (treated with the control siRNA), Con Cacna1b group (treated with the Cacna1b siRNA), Cacna1e siRNA group (treated with the Cacna1e siRNA), Grm1siRNA group (treated with the Grm1siRNA) and Grin2a siRNA group (treated with the Grin2a siRNA); n=8 in all groups.
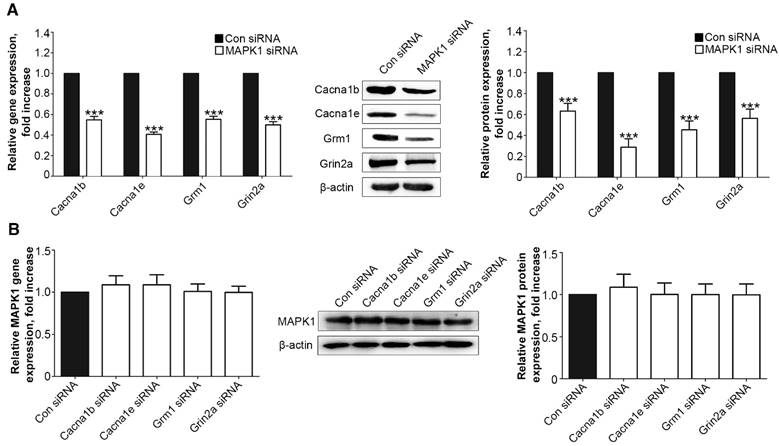
Low-dose Mel suppressed calcium channel and MAPK1 expression in DRG neurons. Real-time PCR and Western blotting from different groups with β-actin as an internal standard: Primary cultured DRG neurons were subjected to different treatments as follows: (A) Vel group (treated with vehicle), 100 μM Mel group (treated with 100 μM Mel) and 100 μM Mel+L group (treated with 100 μM Mel and 100 µM luzindole), ***P<0.001 vs. Vel group, ###P<0.001 vs. 100 µM Mel; (B) Vel group (treated with vehicle) and 1 mM Mel group (treated with 1 mM Mel), ***P<0.001 vs. Vel group; n=8 in all groups
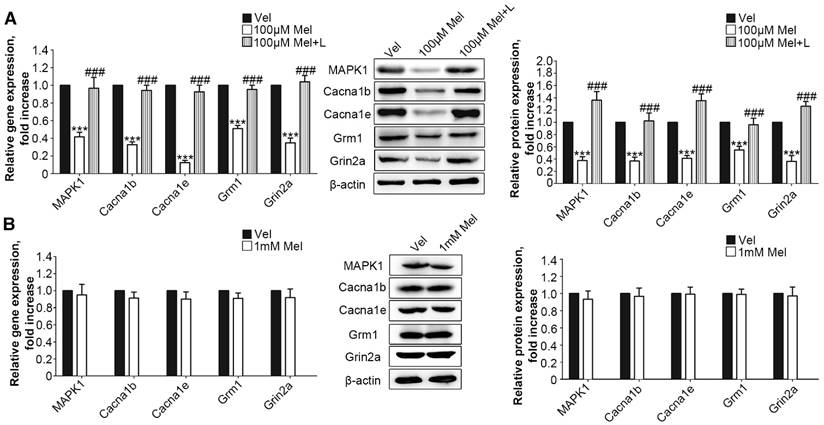
RORα activation by Mel inhibited MAPK1 and calcium channel down-regulation via MT2. Real-time PCR and Western blotting from different groups with β-actin as an internal standard: Primary cultured DRG neurons were subjected to different treatments as follows: (A) 1 mM Mel+Con siRNA group (treated with 1 mM Mel and control siRNA), 1 mM Mel+MAPK1 siRNA group (treated with 1 mM Mel and MAPK1 siRNA), ***P<0.001 vs. 1 mM Mel+Con siRNA; (B) and (C) 1 mM Mel group (treated with 1 mM Mel), 1 mM Mel+200 µM ML-176 group (treated with 1 mM Mel and 200 µM ML-176), 1 mM Mel+RORα siRNA group (treated with 1 mM Mel and RORα siRNA), **P<0.01 vs. 1 mM Mel ***P<0.001 vs. 1 mM Mel; (D) 100 µM Mel group (treated with 100 µM Mel), 100 µM Mel+C group (treated with 100 µM Mel and 200 µM CGP52608), 100 µM Mel+C+RORα siRNA group (treated with 100 µM Mel, 200 µM CGP52608 and RORα siRNA), 100 µM Mel+L group (treated with 100 µM Mel and 200 µM luzindole), ***P<0.001 vs. 100 µM Mel, ###P<0.001 vs. 100 µM Mel+C; (E) 100 µM 8MP group (treated with 100 µM 8MP), 100 µM 8MP+C group (treated with 100 µM 8MP and 200 µM CGP52608), 100 µM 8MP+C+RORα siRNA group (treated with 100 µM 8MP, 200 µM CGP52608 and RORα siRNA), ***P<0.001 vs. 100 µM Mel, ###P<0.001 vs. 100 µM Mel+C; n=8 for all groups.
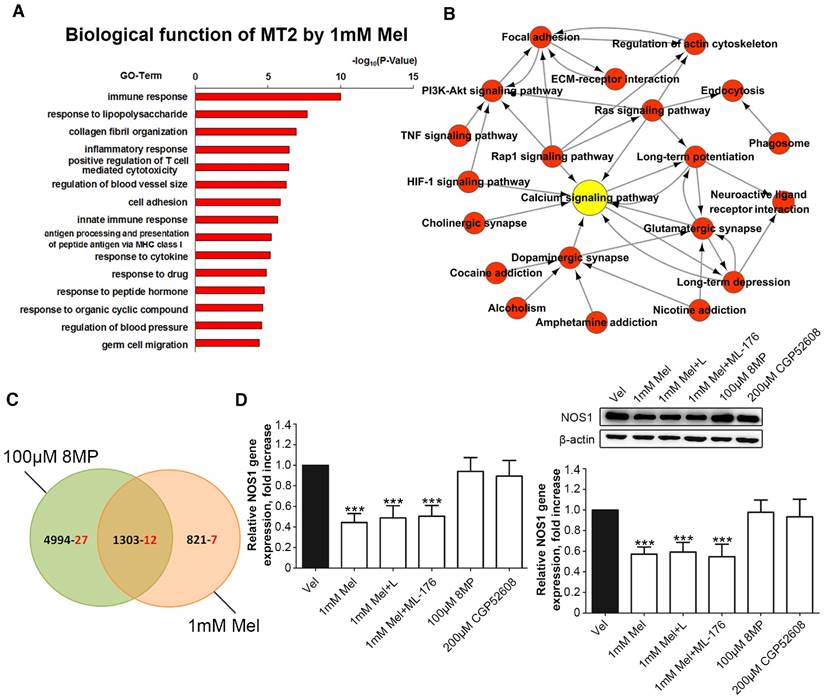
Regulation of NOS1 signaling in the DRG neurons was independent from the MT2 receptor and RORα nuclear receptor. (A) Gene Ontology analysis of MT activation by 1 mM Mel in primary DRG neurons; (B) Pathway network analysis of 19 pain-related DEGs in the 1 mM Mel group; (C) The Venn diagram analysis of the 46 pain-related DEGs in the 1 mM Mel group and 100 μM 8MP group (the black number represents the overall DEGs and the red number represents the pain-related DEGs); (D) Real-time PCR and Western blotting of NOS1 under different treatments in DRG neurons. The primary cultured DRG neurons were treated with different reagents as follows: Vel group (treated with vehicle), 1 mM Mel group (treated with 1 mM Mel), 1 mM Mel+L group (treated with 1 mM Mel and 200 µM luzindole), 100 μM Mel+ML-176 group (treated with 100 μM Mel and 200 µM ML-176), 100 μM 8MP group (treated with 100 μM 8MP), and 200 μM CGP52608 group (treated with 200 µM CGP52608); ***P<0.001 vs. Vel; n=8 for each group.
Regulation of NOS1 signaling by Mel in DRG neurons was independent of MT2 receptor and RORα receptor
Microarray analysis was used in primary cultured DRG neurons treated with 1 mM Mel followed by Gene Ontology annotations of DEGs revealed the enrichment of genes in several pathways, including the immune response, the response to lipopolysaccharide, collagen fibril organization, and the inflammatory response (Fig. 7A). Nineteen pain-related DEGs in the 1 mM Mel group were used to analyze the pathway network. The analysis revealed that the calcium signaling pathway accounted for most of the DEGs involved in the Mel-related pain signaling including P2X purinoceptor 7 (P2rx7), nitric oxide synthase, brain (NOS1), endothelin B receptor (Ednrb), and beta-2 adrenoceptor (Adrb2) (Fig. 7B). We subsequently compared the pain-related DEGs between the 1 mM Mel group and 100 μm 8MP group and observed the existence of 12 common DEGs between the two groups in the Venn diagram (Fig. 7C). The upward/downward trends of the 12 common genes in the 1 mM Mel and 100 μm 8MP group were similar (Table 2). Further examination revealed that NOS1 was the unique gene in the calcium signaling pathway in the 1 mM Mel group.
Trend of alteration in pain related DEGs in the 1mM MT group and 100μM 8MP group
| Gene IDa | Overlap Infob | Style in MT groupc | Style in 8MP groupd | Description e | KeggIDf |
|---|---|---|---|---|---|
| Kcnj10 | MT group | up | MCG4485, isoform CRA_a | mmu:16513 | |
| Gch1 | MT group | up | GTP cyclohydrolase 1 | mmu:14528 | |
| Chrna4 | MT group | down | Neuronal acetylcholine receptor subunit alpha-4 | mmu:11438 | |
| Itga2 | MT group | up | Integrin alpha-2 | mmu:16398 | |
| Cck | MT group | down | Cholecystokinin | mmu:12424 | |
| Nos1 | MT group | down | Nitric oxide synthase, brain | mmu:18125 | |
| Tac1 | MT group | down | Protachykinin-1 | mmu:21333 | |
| Oprd1 | overlap | down | down | Opioid receptor, delta 1 | mmu:18386 |
| Edn1 | overlap | up | up | Endothelin-1 | mmu:13614 |
| Grik1 | overlap | down | down | Glutamate receptor ionotropic, kainate 1 | mmu:14805 |
| Adrb2 | overlap | up | up | Beta-2 adrenoceptor | mmu:11555 |
| Grin2b | overlap | down | down | Glutamate receptor ionotropic, NMDA 2B | mmu:14812 |
| P2rx7 | overlap | up | up | P2X purinoceptor 7 | mmu:18439 |
| Grm3 | overlap | down | down | Grm3 protein | mmu:108069 |
| Ednrb | overlap | up | up | Endothelin B receptor | mmu:13618 |
| Gja4 | overlap | down | down | Gap junction alpha-4 protein | mmu:14612 |
| Crh | overlap | down | down | Corticoliberin | mmu:12918 |
| Tspo | overlap | up | up | Translocator protein | mmu:12257 |
| Vip | overlap | down | down | VIP peptides | mmu:22353 |
| Calca | 8MP group | down | Calcitonin | mmu:12310 | |
| Nlgn2 | 8MP group | down | Neuroligin-2 | mmu:216856 | |
| Cacna1b | 8MP group | down | Voltage-dependent N-type calcium channel subunit alpha-1B | mmu:12287 | |
| Oprl1 | 8MP group | down | Nociceptin receptor | mmu:18389 | |
| Adrb1 | 8MP group | down | Beta-1 adrenergic receptor | mmu:11554 | |
| Oxt | 8MP group | up | Oxytocin-neurophysin 1 | mmu:18429 | |
| Cacna1e | 8MP group | down | Voltage-dependent R-type calcium channel subunit alpha-1E | mmu:12290 | |
| Thbs1 | 8MP group | up | Thrombospondin-1 | mmu:21825 | |
| Thbs4 | 8MP group | up | Thrombospondin 4 | mmu:21828 | |
| Grm1 | 8MP group | down | Metabotropic glutamate receptor 1 | mmu:14816 | |
| Rtn4 | 8MP group | down | Reticulon-4 | mmu:68585 | |
| Crhr2 | 8MP group | up | Corticotropin releasing factor receptor type 2 beta | mmu:12922 | |
| Grin2a | 8MP group | down | Glutamate receptor ionotropic, NMDA 2A | mmu:14811 | |
| Uchl1 | 8MP group | down | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | mmu:22223 | |
| Reln | 8MP group | down | Reelin splicing isoform MR-1A | mmu:19699 | |
| P2ry1 | 8MP group | down | Purinergic receptor P2Y, G-protein coupled 1 | mmu:18441 | |
| Mapk1 | 8MP group | down | Mitogen-activated protein kinase 1 | mmu:26413 | |
| Fyn | 8MP group | down | Tyrosine-protein kinase Fyn | mmu:14360 | |
| Trpa1 | 8MP group | down | Transient receptor potential cation channel subfamily A member 1 | mmu:277328 | |
| Ephx2 | 8MP group | up | Epoxide hydrolase 2C | mmu:13850 | |
| Npy1r | 8MP group | down | Neuropeptide Y receptor type 1 | mmu:18166 | |
| Adra2c | 8MP group | down | Alpha-2C adrenergic receptor | mmu:11553 | |
| Nr2f6 | 8MP group | down | Nuclear receptor subfamily 2 group F member 6 | mmu:13864 | |
| Scn10a | 8MP group | up | Sodium channel protein type 10 subunit alpha | mmu:20264 | |
| Dlg2 | 8MP group | up | Disks large homolog 2 | mmu:23859 | |
| P2rx3 | 8MP group | down | P2X purinoceptor 3 | mmu:228139 | |
| Hoxb8 | 8MP group | down | Homeo box B8 | mmu:15416 |
a: GeneID refers to the specific abbreviation of genes names in the GO database.
b: Overlap Info refers to the location of these pain related genes in the Venn analysis of the Figure 4. The MT group referred to genes expressed only in the 1mM MT group while the 8MP group referred to genes expressed only in the 100μM 8MP group. The overlap refers to the genes expressed in the both 1mM MT group and 100μM 8MP group
c: Style in MT group refers to alteration trend of pain related DEGs in the 1mM MT group.
d: Style in 8MP group refers to alteration trends of pain related DEGs in the 100μM 8MP group.
e: Description refers to the full name of gene of corresponding GeneID.
f: KeggID refers to serial number of specific genes in the KEGG database.
Real-time PCR and Western blotting revealed that 1 mM Mel induced a decrease in NOS1 gene and protein expression, while there was no significant change with 100 μm 8MP (P<0.001, Fig. 7D). Also, addition of 100 µM luzindole or 200 µM ML-176 did not significantly influence the NOS1 expression compared with 1 mM Mel. Similarly, 200 µM CGP52608 also did not induce down-regulation of NOS1 (P<0.001, Fig. 7D).
Expression of c-fos, CGRP, and neuro-inflammatory cytokines could be modulated after MT2 activation by Mel or 8MP in the DRG neurons of the cuff-implanted mice
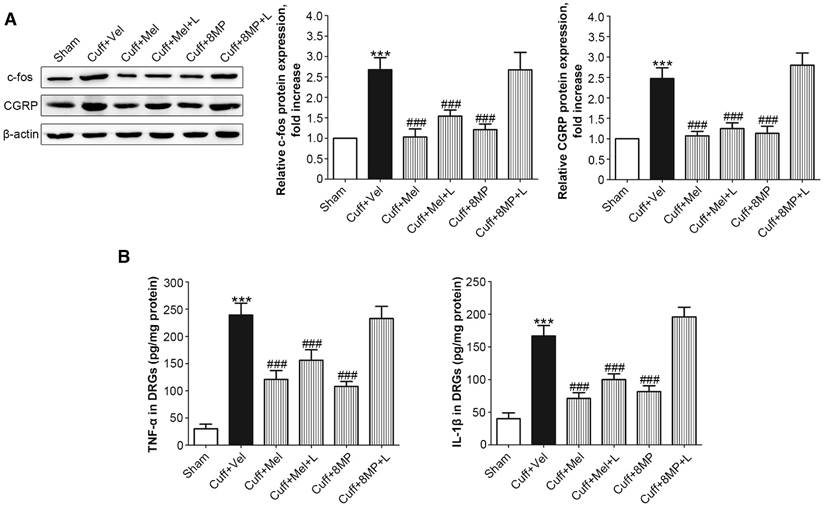
Following the unilateral cuff implantation, we harvested DRG neurons from mice 2 days post-surgery. Unilateral cuff implantation induced an ipsilateral increase in the expression of c-fos, which could be blocked by injection of 100 mg/kg Mel or 50 mg/kg 8MP (P<0.001, Fig. 8A). The addition of 20 mg/kg luzindole did not block the inhibitory effects of 100 mg/kg Mel on c-fos expression, while addition of 20 mg/kg luzindole suppressed the decrease in c-fos expression induced by 50 mg/kg 8MP (P<0.001, Fig. 8A). Furthermore, following the unilateral cuff implantation, the up-regulated ipsilateral expression of CGRP was attenuated by exogenous i.p. injection of 100 mg/kg Mel and 50 mg/kg 8MP. In the Cuff+Mel+L group, CGRP expression did not significantly differ from that in the 100 mg/kg Mel treatment. In contrast, we observed a significant increase in CGRP expression upon injecting 20 mg/kg luzindole and 50 mg/kg 8MP in mice (P<0.001, Fig. 8A). Next, we investigated the neuro-inflammatory response by examining the expression of TNF-α and IL-1β. Results of ELISA assays demonstrated that both TNF-α and IL-1β were increased in the ipsilateral side of cuff implantation in the Cuff+Vel group. The injection of 100 mg/kg Mel or 50 mg/kg 8MP suppressed the expression of TNF-α and IL-1β compared to the Cuff+Vel control group (P<0.001, Fig. 8B). The injection of 20 mg/kg luzindole along with 8MP significantly increased the expression of TNF-α and IL-1β in the 50 mg/kg 8MP group (P<0.001, Fig. 8B). There were no significant differences in the expression of c-fos, CGRP, and cytokines between the Cuff+Vel and Cuff+8MP+L groups.
Real-time PCR and Western blotting revealed that 1 mM Mel induced a decrease in NOS1 gene and protein expression, while there was no significant change with 100 μm 8MP (P<0.001, Fig. 7D). Also, addition of 100 µM luzindole or 200 µM ML-176 did not significantly influence the NOS1 expression compared with 1 mM Mel. Similarly, 200 µM CGP52608 also did not induce down-regulation of NOS1 (P<0.001, Fig. 7D).
Expression of c-fos, CGRP, and neuro-inflammatory cytokines could be modulated after MT2 activation by Mel or 8MP in the DRG neurons of the cuff-implanted mice
Following the unilateral cuff implantation, we harvested DRG neurons from mice 2 days post-surgery. Unilateral cuff implantation induced an ipsilateral increase in the expression of c-fos, which could be blocked by injection of 100 mg/kg Mel or 50 mg/kg 8MP (P<0.001, Fig. 8A). The addition of 20 mg/kg luzindole did not block the inhibitory effects of 100 mg/kg Mel on c-fos expression, while addition of 20 mg/kg luzindole suppressed the decrease in c-fos expression induced by 50 mg/kg 8MP (P<0.001, Fig. 8A). Furthermore, following the unilateral cuff implantation, the up-regulated ipsilateral expression of CGRP was attenuated by exogenous i.p. injection of 100 mg/kg Mel and 50 mg/kg 8MP. In the Cuff+Mel+L group, CGRP expression did not significantly differ from that in the 100 mg/kg Mel treatment. In contrast, we observed a significant increase in CGRP expression upon injecting 20 mg/kg luzindole and 50 mg/kg 8MP in mice (P<0.001, Fig. 8A). Next, we investigated the neuro-inflammatory response by examining the expression of TNF-α and IL-1β. Results of ELISA assays demonstrated that both TNF-α and IL-1β were increased in the ipsilateral side of cuff implantation in the Cuff+Vel group. The injection of 100 mg/kg Mel or 50 mg/kg 8MP suppressed the expression of TNF-α and IL-1β compared to the Cuff+Vel control group (P<0.001, Fig. 8B). The injection of 20 mg/kg luzindole along with 8MP significantly increased the expression of TNF-α and IL-1β in the 50 mg/kg 8MP group (P<0.001, Fig. 8B). There were no significant differences in the expression of c-fos, CGRP, and cytokines between the Cuff+Vel and Cuff+8MP+L groups.
Discussion
NP is an umbrella term that encompasses different conditions caused by a lesion or disease of the nervous system that signals somatosensory information [44]. NP usually presents with several clinical features, including pain in an area with partial or complete sensory loss; different types of evoked pain; specific complaints as burning pain; increased pain after repetitive stimulation; and persisting pain after stimulation [44, 45]. Allodynia (i.e., pain elicited by a stimulus that normally does not cause pain) and hyperalgesia (i.e., an increased pain response produced by a stimulus that normally causes pain) are two prominent symptoms of NP [46, 47]. Allodynia and hyperalgesia are classified according to the sensory modality used to elicit pain—i.e., mechanical (dynamic, punctate, and static) or thermal (cold and heat) stimuli, with their separate mechanisms [46, 47]. Mechanical allodynia is believed to be mediated by low-threshold Aβ fibers, whereas thermal stimuli are conducted via C fibers and Aδ fibers [46, 47]. The molecular mechanisms of mechanical allodynia and thermal hyperalgesia are also distinct [46, 47]. Assessments of mechanical allodynia and thermal hyperalgesia are generally carried out with the use of monofilaments and thermotest equipment, and they are widely applied in fundamental pain research [48, 49].
Mel is called a hormone of night as 80% is secreted by the pineal body with a significant circadian rhythm (low level of secretion during the day and high levels during the night) [23, 26]. The secretion of Mel was variable in different types of chronic pain that induced sleep deprivation and mood disorder [26]. Srinivasan et al. observed a decrease in nighttime Mel secretion in cluster headache patients compared with healthy controls as well as a lower Mel level during the cluster headache attacks compared with the remission period [26]. To this end, here, we investigated the impact of Mel administration on pain behavior in a cuff implantation model.
In our present study, both mechanical allodynia and heat hyperalgesia were relieved with exogenous Mel application in the DRG-friendly cuff models. In agreement with previous results, we observed that the activation of MT2 by 8MP suppressed the cuff implantation-induced pain behaviors similar to Mel, while MT1 activation, using a combination of ramelteon (MT1 and MT2 agonist) and 4PP (MT2 antagonist), did not produce a pronounced impact on pain behavior [26]. Furthermore, it has been suggested that the expression of Mel receptors can respond to pain stimuli in the nervous system [50, 51]. Western blotting demonstrated that only MT2 was up-regulated in the DRGs while the levels of MT2 in the spinal cord and MT1 expression in the DRGs and spinal cord remained similar (Figs. 1B and 9A). More importantly, Mel and MT1/2 exert anti-nociceptive actions by acting at both spinal cord and supraspinal levels [26]. To further understand the role of MT1/2 in the pain signal transduction mechanism, we performed immunostaining in both spinal cord and DRGs samples. We observed that both MT1 and MT2 were widely expressed in the neurons of the anterior angle and dorsal horn, whereas the expression of MT1 and MT2 presented completely different patterns in DRG (Fig. 9B-C). MT2 was more prominently expressed in small and median peptidergic/non-peptidergic (CGRP/IB4 immuno-reactive) neurons (Fig. 9B), which are closely associated with pain transmission [35]. Taken together, these findings indicate is plausible to speculate that MT2 of DRG neurons are critical in mediating the sensory component of NP.
Impact of MT and 8MP treatment on expression of c-fos, CGRP, and neuro-inflammatory cytokines in the DRG of the cuff-implanted mice. (A) Western blotting for c-fos and CGRP obtained from different groups with β-actin as an internal standard: Sham group (sham-operated mice), Cuff+Vel group (i.p. injection of vehicle), Cuff+Mel (i.p. injection of 100mg/kg Mel), Cuff+Mel+L group (i.p. injection of 100 mg/kg Mel and 20 mg/kg luzindole), Cuff+8MP group (i.p. injection of 100mg/kg 8MP) and Cuff+8MP+L group (i.p. injection of 00 mg/kg 8MP and 20 mg/kg luzindole), ***P<0.001 vs. Sham; ###P<0.001 vs. Cuff+Vel; n=8 for each group. (B) Analysis of the inflammatory cytokines TNF-α and IL-1β by ELISA in different DRG treatment groups as above (***P<0.001 vs. Sham; ###P<0.001 vs. Cuff+Vel; n=8 for each group).
MT1 and MT2 were expressed in the neurons of spinal cord. (A) Representative immunoblots of MT1/MT2 in the spinal cord of mice subjected to cuff implantation with β-actin as an internal standard. The baseline represented the 2 days before the implantation. Expression of MT1 and MT2 in the spinal cord was not affected after cuff implantation. (***P<0.001 vs. baseline; n=8 for each group). (B) DAB staining of MT1 and MT2 in the spinal cord in control C57BL/6J mice. Both MT1 and MT2 were expressed in the neurons of spinal cord. (bar=60 μm in the middle panel; bar=18 μm in the right panel). (C) Immunofluorescence staining of MT1/2 and NeuN in the spinal cord of normal C57BL/6J mice. The right panel is a magnified section of pictures in the left panel. (bar=30 μm in the left panel; bar=9 μm in the right panel).
Microarray analysis indicated that both calcium and MAPK signaling pathways were largely inhibited in the primary DRG neurons treated with 100 μM 8MP. Although the action of Mel on the intracellular free Ca2+ concentration varies in different tissues or organs [52], previous studies demonstrated that several types of calcium channels are involved in the inhibition of intracellular Ca2+ by Mel in DRG neurons, including TRPM2, TRPV1, and high-voltage activated calcium channels [20-22, 53]. Similar to Mel treatment, our present work also verified that MT2 activation by 100 μM 8MP also suppressed a series of calcium channels, including Cacna1b, Cacna1e, Grm1, and Grin2a. It is noteworthy that activation of a single G-protein coupled receptor (MT2) could modulate these calcium channels.
MAPKs are important mitogen activated protein kinases that often integrate signals from G protein-coupled receptors [54]. In our transcriptome microarray, not only MAPK1 but also MAPK3\5\7\9\10\11 were inhibited by MT2 activation by 100 μM 8MP. However, only knockdown of MAPK1 could significantly inhibit a series of calcium channels (data not shown). MAPK1 and MAPK2 are often phosphorylated (also referred as pERK2 and pERK1) following noxious stimulation or inflammation in primary afferents, dorsal horn, and brain regions involved in pain processing [55-57]. However, recent studies indicated an isoform-specific role for a MAPK1 prototype in pain processing in both the CNS and PNS [58]. Conditional deletion of MAPK1 in CNS neurons attenuated the development of inflammatory and neuropathic pain [59], and peripheral sensory neuron MAPK1 was necessary for the development of inflammation-induced hypersensitivity and cold sensation [58]. [60]. To the best of our knowledge, this is the first report that demonstrates that MAPK1 expression is regulated by a neurohormone - Mel. Also, we provided a connection between MAPK1 and calcium channels, which will help promote research into the role of MAPK1 in pain sensation. However, the exact mechanism remains to be elucidated in future studies.
Due to the lack of systematic theoretical research on Mel regulation in DRG neurons, we applied a relatively high dose of Mel in order to fully activate MT2 in the DRG neurons. There was a limited number of down-regulated genes in the microarray analysis of the primary DRG neurons group subjected to 1 mM Mel. A total of 6300 and 2124 probe sets representing 6342 and 2148 DEGs were detected in the 100 μM 8MP group and the 1 mM Mel group, respectively. The high dose of Mel (1 mM) was not as effective at suppressing MAPK1 and calcium channel expression as 8MP (100 μM; Fig. 10A). Furthermore, the inhibitory actions of 100 μM Mel and blockade by the MT2 antagonist luzindole suggested a negative regulation with the high dose Mel. Interestingly, when RORα receptor was blocked, the high dose Mel (1 mM) could suppress MAPK1 expression as well as calcium channel expression. Activation of RORα receptor by CGP53608 reversed the down-regulation of MAPK1 in the 100 μM 8MP and 100 μM Mel groups. This effect was inhibited by knocking down the RORα receptor via siRNA. Therefore, RORα receptor activation could reverse the MT2-induced MAPK1 decrease, suggesting the presence of a negative feedback between the membrane receptors and nuclear receptors of Mel in the DRG neurons (Fig. 10B).
Although several types of NOSs were demonstrated in the Mel signaling, the regulation by Mel of NOSs depends on cell type and tissue origin. For example, Mel inhibited iNOS in the heart tissue, while it could promote eNOS in the aorta of rats with metabolic syndrome [61]. Regulation of NOS1 (also named nNOS) by Mel appears to be context dependent. For example, Mel enhances NOS1 expression in the HaCaT cells and heart [62], while it attenuates NOS1 expression in the nodose ganglion of acute hypoxic rats [63]. Therefore, the impact of Mel on the NOSs needs further verification. In our present study, although there was a negative feedback in the high dose Mel (1 mM) group, the microarray analysis identified NOS1 as a unique gene specific to the Mel group. Previous studies demonstrated that NOS1 expression was mainly up-regulated in small- and medium-sized DRG sensory neurons ipsilateral to the injury side starting from day 1 post-lumbar spinal nerve ligation [64, 65]. Furthermore, NOS1 inhibitors could significantly reduce mechanical allodynia and thermal hyperalgesia in chronic constriction injury in rats [66]. Therefore, the decrease in NOS1 expression can be correlated with the Mel-mediated analgesic effect in DRGs. Moreover, NOS expression was not affected by specific drugs for MT2 or RORα receptor, indicating that Mel decreased NOS1 via an MT2-independent pathway in DRG neurons (Fig. 10B).
Graphical representation for Mel regulation in DRG neurons. (A) Overview of altered trends in all DEGs in the MAPK and calcium signaling pathways in the 1 mM Mel group and 100 μM 8MP group on microarray. There were more down-regulated genes in the MAPK and calcium signaling pathways in the 1 mM Mel compared to the 100 μM 8MP group. (B) Diagram of the proposed signaling pathway for Mel in the DRG neurons: 1: Activation of RORα by high-dose Mel could inhibit the biological effects of MT2; 2: Inhibition of MAPK-calcium signaling pathways by MT2 activation could suppress c-fos, CGRP, TNF-1α, and IL-1β; and 3: Inhibition of NOS1 by Mel was independent of MT2, and RORα was also involved in the pain-related regulation.
Both MT2-dependent (MAPK-calcium channels) and MT2-independent (NOS1) pathways have well-documented roles in regulating neuronal activation, peptidergic neurons and neuro-inflammation [67-69]. To understand these regulations in the mouse DRG, we measured expression of c-fos, CGRP, and inflammatory cytokines in the cuff models. The expression of c-fos and activation of its protein FOS was correlated with neuronal activity in the DRG, which is often considered as a reflection of pain sensation [70, 71]. CGRP is a pain-transmitting neuropeptide present in the DRG, which is always up-regulated in pain conditions [72]. TNF-α and IL-1β are critical Mel-mediated molecules in neuro-inflammation [53, 73], and their expression is associated with neuronal excitability and sensitization [74, 75]. Our results indicated that cuff implantation increased the expression of c-fos, CGRP, and the inflammatory cytokines TNF-1α and IL-1β, which indicated pain sensation after cuff implantation. The increase in the expression of various cytokines could be suppressed by exogenous 100 mg/kg Mel or 50 mg/kg 8MP. However, the addition of 20 mg/kg luzindole could only reverse the analgesic effect of MT2 activation in the 8MP-treated group but not in the Mel-treated group. These results demonstrated that Mel could also suppress NP via MT2-independent pathways in DRG neurons. Taken together, our results indicate that Mel suppresses NP via MT2-dependent and -independent pathways in DRG neurons of mice.
Supplementary Material
Supplementary figure S1.
Acknowledgements
We thank Dr. Guang Hu in the Epigenetics and Stem Cell Biology Laboratory of the National Institute of Environmental Health and Sciences for his constructive suggestions for this microarray analysis.
We acknowledge the technical support from Novel Bioinformatics Co., Ltd, Shanghai, China.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31270952, 81571074, 81102217, 81571218).
Author contributions
ZY.L. and JL.L. participated in the study design and interpretation of the results.
JJ.L., Y.L. and TZ.Z. drafted the manuscript and performed the data analysis.
CK.Z and T.Z. carried out the animal experiment and cell culture.
XL.C., DD.Z., JQ.D., T.C., Z.Z. and S.C. conducted the biochemical study.
JL.Z. provided technical and supervisory support.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Woolf CJ, Mannion RJ. Neuropathic pain: aetiology, symptoms, mechanisms, and management. Lancet. 1999;353(9168):1959-64
2. Baron R. Neuropathic pain: a clinical perspective. Handb Exp Pharmacol. 2009(194):3-30
3. Baron R. Mechanisms of disease: neuropathic pain-a clinical perspective. Nat Clin Pract Neurol. 2006;2(2):95-106
4. Nahin RL. Estimates of pain prevalence and severity in adults: United States, 2012. J Pain. 2015;16(8):769-80
5. Bouhassira D, Lantéri-Minet M, Attal N. et al. Prevalence of chronic pain with neuropathic characteristics in the general population. Pain. 2008;136(3):380-7
6. Torrance N, Smith BH, Bennett MI. et al. The epidemiology of chronic pain of predominantly neuropathic origin. Results from a general population survey. J Pain. 2006;7(4):281-9
7. Dubocovich ML, Delagrange P, Krause DN. et al. International Union of Basic and Clinical Pharmacology. LXXV. Nomenclature, classification, and pharmacology of G protein-coupled melatonin receptors. Pharmacol Rev. 2010;62(3):343-80
8. Manchester LC, Coto-Montes A, Boga JA. et al. Melatonin: an ancient molecule that makes oxygen metabolically tolerable. J Pineal Res. 2015;59(4):403-19
9. Reiter RJ, Mayo JC, Tan D. et al. Melatonin as an antioxidant: under promises but over delivers. J Pineal Res. 2016;61(3):253-78
10. Wang T, Li SR, Dai X. et al. Effects of melatonin on orphanin FQ/nociceptin-induced hyperalgesia in mice. Brain Res. 2006;1085(1):43-8
11. Ambriztututi M. and V. Granadossoto, Oral and spinal melatonin reduces tactile allodynia in rats via activation of MT2 and opioid receptors. Pain. 2007;132(3):273-280
12. Lin TB, Hsieh MC, Lai CY. et al. Melatonin relieves neuropathic allodynia through spinal MT2-enhanced PP2Ac and downstream HDAC4 shuttling-dependent epigenetic modification of hmgb1 transcription. J Pineal Res. 2016;60(3):263-76
13. Sapunar D, Ljubkovic M, Lirk P. et al. Distinct membrane effects of spinal nerve ligation on injured and adjacent dorsal root ganglion neurons in rats. Anesthesiology. 2005;103(2):360-76
14. Wu G, Ringkamp M, Murinsonet BB. et al. Degeneration of myelinated efferent fibers induces spontaneous activity in uninjured C-fiber afferents. J Neurosci. 2002;22(17):7746-53
15. Wall P.D. and M. Devor. Sensory afferent impulses originate from dorsal root ganglia as well as from the periphery in normal and nerve injured rats. Pain. 1983;17(4):321-39
16. Sukhotinsky I, Ben-Dor E, Raber P. et al. Key role of the dorsal root ganglion in neuropathic tactile hypersensibility. Eur J Pain. 2004;8(2):135-43
17. Watkins LR, Milligan ED, Maier SF. Glial activation: a driving force for pathological pain. Trends Neurosci. 2001;24(8):450-5
18. DeLeo JA, Tanga FY, Tawfik VL. Neuroimmune activation and neuroinflammation in chronic pain and opioid tolerance/hyperalgesia. Neuroscientist. 2004;10(1):40-52
19. Krames ES. The dorsal root ganglion in chronic pain and as a target for neuromodulation: a review. Neuromodulation. 2015;18(1):24-32
20. Ayar A, Martin DJ, Ozcan M. et al. Melatonin inhibits high voltage activated calcium currents in cultured rat dorsal root ganglion neurones. Neurosci Lett. 2001;313(1-2):73-7
21. Kahya M.C, M. Naziroglu, and I.S. Ovey, Modulation of Diabetes-Induced Oxidative Stress, Apoptosis, and Ca2+ Entry Through TRPM2 and TRPV1 Channels in Dorsal Root Ganglion and Hippocampus of Diabetic Rats by Melatonin and Selenium. Mol Neurobiol. 2016 DOI: 10.1007/s12035-016-9727-3
22. Naziroglu M, Çelik Ö, Özgül C. et al. Melatonin modulates wireless (2.45 GHz)-induced oxidative injury through TRPM2 and voltage gated Ca(2+) channels in brain and dorsal root ganglion in rat. Physiol Behav. 2012;105(3):683-92
23. Danilov A, Kurganova J. Melatonin in Chronic Pain Syndromes. Pain Ther. 2016;5(1):1-17
24. Laste G, Macedo ICD, Roziskyet JR. et al. Melatonin administration reduces inflammatory pain in rats. J Pain Res. 2012;5:359-62
25. Lopez-Canul M, Palazzo E, Dominguez-Lopez S. et al. Selective melatonin MT2 receptor ligands relieve neuropathic pain through modulation of brainstem descending antinociceptive pathways. Pain. 2015;156(2):305-17
26. Srinivasan V, Zakaria R, Jeet SH. et al. Melatonin, its agonists in pain modulation: clinical application. Archives Italiennes de Biologie. 2012;150:274-89
27. Wilhelmsen M, Amirian I, Reiter RJ. et al. Analgesic effects of melatonin. a review of current evidence from experimental and clinical studies. J Pineal Res. 2011;51(3):270-7
28. Ulugol A, Dokmeci D, Guray G. et al. Antihyperalgesic, but not antiallodynic, effect of melatonin in nerve-injured neuropathic mice: Possible involvements of the L-arginine-NO pathway and opioid system. Life Sci. 2006;78(14):1592-7
29. Zurowski D, Nowak L, Machowska A. et al. Exogenous melatonin abolishes mechanical allodynia but not thermal hyperalgesia in neuropathic pain. The role of the opioid system and benzodiazepine-gabaergic mechanism. J Physio Phar. 2012;63(6):641-7
30. Challa SR. Surgical animal models of neuropathic pain: Pros and Cons. Int J Neurosci. 2015;125(3):170-4
31. Benbouzid M, Pallage V, Rajalu M. et al. Sciatic nerve cuffing in mice: a model of sustained neuropathic pain. Eur J Pain. 2008;12(5):591-9
32. Chaplan SR, Bach FW, Pogrel JW. et al. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods. 1994;53(1):55-63
33. Hargreaves K, Dubner R, Brownet F. et al. A new and sensitive method for measuring thermal nociception in cutaneous hyperalgesia. Pain. 1988;32(1):77-88
34. Gandini MA, Sandoval A, Felix R. Whole-cell patch-clamp recordings of Ca2+ currents from isolated neonatal mouse dorsal root ganglion (DRG) neurons. Cold Spring Harb Protoc. 2014;2014(4):389-95
35. Lin JJ, Du Y, Cai WK. et al. Toll-like receptor 4 signaling in neurons of trigeminal ganglion contributes to nociception induced by acute pulpitis in rats. Sci Rep. 2015;5:12549
36. Norton AJ, Jordan S, Yeomans P. Brief, high-temperature heat denaturation (pressure cooking): a simple and effective method of antigen retrieval for routinely processed tissues. J Pathol. 1994;173(4):371-9
37. Pawitan Y, Michiels S, Koscielny S. et al. False discovery rate, sensitivity and sample size for microarray studies. Bioinformatics. 2005;21(13):3017-24
38. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:322-6
39. Ashburner M, Ball CA, Blake JA. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25-9
40. Jansen R, Greenbaum D, Gerstein M. Relating whole-genome expression data with protein-protein interactions. Genome Res. 2002;12(1):37-46
41. Li C, Li H. Network-constrained regularization and variable selection for analysis of genomic data. Bioinformatics. 2008;24(9):1175-82
42. Zhang JD, Wiemann S. KEGGgraph: a graph approach to KEGG PATHWAY in R and bioconductor. Bioinformatics. 2009;25(11):1470-1
43. Shannon P, Markiel A, Ozier O. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498-504
44. Jensen TS, Baron R, Haanpää M. et al. A new definition of neuropathic pain. Pain. 2011;152(10):2204-5
45. Jensen TS, Baron R. Translation of symptoms and signs into mechanisms in neuropathic pain. Pain. 2003;102(1-2):1-8
46. Jensen T, Finnerup NB. Allodynia and hyperalgesia in neuropathic pain: clinical manifestations and mechanisms. Lancet Neurol. 2014;13(9):924-35
47. Truini A, Garcia-Larrea L, Cruccu G. Reappraising neuropathic pain in humans-how symptoms help disclose mechanisms. Nat Rev Neurol. 2013;9(10):572-82
48. Haanpää M, Attal N, Backonja M. et al. NeuPSIG guidelines on neuropathic pain assessment. Pain. 2011;152(1):14-27
49. Backonja MM, Attal N, Baron R. et al. Value of quantitative sensory testing in neurological and pain disorders: NeuPSIG consensus. Pain. 2013;154(9):1807-19
50. Bahna SG, Sathiyapalan A, Foster JA. et al. Regional upregulation of hippocampal melatonin MT2 receptors by valproic acid: therapeutic implications for Alzheimer's disease. Neurosci Lett. 2014;576:84-7
51. Niles LP, Sathiyapalan A, Bahna S. et al. Valproic acid up-regulates melatonin MT1 and MT2 receptors and neurotrophic factors CDNF and MANF in the rat brain. Int J Neuropsychopharmacol. 2012;15(9):1343-50
52. Agil A, Elmahallawy EK, Rodríguez-Ferrer JM. et al. Melatonin increases intracellular calcium in the liver, muscle, white adipose tissues and pancreas of diabetic obese rats. Food Funct. 2015;6(8):2671-8
53. Yuruker V, Naziroglu M, Senol N. Reduction in traumatic brain injury-induced oxidative stress, apoptosis, and calcium entry in rat hippocampus by melatonin: Possible involvement of TRPM2 channels. Metab Brain Dis. 2015;30(1):223-31
54. Roskoski R. ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res. 2012;66(2):105-43
55. Carrasquillo Y, Gereau RW. Activation of the extracellular signal-regulated kinase in the amygdala modulates pain perception. J Neurosci. 2007;27(7):1543-51
56. Obata K, Yamanaka HY, Mizushima T. et al. Activation of extracellular signal-regulated protein kinase in the dorsal root ganglion following inflammation near the nerve cell body. Neuroscience. 2004;126(4):1011-21
57. Dai Y, Iwata K, Fukuoka T. et al. Phosphorylation of extracellular signal-regulated kinase in primary afferent neurons by noxious stimuli and its involvement in peripheral sensitization. J Neurosci. 2002;22(17):7737-45
58. O'Brien DE, Alter BJ, Satomoto M. et al. ERK2 Alone Drives Inflammatory Pain But Cooperates with ERK1 in Sensory Neuron Survival. J Neurosci. 2015;35(25):9491-507
59. Otsubo Y, Satoh Y, Kodama M. et al. Mechanical allodynia but not thermal hyperalgesia is impaired in mice deficient for ERK2 in the central nervous system. Pain. 2012;153(11):2241-52
60. Reyesgibby CC, Wang J, Silvas MRT. et al. MAPK1/ERK2 as novel target genes for pain in head and neck cancer patients. BMC Genet. 2016;17:40
61. Klimentova J, Cebova M, Barta A. et al. Effect of melatonin on blood pressure and nitric oxide generation in rats with metabolic syndrome. Physiol Res. 2016;65(3):373-80
62. Arese M, Magnifico MC, Mastronicola D. et al. Nanomolar melatonin enhances nNOS expression and controls HaCaT-cells bioenergetics. IUBMB Life. 2012;64(3):251-8
63. Chang HM, Ling EA, Chen CF. et al. Melatonin attenuates the neuronal NADPH-d/NOS expression in the nodose ganglion of acute hypoxic rats. J Pineal Res. 2002;32(2):65-73
64. Luo ZD, Chaplan SR, Scott BP. et al. Neuronal nitric oxide synthase mRNA upregulation in rat sensory neurons after spinal nerve ligation: lack of a role in allodynia development. J Neurosci. 1999;19(21):9201-8
65. Mika J, Rojewska E, Makuch W. et al. The effect of botulinum neurotoxin A on sciatic nerve injury-induced neuroimmunological changes in rat dorsal root ganglia and spinal cord. Neuroscience. 2011;175:358-66
66. Wioletta M, Joanna M, Ewelina R. et al. Effects of selective and non-selective inhibitors of nitric oxide synthase on morphine- and endomorphin-1-induced analgesia in acute and neuropathic pain in rats. Neuropharmacology. 2013;75:445-57
67. Li Q, Chen M, Liu H. et al. The dual role of ERK signaling in the apoptosis of neurons. Front Biosci (Landmark Ed). 2014;19:1411-7
68. Forstermann U, Sessa WC. Nitric oxide synthases: regulation and function. Eur Heart J. 2012;33(7):829-37
69. Jones BL, Smith SM. Calcium-Sensing Receptor: A Key Target for Extracellular Calcium Signaling in Neurons. Front Physiol. 2016;7:116
70. Cai W, Cao J, Ren X. et al. shRNA mediated knockdown of Nav1.7 in rat dorsal root ganglion attenuates pain following burn injury. BMC Anesthesiol. 2016;16(1):59
71. Rigon F, Rossato D, Auler VB. et al. Effects of sciatic nerve transection on ultrastructure, NADPH-diaphorase reaction and serotonin-, tyrosine hydroxylase-, c-Fos-, glucose transporter 1- and 3-like immunoreactivities in frog dorsal root ganglion. Braz J Med Biol Res. 2013;46(6):513-20
72. Amara SG, Jonas V, Rosenfeld MG. et al. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982;298(5871):240-4
73. Kahya MC, Naziroglu M, Cig B. Melatonin and selenium reduce plasma cytokine and brain oxidative stress levels in diabetic rats. Brain Inj. 2015;29(12):1490-6
74. Ren K, Torres R. Role of interleukin-1beta during pain and inflammation. Brain Res Rev. 2009;60(1):57-64
75. Sorkin LS, Xiao WH, Wagner R. et al. Tumour necrosis factor-alpha induces ectopic activity in nociceptive primary afferent fibres. Neuroscience. 1997;81(1):255-62
Author contact
![]() Corresponding authors: Zhu-Yi Li and Jin-Lian Li. Zhu-Yi Li: lizhuyiedu.cn; #1 XinSi Road, Xi'an, 710000, PR China; Tel: 86-29-84774130; Fax: 86-29-83552982; Jin-Lian Li: jinlianedu.cn; #169 West Changle Road, Xi'an, 710032, PR China; Tel: 86-29-84773087; Fax: 86-29-83283229.
Corresponding authors: Zhu-Yi Li and Jin-Lian Li. Zhu-Yi Li: lizhuyiedu.cn; #1 XinSi Road, Xi'an, 710000, PR China; Tel: 86-29-84774130; Fax: 86-29-83552982; Jin-Lian Li: jinlianedu.cn; #169 West Changle Road, Xi'an, 710032, PR China; Tel: 86-29-84773087; Fax: 86-29-83283229.
 Global reach, higher impact
Global reach, higher impact