13.3
Impact Factor
Theranostics 2017; 7(19):4836-4849. doi:10.7150/thno.20942 This issue Cite
Research Paper
Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression
1. State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, Guangzhou, PR China;
2. Department of Oncology, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, PR China;
3. Department of Molecular and Cellular Oncology, the University of Texas MDAnderson Cancer Center, Houston, TX77030, USA.
* These authors contributed equally to this work
Received 2017-5-9; Accepted 2017-8-20; Published 2017-10-17
Abstract
Long non-coding RNAs (lncRNAs) are involved in the pathology of various tumors, including colorectal cancer (CRC). However, the role of lncRNA in CRC liver metastasis remains unclear. Methods: a microarray was performed to identify the differentially expressed lncRNAs between CRC tissues with and without liver metastasis. Survival analysis was evaluated using the Kaplan-Meier method and assessed using the log-rank test. In vitro and in vivo assays were preformed to explore the biological effects of the differentially expressed lncRNA in CRC cells. Results: the lncRNA UICLM (up-regulated in colorectal cancer liver metastasis) was significantly up-regulated in cases of CRC with liver metastasis. Moreover, UICLM expression was higher in CRC tissues than in normal tissues, and UICLM expression was associated with poor patient survival. Knockdown of UICLM inhibited CRC cell proliferation, invasion, epithelial-mesenchymal transition (EMT) and CRC stem cell formation in vitro as well as tumor growth and liver metastasis in vivo. Ectopic expression of UICLM promoted CRC cell proliferation and invasion. Mechanistic investigations revealed that UICLM induced its biological effects by regulating ZEB2, as the oncogenesis facilitated by UICLM was inhibited by ZEB2 depletion. Further study indicated that UICLM acted as a competing endogenous RNA (ceRNA) for miR-215 to regulate ZEB2 expression. Conclusions: taken together, our findings demonstrate how UICLM induces CRC liver metastasis and may offer a novel prognostic marker and therapeutic target for this disease.
Keywords: long non-coding RNA, CRC, lncRNA UICLM, liver metastasis.
Introduction
Colorectal cancer (CRC) is the second most common and the third leading cause of cancer-related deaths worldwide [1]. Despite tremendous progress in the treatment of CRC in recent decades, the prognosis remains unsatisfactory, especially in advanced-stage tumors with distant metastasis [2]. The most common site for distant metastasis in CRC is the liver. It is reported that approximately 25% of CRC patients present with synchronous liver metastases at diagnosis, and approximately 50% of CRC patients develop metachronous liver metastases 3 years after treatment [3]. As most CRC patients with liver metastases are not able to undergo surgery, the prognosis is dismal, with a 5-year survival rate less than 10% after the diagnosis of liver metastasis [4]. Therefore, it is vital to identify the molecular mechanisms and genetic alterations of CRC metastases to develop effective therapies.
Recently, integrative genomic studies have revealed that more than 90% of the DNA sequence is actively transcribed, with only 2% of these transcripts encoding protein, while most of the transcripts are referred to as non-coding RNAs (ncRNAs) [5]. Among these ncRNAs, long non-coding RNAs (lncRNAs) are a class of transcripts with lengths greater than 200 nucleotides. lncRNAs have been found to be abundantly transcribed in mammalian cells as well as plant cells [6, 7]. These lncRNAs can act as guides, scaffolds, decoys and tethers of other biological molecules and are involved in various biological processes, such as cell growth, cell differentiation, cell apoptosis, cell invasion and stem cell pluripotency [8-10]. Increasing evidence has indicated that lncRNA expression profiling can be used as a diagnostic and prognostic tool for different tumors, such as prostate cancer, breast cancer, gastric cancer and CRC [11-14]. Although a growing number of lncRNAs have been annotated, the role and molecular regulatory mechanisms of lncRNAs in CRC liver metastasis are still unclear.
Materials and Methods
Human tissue samples
This study was approved by the institutional ethics review board of Sun Yat-sen University Cancer Center (SYSUCC) (Guangzhou, China), and written informed consent was obtained from all patients enrolled in this study. None of the patients receive any treatment (chemotherapy or radiotherapy) before surgery. For microarray analysis, a total of 15 CRC tissue samples (7 with liver metastasis at the time of diagnosis and 8 stage III patients who did not have metastasis at the time of diagnosis or during the follow-up period of more than 2 years after resection) were obtained from SYSUCC between May 2005 and September 2012 (Table S1). For validation, another larger cohort of patients including 55 patients with liver metastases and 67 patients without liver metastases who received surgery between March 2004 and October 2013 in SYSUCC was also included in this study. Their primary CRC tissues (n = 122), adjacent non-tumor tissues (n = 122) and corresponding matched liver metastatic tissues (n = 55) were obtained immediately upon surgery and stored at -80°C or paraffin-embedded. The clinical stage was classified based on the 7th edition of the International Union Against Cancer (UICC) on Tumor-Node-Metastasis (TNM) staging system. All of the patients were followed-up regularly at 3-month intervals after surgery. The median follow-up period was 55.7 months (range, 24.5 - 156.3). The clinical data of these patients were obtained from their medical records.
RNA extraction and microarray analysis
Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The RNA quality was confirmed by formaldehyde agarose gel electrophoresis and quantified by NanoDrop ND-1000. The samples (CRC tissues with or without liver metastasis, Table S1) were used to synthesize double-stranded cDNA, and the cDNA was then labeled and hybridized to the LncRNA Expression Microarray (Arraystar 8x60Kv3.0, Rockville, USA) according to the manufacturer's protocol. After hybridization, the arrays were washed, and the slides were scanned with an Agilent Microarray Scanner (Agilent p/n G2565BA). Raw data were extracted as pair files using the Agilent Feature Extraction. The random variance model was used to identify the differentially expressed genes. The paired t-test was used to calculate the P-value. The threshold set for up- and down-regulated genes was a fold change >=4.0 and a P-value <= 0.05, respectively. The microarray data have been deposited into the NCBI Gene Expression Omnibus (GEO) database and are available through GEO with the accession number GSE95423.
Real-time PCR analysis
The High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) was used for UICLM and ZEB2 reverse transcription. Quantitative real-time PCR was performed using the TaqMan Universal Master Mix II, and β-actin and GAPDH were used as internal controls. To measure the level of miR-215, a Bulge-LoopTM miRNA qRT-PCR Primer Set (Ribobio, Guangzhou, China) was used according to the manufacturer's instructions, and U6 snRNA was used as the endogenous control. The primers used are listed in Table S2. All real-time PCR analysis was performed with the Bio-Rad CFX96 qPCR system, and fold changes were determined using the relative quantification 2-△△CT method.The nuclear and cytoplasm fractions of SW620 or DLD-1 cells were separated using the PARIS Kit (Life Technologies) according to the manufacturer's instructions. RNA was extracted from both fractions. Then, real-time PCR was performed to determine the expression ratios of specific RNA molecules between the nuclear and cytoplasm fractions. GAPDH served as the cytoplasm marker, and U6 served as the nuclear marker.
In situ hybridization (ISH) analysis
ISH analysis was performed according to a previously described method [15].
Cell culture and transfections
Human CRC cell lines (SW620, SW480, LoVo, HT-29, HCT116, DLD-1, and RKO), the immortalized colon epithelial cell line CCD-112CoN, and the human embryonic kidney cell line HEK293a were purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured and stored according to their instructions. The miR-215 mimic and negative control (NC) oligonucleotides, miR-215 inhibitor and scramble oligonucleotides were obtained from Ribobio (Guangzhou, China). The small interfering RNA (siRNA) duplex oligonucleotides targeting human ZEB2 mRNA and UICLM (Accession number: NR_033841) were obtained from GenePharma (Shanghai, China). Cell transfections were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions.
Cell proliferation assays
The CCK-8 assay and the colony formation assay were performed to test cell proliferation. The details were described in our previous study [16]. Briefly, for CCK-8 assay, 1×103 cells were cultured in a 96-well plate at 37°C. After 10 μL CCK-8 solution was added to each well, plates were incubated at 37˚C for 2 h. Finally, the spectrophotometric absorbance at 570 nm was measured for each sample. All the experiments were repeated 3 times in triplicate and the mean was calculated. For colony formation assay, cells were trypsinized and suspended in RPMI 1640 medium (GIBCO) with 10% FBS. The cells were seeded in 6-well plates and cultured in a humidified atmosphere containing 5% CO2 at 37°C for 2 weeks. Cell colonies were washed with PBS, fixed with methanol, and stained with 0.1% crystal violet (1 mg/mL). Colonies containing more than 50 cells were counted and the mean colony numbers were calculated.
In vitro cell wound healing, migration and invasion assays
Wound healing assays and transwell assays were performed to detect cell migration and invasion. The details were described in our previous study [17].
Flow cytometric sorting of side population (SP) and non-SP cells
Cells were trypsinized, washed and resuspended at a density of 1.0 × 106 cells/mL in RPMI 1640 (pre-warmed) medium with 2% FBS. Cell staining was performed using a method described previously [18]. The cells were then incubated with Hoechst 33342 at a concentration of 5 mg/mL with or without the ABC transporter inhibitor verapamil (50 mM) at 37°C for 90 min and kept in the dark with intermittent shaking. After being washed and resuspended with PBS, the cells were stored at 4°C for flow cytometry and sorting. Cell sorting and analysis was performed with a MoFlo XDP Cell Sorter (Beckman Coulter, Brea, USA).
Sphere-forming assays
A sphere-forming assay was performed according to a published method with minor modifications. Briefly, cell suspensions (1.0 × 103 cells/well) were seeded in 6-well ultralow attachment plates (Corning Inc. Corning, USA) using serum-free DMEM/F12 (Invitrogen) containing 20 ng/mL of basic fibroblast growth factor (Miltenyi Biotec), 20 ng/mL of epidermal growth factor (Miltenyi Biotec, Auburn, USA), and 2 mM L-glutamine (Mediatech Inc.). After culturing for 7 days, the size and number of tumor spheres were evaluated using microscopy.
Lentivirus production and transduction
Short hairpin RNA (shRNA) directed against human UICLM or scrambled oligonucleotides were ligated into the LV-3 (pGLVH1/GFP+Puro) vector (GenePharma, Shanghai, China). HEK293a cells were co-transfected with Lenti-Pac HIV Expression Packaging Mix and the lentiviral vectors (or the control lentivirus vectors) using Lipofectamine 2000 (Life Technologies Corporation, Carlsbad, CA, USA). 48 h later, lentiviral particles in the supernatant were harvested and filtered by centrifugation at 500 × g for 10 min. Cells were then transfected with lentivirus or control virus (NC). To select the stably transfected cells, the cells were treated with puromycin (2 μg/mL) for two weeks. GFP-positive cells were picked as sh-UICLM and sh-NC and then used for subsequent assays.
In vivo proliferation and metastasis assays
All animal experiments were performed under the experimental animal use guidelines of the National Institutes of Health. Female BABL/c athymic nude mice (aged 5-6 weeks) purchased from the Animal Center of Guangdong Province (Guangzhou, China) were used. The details were described in the previous study [17].
Western blotting analyses and immunofluorescence analyses
Western blotting analyses and immunofluorescence analyses were conducted according to the method described previously [19]. The following antibodies were used in this study: ZEB2 (#abs116801), GAPDH (#2118L), E-cadherin (#3199S), N-cadherin (#14215S).
RNA sequencing analysis
Total RNA was isolated from cells/tissues using Trizol (invitrogen) according to the manufacturer's protocol. RNA purity was assessed using the Qubit®. Each RNA sample had an A260:A280 ratio above 1.8 and A260:A230 ratio above 2.0. RNA integrity was evaluated using the Agilent 2200 TapeStation (Agilent Technologies, USA) and each sample had a RINe above 7.0. Briefly, rRNAs were removed from Total RNA using Epicentre Ribo-Zero rRNARemoval Kit (illumina, USA) and fragmented to approximately 200 bp. Subsequently, the purified RNAs were subjected to first strand and second strand cDNA synthesis followed by adaptor ligation and enrichment with a low-cycle according to instructions of TruSeq® RNA LT/HT Sample Prep Kit (Illumina, USA). The purified library products were evaluated using the Agilent 2200 TapeStation and Qubit®2.0 (Life Technologies, USA) and then diluted to 10 pM for cluster generation in situ on the HiSeq3000 pair-end flow cell followed by sequencing (2×150 bp) on HiSeq3000. The data have been deposited into the NCBI Gene Expression Omnibus (GEO) database and are available through GEO with the accession number GSE101081.
Vector constructs, transfections and assays
For overexpression of UICLM in CRC cells, the full-length UICLM cDNA was synthesized and cloned into the expression vector pcDNA3.1 (+) (Invitrogen, Carlsbad, CA, USA). The fragment from UICLM containing the predicted miR-215 binding site was amplified by PCR and cloned into a pmirGLO Dual-luciferase Target Expression Vector (Promega, Madison, WI, USA) to form the reporter vector UICLM-wild-type (UICLM-wt). To test the binding specificity, the corresponding mutant was created by mutating the miR-215 seed region binding site (seed sequence binding fragment 5'-CAAUUCAAUG-3' changed to 5'-GGACCGAAUG-3'), which was named UICLM-mt. HEK 293Ta cells were co-transfected with the pmirGLO vector with either wild-type fragments or mutation fragments and indicated miRNAs/siRNAs using Lipofectamine 2000. 48 h later, a luciferase reporter assay was performed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). The firefly luciferase activity was measured and normalized based on Renilla luciferase activity. To confirm the direct regulating relationship between miR-215 and ZEB2, the full-length 3'-UTR of ZEB2 mRNA and a mutant variant were amplified by PCR and cloned into the XbaI site of a pGL3-basic vector (Promega) and termed ZEB2-wt-3'UTR and ZEB2-mt-3'UTR, respectively. The miR-215 expression plasmid (pcDNA-miR-215) was generated using synthetic oligonucleotides and the pcDNA6.2-GW/EmGFP vector. Cells were cultured in a 6-well plate and then transfected with the pcDNAmiR-215 or the negative control (NC) (750 ng/well), the pGL3 reporter vector (250 ng/well) and the pRL-TK luciferase reporters (25 ng/well) using Lipofectamine 2000 (Invitrogen). Luciferase activity levels were measured using the Dual-Luciferase Reporter Assay Kit (Promega) following the manufacturer's instructions.
RNA immunoprecipitation (RIP) assay
RIP was performed using a Magna RNA-binding protein immunoprecipitation kit (Millipore, Bedford, MA) according to the manufacturer's instructions. Briefly, cell lysates were incubated with RIP buffer containing magnetic beads conjugated with negative control normal mouse IgG or human anti-Ago2 antibody. The samples were then incubated with Proteinase K to isolate the immunoprecipitated RNA. Finally, purified RNAs were extracted and determined by real-time PCR to confirm the presence of the binding targets.
Immunohistochemistry (IHC) assays
The paraffin-embedded tissue blocks were cut into 4 μm slides. A ZEB2 rabbit antibody (Millipore, Bedford, MA) was used. IHC analysis and qualification of ZEB2 expression were performed according to a previously described method [20].
Statistical analyses
Each experiment was repeated for three times or more. Unless otherwise noted, data are presented as the mean ± SD, and analyzed by student's t-test (unpaired, two-tailed) using the SPSS software package (version 16.0, SPSS Inc.) or GraphPad Prism 5.0. The correlations between UICLM and miR-215 as well as ZEB2 in human tissues were analyzed by Spearman's rank test. Survival analysis was evaluated using the Kaplan-Meier method and assessed using the log-rank test. P < 0.05 was considered statistically significant.
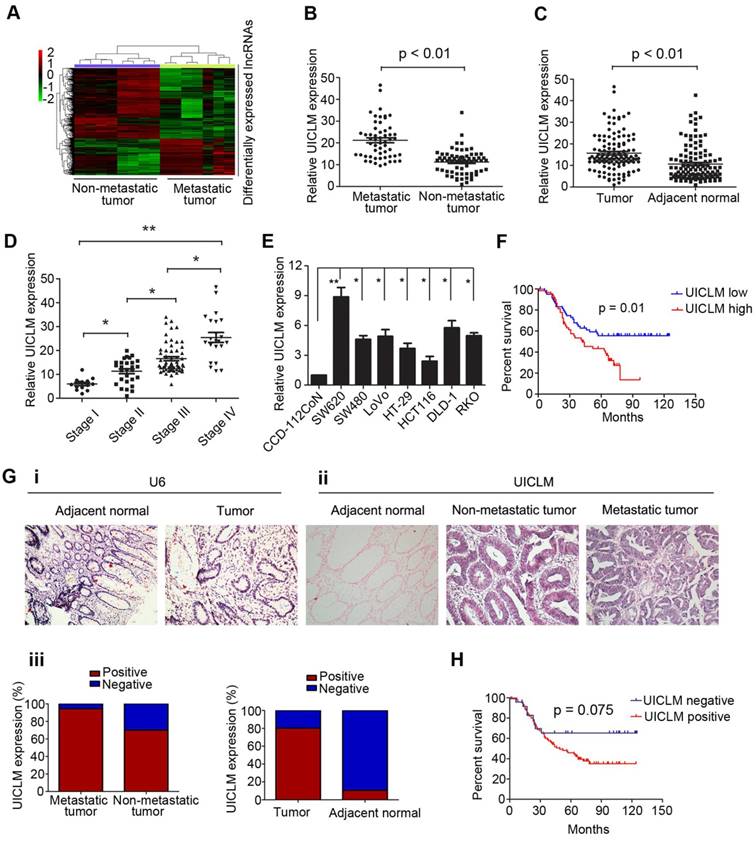
The lncRNA UICLM is up-regulated in CRC with liver metastasis. (A) Differentially expressed lncRNAs between CRC tissues with and without liver metastases. A lncRNA microarray was conducted in CRC samples with (n = 7) and without (n = 8) liver metastases. (B) The relative UICLM expression levels in CRC with liver metastases (n = 55) were significantly higher than in CRC tissues without liver metastases (n = 67; P < 0.01). (C) The relative UICLM expression levels in tumor tissues (n = 122) were significantly higher than in adjacent normal tissues (n = 122; P < 0.01). (D) Relative UICLM expression levels in different clinical stages (*P < 0.05, **P < 0.01). (E) Kaplan-Meier analysis of progression-free survival based on UICLM levels (determined by real-time PCR) in 122 CRC patients showed that patients with high UICLM levels (n = 61) had a worse clinical outcome than patients with low UICLM levels (n = 61) (P = 0.01). (F) Relative UICLM expression levels in CRC cell lines (SW620, SW480, LoVo, HT-29, HCT116, DLD-1, and RKO) and the immortalized colon epithelial cell line CCD-112CoN. (G) i) Analyses of UICLM expression in adjacent normal tissues, tumor tissues and liver metastasis by ISH, with U6 used as the internal control (blue, positive staining; red, negative staining). (ii) and (iii) The percent of ISH-positive samples in different groups. (H) Kaplan-Meier analysis of progression-free survival based on UICLM expression (determined by ISH) showed that patients who were UICLM-positive (n = 98) had significantly worse survival than patients who were UICLM-negative (n = 24) (P = 0.075).
Results
lncRNA UICLM is significantly up-regulated in CRC with liver metastasis
To investigate the lncRNA expression profile involved in CRC liver metastasis, we performed a lncRNA microarray analysis on 15 CRC tissues (7 with liver metastasis and 8 without liver metastasis at the time of diagnosis and during the follow-up period of 2 years). We identified 493 differentially expressed lncRNAs between the two groups, including up-regulated and down-regulated lncRNAs (Figure 1A). To confirm the microarray results, we randomly selected top 12 lncRNAs that were up-regulated in CRC with liver metastasis for validation. The real-time PCR results confirmed that 9 of these lncRNAs were significantly overexpressed in CRC with liver metastasis compared to their expression in CRC without liver metastasis (Figure S1). To further select the lncRNAs that play a critical role in CRC liver metastasis, the expression of the above 9 lncRNAs was detected in another larger cohort of 122 CRC patients. As demonstrated in Figure 1B, only lncRNA LOC200772 (Accession: NR_033841, a 2274 bp transcript with 9 exon and localizes in human chromosome 2q37.3) was significantly up-regulated in CRC with liver metastasis. As its function is still uncharacterized in CRC, we therefore designated it as lncRNA UICLM (up-regulated in colorectal cancer liver metastasis). Moreover, UICLM expression was markedly increased in tumor tissues compared with its expression in adjacent non-tumor tissues, and increased expression was also observed in advanced tumor stage patients (*P < 0.05, **P < 0.01, Figure 1C and D). Likewise, the expression of UICLM was higher in CRC cells than in the immortalized colon epithelial cell line CCD-112CoN (*P < 0.05, **P < 0.01, Figure 1E). We verified that UICLM was indeed a non-coding RNA with an in vitro translation assay (Figure S2) and online protein coding potential assessment software (data not shown). Subcellular fractionation and real-time PCR analysis showed that UICLM is mainly located in the cytoplasm (Figure S3). ISH analysis confirmed the expression pattern of UICLM in tissues (Figure 1g i-iii). Then, to analyze the clinical and pathological association of UICLM in CRC patients, the patients were divided into two groups (high expression and low expression) based on the mean UICLM expression level from real-time PCR. As shown in Table S3, UICLM expression was significantly associated with tumor size (P = 0.004), liver metastasis (P = 0.002) and TNM stage (P = 0.008). However, no correlation was observed between UICLM and age, gender, tumor depth, histological grade or lymph node invasion. Kaplan-Meier survival curves showed that patients with high UICLM levels presented with worse progression-free survival (P = 0.01 and P = 0.075, respectively, Figure 1F and H). In addition, univariate analysis revealed that tumor size, lymph node invasion, liver metastasis and TNM stage were also associated with overall survival (Table S4). Finally, multivariate analysis demonstrated that UICLM expression and liver metastasis were independent prognostic indicators for CRC patients (P = 0.009 and P = 0.017, respectively; Table S4).
lncRNA UICLM is required for efficient proliferation and tumor growth of CRC cells
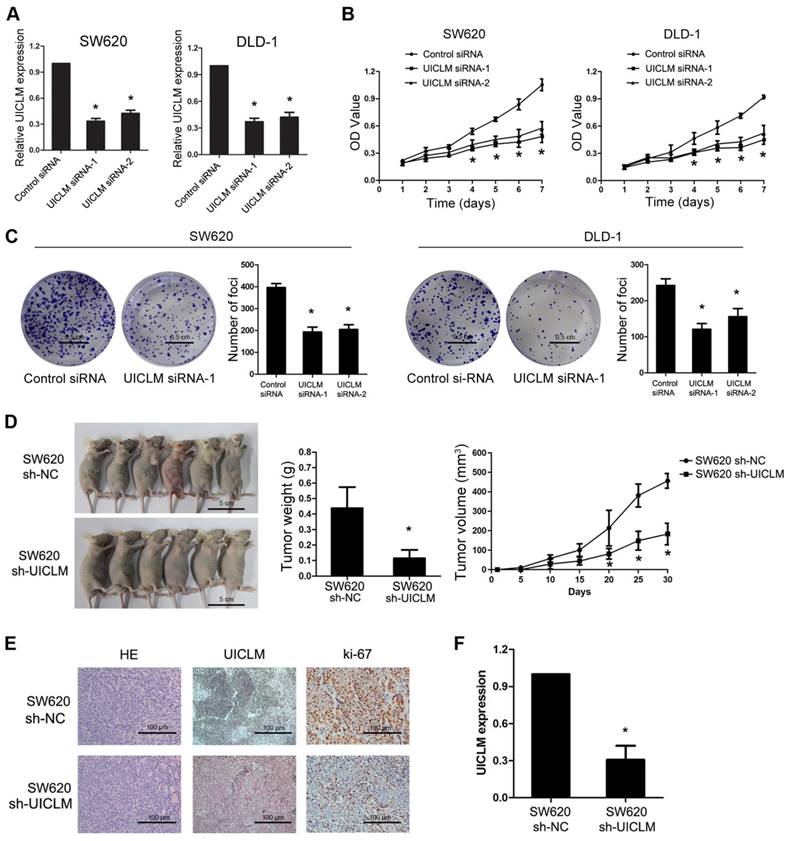
Specific siRNAs were used to knock down UICLM expression in two CRC cell lines, SW620 and DLD-1, which have relatively high expression of UICLM. The CCK-8 assay showed that knockdown of UICLM significantly inhibited the proliferation of SW620 and DLD-1 cells, while ectopic expression of UICLM increased the proliferation of HCT116 and HT-29 cells (*P < 0.05, Figure 2A and B, Figure S4A-B). A colony formation assay also confirmed that UICLM knockdown markedly reduced the colony formation number in SW620 and DLD-1 cells, whereas ectopic expression of UICLM increased the colony formation number in HCT116 and HT-29 cells (*P < 0.05, Figure 2C, Figure S4C-D). To further investigate the in vivo effect of UICLM on CRC cells, we constructed two stable cell lines using a lentivirus vector to mediate UICLM suppression in SW620 cells, designated SW620 sh-NC and SW620 sh-UICLM. The knockdown efficiency was confirmed by real-time PCR and the observation of green fluorescence (Figure S5). The results indicated that the tumor volume and tumor weight of tumors formed by SW620 sh-UICLM cells (n = 10) were significantly reduced compared with the tumor volume and weight of those formed by SW620 sh-NC cells (n =10) (*P < 0.05, Figure 2D). The ISH analysis confirmed that knockdown of UICLM reduced UICLM expression in vivo, and IHC analysis showed that knockdown of UICLM significantly reduced Ki-67 expression (*P < 0.05, Figure 2E and F).
lncRNA UICLM is required for cell migration, invasion, EMT and liver metastasis of CRC cells
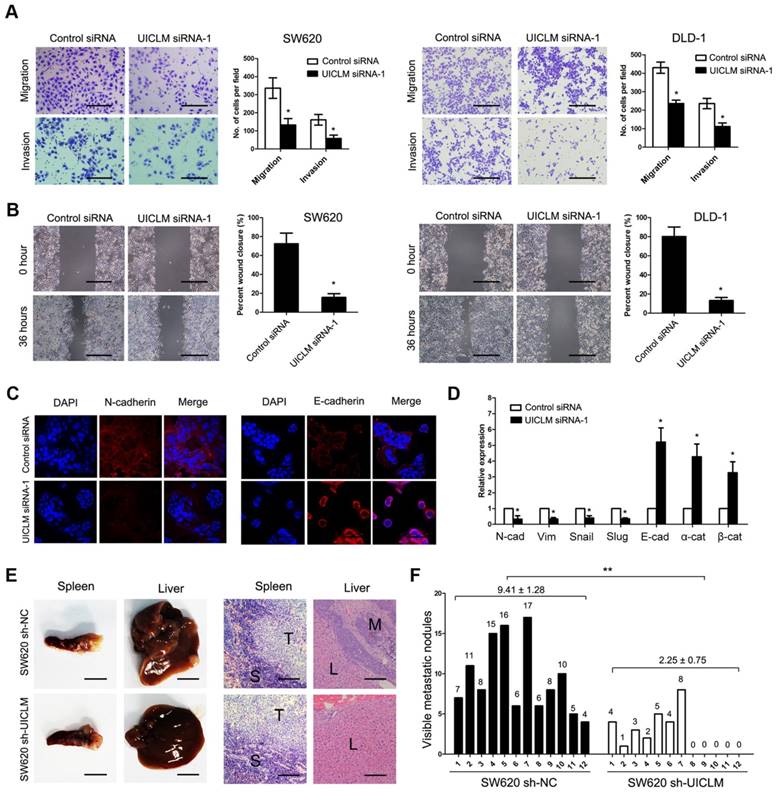
A transwell assay demonstrated that knockdown of UICLM significantly inhibited cell migration and invasion in SW620 and DLD-1 cells (*P < 0.05, Figure 3A), while ectopic expression of UICLM markedly increased cell invasion in HCT116 and HT-29 cells (P < 0.05, Figure S6A). A wound healing assay also indicated that knockdown of UICLM markedly reduced cell migration in SW620 and DLD-1 cells (*P < 0.05, Figure 3B). As EMT plays an important role in cell migration and invasion, we tested whether UICLM affects EMT in CRC cells. Immunofluorescence analysis showed that knockdown of UICLM decreased the expression of N-cadherin, whereas the expression of E-cadherin was increased (Figure 3C). The real-time PCR results confirmed that knockdown of UICLM significantly reduced the expression of mesenchymal markers (N-cadherin, Vimetin, Snail, Slug) while increasing the expression of epithelial markers (E-cadherin, α-catenin, β-ctenin) (*P < 0.05, Figure 3D). In contrast, ectopic expression of UICLM markedly promoted cell invasion in HCT116 and HT-29 cells and increased the expression of mesenchymal markers (N-cadherin, Vimetin, Snail, Slug) while decreasing the expression of epithelial markers (E-cadherin, α-catenin, β-catenin) (*P < 0.05, Figure S6B). To investigate the in vivo effect of UICLM on liver metastasis, cells (SW620 sh-NC and SW620 sh-UICLM) were injected into the distal tip of the spleen using a Hamilton syringe. The results showed that all of the mice formed tumors in the spleen, and 100% of the mice injected with SW620 sh-NC cells formed liver metastases. In contrast, only approximately 58% of the mice injected with SW620 sh-UICLM cells formed liver metastases (Figure 3E). Moreover, the numbers of metastatic nodules in the livers were significantly reduced in mice injected with SW620 sh-UICLM cells compared with the numbers in those injected with SW620 sh-NC cells (**P < 0.01, Figure 3F).
Knockdown of UICLM suppresses CRC cell proliferation and colony formation in vitro as well as tumorigenesis in vivo. (A) Relative expression levels of UICLM in SW620 and DLD-1 cells after transfection with UICLM siRNAs (*P < 0.05). (B) Knockdown of UICLM expression significantly inhibited cell viability in SW620 and DLD-1 cells (*P < 0.05). (C) Knockdown of UICLM expression significantly inhibited colony formation in SW620 and DLD-1 cells (*P < 0.05) (Scale bar: 0.5 cm). (D) Knockdown of UICLM expression significantly inhibited CRC cell growth in nude mice, and the tumor weight and tumor volume were significantly reduced in the sh-UICLM group compared to that in the sh-NC group (*P < 0.05) (Scale bar: 5 cm). (E) and (F) Knockdown of UICLM significantly reduced UICLM and Ki-67 expression in vivo (*P < 0.05) (Scale bar: 100μm).
Knockdown of UICLM inhibits CRC cell migration and invasion in vitro as well as liver metastasis in vivo. (A) Knockdown of UICLM expression significantly inhibited cell migration and invasion in SW620 and DLD-1 cells as demonstrated by transwell assay (*P < 0.05) (Scale bar:100μm). (B) Knockdown of UICLM expression markedly suppressed cell motility in SW620 and DLD-1 cells as demonstrated by wound healing assay (*P < 0.05) (Scale bar: 100 μm). (C) Knockdown of UICLM significantly inhibited the expression of N-cadherin while increasing the expression of E-cadherin as demonstrated by Immunofluorescence analysis (Scale bar: 50 μm). (D) Real-time PCR analysis showed that knockdown of UICM significantly reduced the expression of mesenchymal markers (N-cadherin, Vimetin, Snail, Slug) while increasing the expression of epithelial markers (E-cadherin, α-catenin, β-catenin) (*P < 0.05). (E) All of the mice formed tumors in the spleen, 12 of 12 mice injected with SW620 sh-NC cells formed liver metastases, and 7 of 12 mice injected with SW620 sh-UICLM cells formed liver metastases (Scale bar, left: 5 mm; right: 100 μm). (F) The numbers of metastatic nodules in the livers were significantly reduced in mice injected with SW620 sh-UICLM cells (2.25 ± 0.75) compared with the numbers in those injected with SW620 sh-NC cells (9.41 ± 1.28) (**P < 0.01).
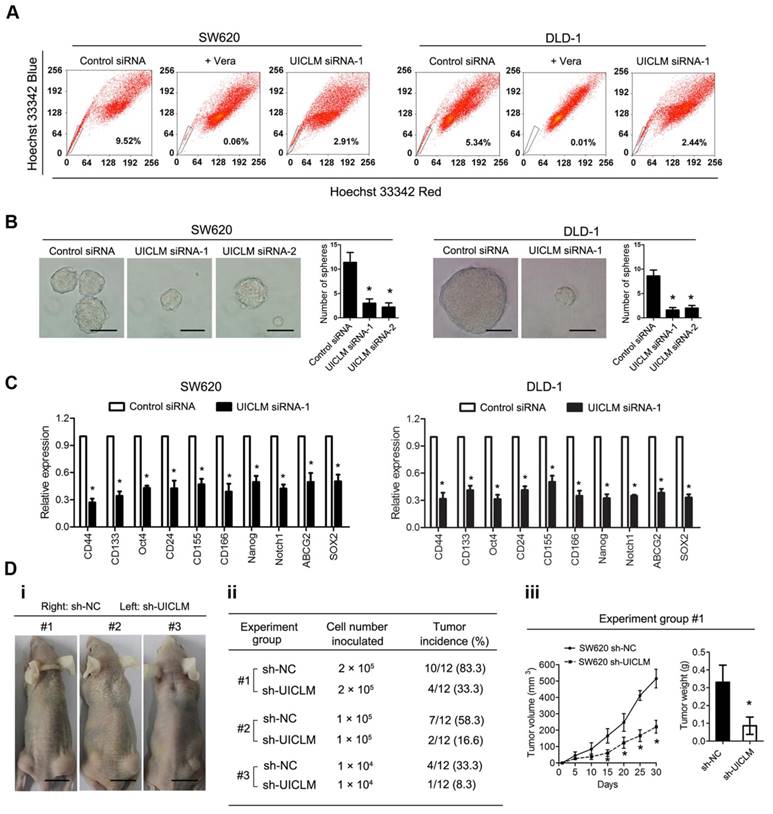
lncRNA UICLM regulates the stemness of CRC cells
To assess the role of UICLM in regulating the stemness of CRC cells, a fluorescence-activated cell sorting (FACS) technique that can exclude Hoechst 33342 dye was used to isolate CRC side population (SP) cells. The results showed that knockdown of UICLM could significantly reduce the SP cell proportion in SW620 and DLD-1 cells (Figure 4A). In addition, functional assays were performed to test whether UICLM could increase the stemness of CRC cells. The results showed that knockdown of UICLM significantly reduced sphere formation in SW620 and DLD-1 cells (*P < 0.05, Figure 4B). Furthermore, knockdown of UICLM could markedly down-regulate many stemness-associated genes (Nanog, Oct-4, SOX2, and Notch1), multiple drug-resistant transporter genes (ABCG2) and surface antigens associated with cancer stem cells (CD24, CD44, CD133, CD155 and CD166) (*P < 0.05, Figure 4C). Finally, a tumor formation assay was used to assess whether UICLM could increase in vivo tumorigenicity. The results demonstrated that more than 80% of the mice developed tumors when injected with 2.0 × 105 SW620 sh-NC cells, whereas the tumor incidence was reduced to 33.3% when mice were injected with SW620 sh-UICLM cells (Group #1) (Figure 4D i-ii). The tumor incidence was 58.3% in the control group when the mice were injected with 1.0 × 105 cells, and the tumor incidence was reduced to 16.6% in the sh-UICLM group (Group #2) (Figure 4D i-ii). Likewise, knockdown of UICLM dramatically reduced the tumor incidence from 33.3% to 8.3% when the mice were injected with 1.0 × 104 cells (Group #3) (Figure 4D i-ii). Consistently, compared with the sh-UICLM group, in the sh-NC group, the tumor volume and tumor weight were significantly decreased (*P < 0.05, Figure 4D iii).
UICLM regulates the stemness of CRC cells. (A) Knockdown of UICLM significantly reduced the SP cell proportion in SW620 (reduce from 9.52% to 2.91%) and DLD-1 (reduced from 5.34% to 2.44%) cells. (B) Knockdown of UICLM significantly reduced sphere formation in SW620 and DLD-1 cells (*P < 0.05) (Scale bar: 100 μm). (C) Knockdown of UICLM markedly reduced the expression of stemness-associated genes (Nanog, Oct-4, SOX2, Notch1), multiple drug-resistant transporter genes (ABCG2) and surface antigens associated with cancer stem cells (CD24, CD44, CD133, CD155 and CD166) in both SW620 and DLD-1 cells (*P < 0.05). (D)i-ii) 83.3% of the mice developed tumors when injected with 2.0 × 105 SW620 sh-NC cells, whereas the tumor incidence was reduced to 33.3% when mice were injected with SW620 sh-UICLM cells (Group #1). The tumor incidence was 58.3% in the control group when the mice were injected with 1.0 × 105 cells, and the tumor incidence was reduced to 16.6% in the sh-UICLM group (Group #2). The tumor incidence was 33.3% in the control group when the mice were injected with 1.0 × 104 cells, and the tumor incidence was reduced to 8.3% in the sh-UICLM group (Group #3). iii) The tumor volume and tumor weight were significantly decreased in the sh-NC group compared with that in the sh-UICLM group (*P < 0.05) (Scale bar: 1.5 cm).
UICLM interacts with ZEB2 to promote cell proliferation and invasion in CRC cells. (A) Schematic flowchart showing the results of a transcriptome sequencing study on UICLM-associated pathways. CRC cells were treated with siRNAs for UICLM or control siRNAs, and the mRNA expression profiles were determined. The combination of GSEA and gene expression correlation analysis identified ZEB2 in the cell migration pathway as a potential regulatory target of UICLM. (B) Knockdown of GAPLINC caused alterations in multiple pathways, and the “cellular migration and invasion” pathway showed the highest significance. This result was based on the following principle: when most genes in a defined pathway (gene set) are affected, a higher enrichment score is assigned to that pathway. (C) i)ZEB2 was significantly overexpressed in CRC with liver metastasis compared to CRC without liver metastasis, ii) A positive correlation was found between the expression level of ZEB2 and UICLM (r = 0.75, P < 0.001) (Scale bar: 100 μm). (D) The relative ZEB2 mRNA level in CRC cell lines (SW620, SW480, LoVo, HT-29, HCT116, DLD-1, RKO) and the immortalized colon epithelial cell line CCD-112CoN (*P < 0.05). (E) i) Knockdown of UICLM significantly reduced the expression of ZEB2 mRNA in SW620 and DLD-1 cells (*P < 0.05), ii) Ectopic expression of UICLM significantly increased the expression of ZEB2 mRNA (*P < 0.05), iii) Knockdown of UICLM significantly reduced the expression of ZEB2 protein, and ectopic expression of UICLM increased the ZEB2 protein level in SW620 and DLD-1 cells. (F) i-ii) The cell invasion stimulated by ectopic expression of UICLM could be repressed by knockdown of ZEB2 (*P < 0.05) (Scale bar: 100 μm).
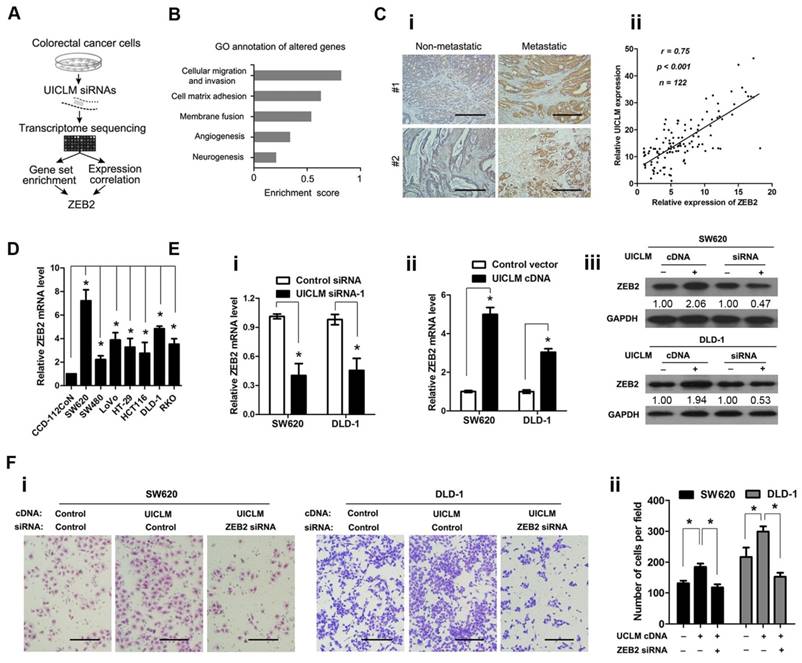
lncRNA UICLM interacts with ZEB2 to promote cell proliferation and invasion in CRC cells
To investigate the molecular mechanism by which UICLM is associated with CRC progression and metastasis, we explored the gene expression profiles of CRC cells with knocked down UICLM expression (schematic shown in Figure 5A). The SW620 cells were treated with specific siRNAs for UICLM, and the expression levels of all mRNAs were measured by RNA sequencing. The UICLM-associated pathways were determined by gene set enrichment analysis (GSEA), which determines whether different pathways show statistically significant differences between two groups. As shown in Figure 5B, the pathway “cellular migration and invasion” was assigned the highest enrichment score. By analysis of the correlation between UICLM and mRNAs from our lncRNA/mRNA microarray dataset, we found that ZEB2 was assigned the highest correlation coefficient in this pathway. Moreover, ZEB2 was one of the most down-regulated genes after knockdown of UICLM in SW620 cells in the RNA sequencing assay (Table S5). Based on these results, we supposed that a correlation might exist between UICLM and ZEB2. We used IHC and real-time PCR to measure the protein and mRNA levels of ZEB2, respectively, in 122 CRC patients. The results showed that ZEB2 was significantly overexpressed in CRC with liver metastasis compared to its expression in CRC without liver metastasis, and a positive correlation was found between the expression level of ZEB2 and UICLM (Figure 5C i-ii). Moreover, the expression level of ZEB2 was higher in CRC cell lines than in the immortalized colon epithelial cell line CCD-112CoN (*P < 0.05, Figure 5D). Knockdown/ectopic expression of UICLM could significantly reduce/increase the mRNA and protein level of ZEB2 in CRC cells (*P < 0.05, Figure 5E i-iii). Further functional study revealed that the cell invasion ability stimulated by UICLM overexpression could be suppressed by knockdown of ZEB2 (*P < 0.05, Figure 5F i-ii).
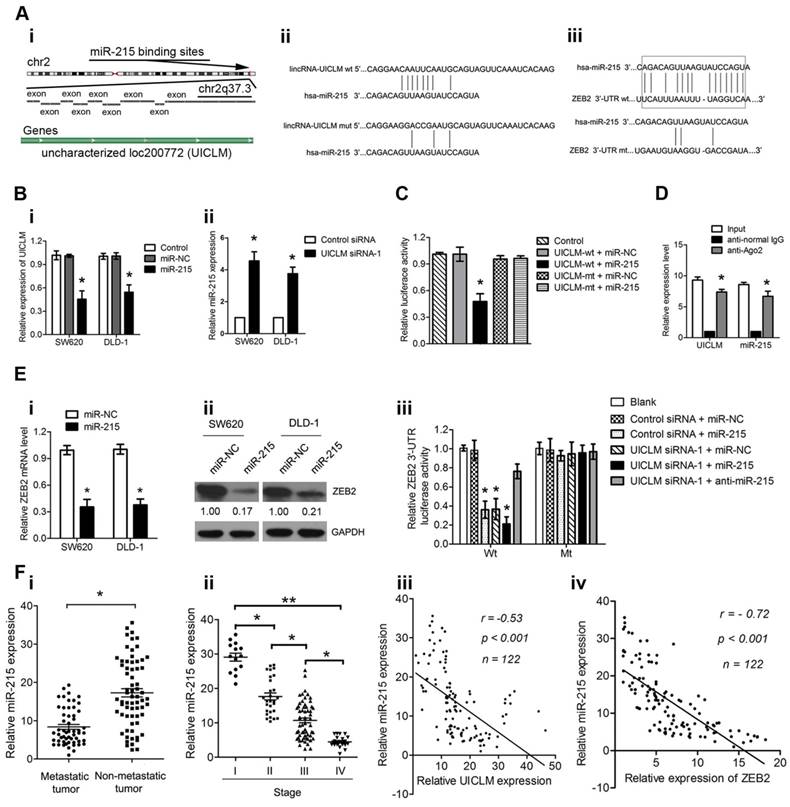
LncRNA UICLM regulates ZEB2 expression by competing for miR-215
It is known that lncRNAs can function as competing endogenous RNAs (ceRNAs) to protect mRNAs by competing for their targeting microRNAs. Therefore, we investigated whether UICLM played such a role. Using bioinformatics (miRcode Starbase v2.0, and RNAhybrid) tools, we found 7 miRNAs that putatively bound to UICLM (Table S6). Among these candidate miRNAs, miR-215 could directly bind to both UICLM and ZEB2 (Figure 6A i-iii). We found that overexpression of miR-215 significantly inhibited UICLM expression, while knockdown of UICLM significantly increased miR-215 expression (*P < 0.05, Figure 6B i-ii). We also performed experiments to determine whether UILCM could affect the processing of miR-215. As UICLM is located in the cytoplasm, we looked for whether UICLM could regulate the expression of pre-miR-215. The results showed that knockdown of UICLM in SW620 and DLD-1 cells did not affect the expression of pre-miR-215 (Figure S7). Dual-luciferase assays indicated a significant reduction in luciferase activities after co-transfection of miR-215 and a wild-type UICLM reporter vector, but not in mutant-type UICLM (*P < 0.05, Figure 6C). To determine whether UICLM and miR-215 are in the same RNA-induced silencing complex (RISC), we performed a RIP assay. The level of UICLM and miR-215 was higher in the anti-Ago2 group than that in the anti-normal IgG group (*P < 0.05, Figure 6D). To confirm the effect of miR-215 on ZEB2 expression, SW620 and DLD-1 cells were transfected with miR-215 mimics, and the ZEB2 mRNA and protein levels were determined. The results showed that ectopic expression of miR-215 significantly decreased the ZEB2 mRNA and protein levels in CRC cells. A luciferase activity assay further demonstrated that ectopic expression of miR-215 or knockdown of UICLM could significantly reduce the luciferase activity of wild-type but not mutant-type ZEB2 3'-UTR, and the reduction of luciferase activity caused by UICLM suppression could be restored by miR-215 inhibition (*P < 0.05, Figure 6E i-iii). Finally, we determined the expression of miR-215 in CRC patient tissues. Compared with tissues without metastasis, in tissues with metastasis, the miR-215 expression level was significantly down-regulated, and the miR-215 expression level showed a significant decrease with advancing clinical stage (*P < 0.05, **P < 0.01, Figure 6F i-ii). Moreover, an inverse association between miR-215 and UICLM as well as ZEB2 was found (both P < 0.001, r = -0.53, and r = -0.72, respectively, Figure 6F iii-iv).
Discussion
The development of distant metastasis is a complex process involving the accumulation of multiple genetic and epigenetic alterations that lead to the activation of oncogenes or the inactivation of tumor suppressor genes [21]. Increasing evidence has indicated that lncRNAs play an important role in tumor progression and metastasis [22]. In this study, we identified a set of lncRNAs involved in the pathology of CRC liver metastasis, among these lncRNAs, UICLM was one of the most differentially expressed lncRNAs between CRCs with and without liver metastasis, suggesting that it plays a critical role in the biological process of CRC liver metastasis. To the best of our knowledge, this is the first study to systematically evaluate the role of lncRNAs in CRC liver metastasis.
Increasing studies have suggested that lncRNAs are active biological molecules rather than “transcriptional noise” [23]. Given that UICLM was up-regulated in CRC with liver metastasis, we speculated that UICLM might affect the tumor phenotype of CRC. Our results showed that UICLM had a strong pro-malignant effect in CRC, with knockdown of UICLM inhibiting cell proliferation, colony formation, cell migration and invasion in vitro as well as tumor growth and liver metastasis in vivo. As EMT plays a key role in cell invasion and tumor metastasis [24, 25], we explored whether the EMT pathway was involved in UICLM-mediated cell motility. Interestingly, knockdown of UICLM lead to increased expression of many epithelial genes and decreased expression of mesenchymal genes. More importantly, we found that UICLM could affect the stemness of CRC cells. Knockdown of UICLM decreased the percentage of SP cells, leading to a reduction of self-renewal and tumor formation. This is in accordance with previous studies that indicated cancer stem cells are a source of cancer metastasis and progression, and there existed a direct link between the EMT and the gain of epithelial stem cell properties [26, 27]. By suppressing UICLM in CRC cells and performing RNA sequencing, we identified a close association between UICLM and ZEB2, a well-defined gene involved in cell migration, invasion and EMT [28, 29]. Knockdown of UICLM could reduce ZEB2 expression in CRC cells, whereas ectopic expression of UICLM enhanced ZEB2 expression. The pro-malignant effect stimulated by UICLM could be neutralized by ZEB2 suppression. These results demonstrated that UICLM modulated ZEB2 expression to regulate EMT and stem cell properties, finally affecting cell invasion and metastasis.
UICLM regulates ZEB2 expression by competing for miR-215. (A) i) Conservation of UICLM in the binding site of miR-215 predicted based on the human genome sequence in the UCSC Genome Browser, ii) Schematic representation of the predicted target site for miR-215 in UICLM, iii) The predicted miR-215 binding sites in ZEB2 mRNA 3'-UTR as predicted by the Targetscan algorithm.(B) i) Ectopic expression of miR-215 significantly reduced the level of UICLM in SW620 and DLD-1 cells (*P < 0.05), ii) Knockdown of UICLM significantly increased the expression of miR-215 in SW620 and DLD-1 cells (*P < 0.05). (C) Dual-luciferase assays indicated a significant reduction in luciferase activities after co-transfection of miR-215 and the wild-type UICLM reporter vector, but not the mutant-type UICLM (*P < 0.05). (D) A RIP assay was performed using input from cell lysates, anti-normal mouse IgG or anti-Ago2. The relative expression levels of UICLM and miR-215 were detected by quantitative real-time PCR (*P < 0.05). (E) i) Ectopic expression of miR-215 significantly reduced the expression of ZEB2 mRNA in SW620 and DLD-1 cells (*P < 0.05). ii) Ectopic expression of miR-215 significantly reduced ZEB2 protein levels in SW620 and DLD-1 cells. iii) The luciferase activity assay demonstrated that ectopic expression of miR-215 or knockdown of UICLM could significantly reduce the luciferase activity of wild-type but not mutant-type ZEB2 3'-UTR, and reduction of luciferase activity caused by UICLM suppression could be restored by miR-215 inhibition (*P < 0.05). (f) i) The relative expression of miR-215 in CRC tissues with and without liver metastasis (*P < 0.05). ii) The relative expression of miR-215 in different clinical stages; miR-215 was decreased in advanced stages compared with early stages (*P < 0.05, *P < 0.01). iii) An inverse correlation was found between the expression level of miR-215 and UICLM (r = -0.53, P < 0.001). iv)An inverse correlation was found between the expression level of miR-215 and ZEB2 (r = -0.72, P < 0.001).
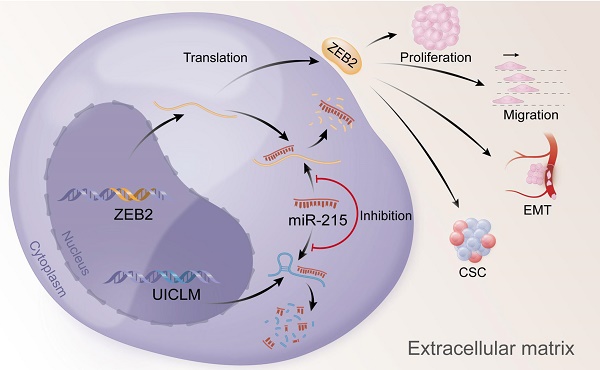
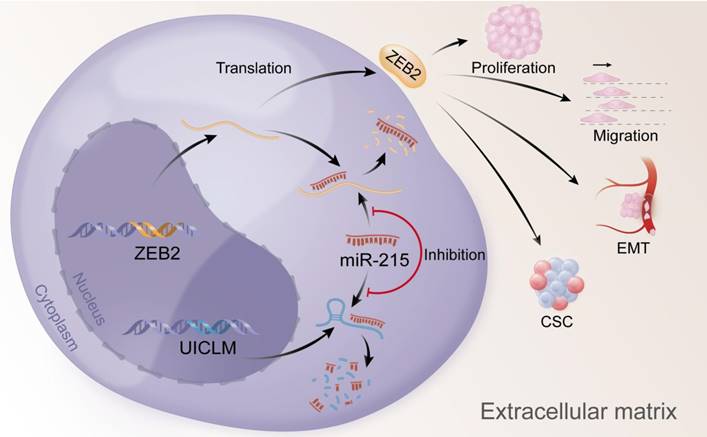
Overexpression of UICLM reduces the level of miR-215, which increases the expression level of ZEB2, thus resulting in stimulated cell proliferation, migration, EMT, and CSC formation in CRC.
Next, we explored the mechanism by which UICLM regulates ZEB2. Recently, increasing evidence showed that a novel regulatory mechanism exists between lncRNAs and miRNAs. lncRNAs can act as endogenous molecular sponges to compete for miRNAs, thereby negatively regulating miRNA expression [30]. For example, the lncRNA GAS5 can function as a ceRNA for miR-21 [31]; the lncRNA H19 promotes glioma cell invasion by deriving miR-675 [32]; and the lncRNA MEG3 functions as a competing endogenous RNA of miR-181s to regulate gastric cancer progression [33]. Using bioinformatics databases (Starbase v2.0, miRcode and RNAhybrid), we identified 7 miRNAs that may interact with UICLM. Among these miRNAs, miR-215 could bind to both UICLM and ZEB2. We confirmed the regulating relationship between UICLM and miR-215 based on the following: 1) knockdown of UICLM significantly increased miR-215 expression; 2) the direct binding ability of the predicted miR-215 binding site on UICLM was validated by luciferase activity assay; 3) Using RIP assays, we found that UICLM and miR-215 were in the same RISC; 4) an inverse correlation was found between the expression of UICLM and miR-215 in CRC tissues. In addition, we confirmed that miR-215 could directly target ZEB2 in CRC cells, which is in agreement with that in other tumor types [34, 35]. Consistent with our results, miR-215 has been identified as a tumor suppressor in many tumor types, such as pancreatic cancer [34], non-small cell lung cancer and colon cancer [36, 37], these data strongly suggest that UICLM acts as a ceRNA for miR-215 to regulate ZEB2 expression in CRC cells.
In conclusion, our findings offer insight into lncRNA-miRNA-mediated CRC liver metastasis and suggest that UICLM might be used as a new biomarker and therapeutic target for patients with CRC liver metastasis (Figure 7).
Abbreviations
CRC, colorectal cancer; lncRNA, long non-coding RNA; UICLM, up-regulated in colorectal cancer liver metastasis; IHC, immunohistochemistry; RT-PCR, real-time quantitative polymerase chain reaction; ISH, in situ hybridization.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No.81372570; No.81602053), the Natural Science Foundation of Guangdong Province (No. 2014A030312015; No. 2016A030310195; No. 2017A030313640), and the fourth outstanding young talents training plan of Sun Yat-sen University Cancer Center (No. PT04141001).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
2. Goldstein DA, Zeichner SB, Bartnik CM, Neustadter E, Flowers CR. Metastatic Colorectal Cancer: A Systematic Review of the Value of Current Therapies. Clin Colorectal Cancer. 2016;15:1-6
3. Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, Bouvier AM. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 2006;244:254-9
4. Chang GJ, Rodriguez-Bigas MA, Skibber JM, Moyer VA. Lymph node evaluation and survival after curative resection of colon cancer: systematic review. J Natl Cancer Inst. 2007;99:433-41
5. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861-74
6. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904-14
7. Liu Y, Ferguson JF, Xue C, Ballantyne RL, Silverman IM, Gosai SJ. et al. Tissue-specific RNA-Seq in human evoked inflammation identifies blood and adipose LincRNA signatures of cardiometabolic diseases. Arterioscler Thromb Vasc Biol. 2014;34:902-12
8. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145-66
9. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F. et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689-93
10. Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y. et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120-4
11. Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M. et al. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. 2014;5:e1008
12. Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS. et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446-61
13. Du Z, Sun T, Hacisuleyman E, Fei T, Wang X, Brown M. et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun. 2016;7:10982
14. Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D. et al. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299-310
15. Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X. et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370-81
16. Ju HQ, Lu YX, Chen DL, Tian T, Mo HY, Wei XL. et al. Redox Regulation of Stem-like Cells Though the CD44v-xCT Axis in Colorectal Cancer: Mechanisms and Therapeutic Implications. Theranostics. 2016;6:1160-75
17. Chen DL, Wang ZQ, Zeng ZL, Wu WJ, Zhang DS, Luo HY. et al. Identification of microRNA-214 as a negative regulator of colorectal cancer liver metastasis by way of regulation of fibroblast growth factor receptor 1 expression. Hepatology. 2014;60:598-609
18. Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797-806
19. Lu YX, Ju HQ, Wang F, Chen LZ, Wu QN, Sheng H. et al. Inhibition of the NF-kappaB pathway by nafamostat mesilate suppresses colorectal cancer growth and metastasis. Cancer Lett. 2016;380:87-97
20. Chen DL, Wang DS, Wu WJ, Zeng ZL, Luo HY, Qiu MZ. et al. Overexpression of paxillin induced by miR-137 suppression promotes tumor progression and metastasis in colorectal cancer. Carcinogenesis. 2013;34:803-11
21. Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449-60
22. Jiang C, Li X, Zhao H, Liu H. Long non-coding RNAs: potential new biomarkers for predicting tumor invasion and metastasis. Mol Cancer. 2016;15:62
23. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26-46
24. Kraljevic Pavelic S, Sedic M, Bosnjak H, Spaventi S, Pavelic K. Metastasis: new perspectives on an old problem. Mol Cancer. 2011;10:22
25. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871-90
26. Chen T, You Y, Jiang H, Wang ZZ. Epithelial-mesenchymal transition (EMT): A biological process in the development, stem cell differentiation, and tumorigenesis. J Cell Physiol. 2017;232:3261-72
27. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY. et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704-15
28. Li X, Roslan S, Johnstone CN, Wright JA, Bracken CP, Anderson M. et al. MiR-200 can repress breast cancer metastasis through ZEB1-independent but moesin-dependent pathways. Oncogene. 2014;33:4077-88
29. Lee JY, Park MK, Park JH, Lee HJ, Shin DH, Kang Y. et al. Loss of the polycomb protein Mel-18 enhances the epithelial-mesenchymal transition by ZEB1 and ZEB2 expression through the downregulation of miR-205 in breast cancer. Oncogene. 2014;33:1325-35
30. Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M. et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358-69
31. Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M. et al. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013;20:1558-68
32. Shi Y, Wang Y, Luan W, Wang P, Tao T, Zhang J. et al. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One. 2014;9:e86295
33. Peng W, Si S, Zhang Q, Li C, Zhao F, Wang F. et al. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J Exp Clin Cancer Res. 2015;34:79
34. Li QW, Zhou T, Wang F, Jiang M, Liu CB, Zhang KR. et al. MicroRNA-215 functions as a tumor suppressor and directly targets ZEB2 in human pancreatic cancer. Genet Mol Res. 2015;14:16133-45
35. Hou Y, Zhen J, Xu X, Zhen K, Zhu B, Pan R. et al. miR-215 functions as a tumor suppressor and directly targets ZEB2 in human non-small cell lung cancer. Oncol Lett. 2015;10:1985-92
36. Ye M, Zhang J, Miao Q, Yao L. Curcumin promotes apoptosis by activating the p53-miR-192-5p/215-XIAP pathway in non-small cell lung cancer. Cancer Lett. 2015;357:196-205
37. Jones MF, Hara T, Francis P, Li XL, Bilke S, Zhu Y. et al. The CDX1-microRNA-215 axis regulates colorectal cancer stem cell differentiation. Proc Natl Acad Sci U S A. 2015;112:E1550-8
Author contact
![]() Corresponding authors: Prof. Rui-hua Xu, M.D. Ph.D. or Prof. Huai-qiang Ju, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Department of Medical Oncology, Sun Yat-sen University Cancer Center, No. 651 Dong Feng East Road, Guangzhou 510060, China. Phone: +86-20-87343228; Fax: +86-20-87343392; E-mail: xurhorg.cn or Huai-qiang Ju, E-mail: juhqorg.cn.
Corresponding authors: Prof. Rui-hua Xu, M.D. Ph.D. or Prof. Huai-qiang Ju, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Department of Medical Oncology, Sun Yat-sen University Cancer Center, No. 651 Dong Feng East Road, Guangzhou 510060, China. Phone: +86-20-87343228; Fax: +86-20-87343392; E-mail: xurhorg.cn or Huai-qiang Ju, E-mail: juhqorg.cn.
 Global reach, higher impact
Global reach, higher impact