13.3
Impact Factor
Theranostics 2018; 8(1):277-291. doi:10.7150/thno.22065 This issue Cite
Research Paper
Tiresias: Context-sensitive Approach to Decipher the Presence and Strength of MicroRNA Regulatory Interactions
1. Electrical and Computer Engineering, Purdue University, West Lafayette, IN, USA;
2. Computer Science, Columbia University, New York, NY, USA;
3. Computer Science, Purdue University, West Lafayette, IN, USA.
Abstract
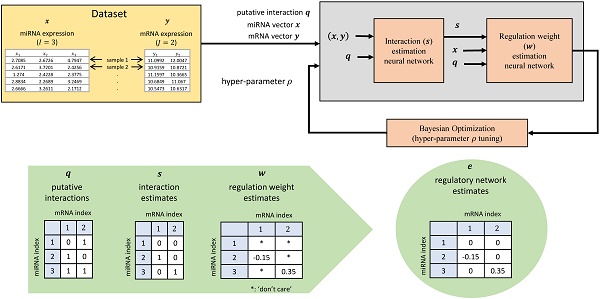
MicroRNAs (miRNAs) are short non-coding RNAs that regulate expression of target messenger RNAs (mRNAs) post-transcriptionally. Understanding the precise regulatory role of miRNAs is of great interest since miRNAs have been shown to play an important role in development, diseases, and other biological processes. Early work on miRNA target prediction has focused on static sequence-driven miRNA-mRNA complementarity. However, recent research also utilizes expression-level data to study context-dependent regulation effects in a more dynamic, physiologically-relevant setting.
Methods: We propose a novel artificial neural network (ANN) based method, named Tiresias, to predict such targets in a context-dependent manner by combining sequence and expression data. In order to predict the interacting pairs among miRNAs and mRNAs and their regulatory weights, we develop a two-stage ANN and present how to train it appropriately. Tiresias is designed to study various regulation models, ranging from a simple linear model to a complex non-linear model. Tiresias has a single hyper-parameter to control the sparsity of miRNA-mRNA interactions, which we optimize using Bayesian optimization.
Results: Tiresias performs better than existing computational methods such as GenMiR++, Elastic Net, and PIMiM, achieving an F1 score of >0.8 for a certain level of regulation strength. For the TCGA breast invasive carcinoma dataset, Tiresias results in the rate of up to 82% in detecting the experimentally-validated interactions between miRNAs and mRNAs, even if we assume that true regulations may result in a low level of regulation strength.
Conclusion: Tiresias is a two-stage ANN, computational method that deciphers context-dependent microRNA regulatory interactions. Experiment results demonstrate that Tiresias outperforms existing solutions and can achieve a high F1 score. Source code of Tiresias is available at
Keywords: MicroRNA, messenger RNA, machine learning, artificial neural networks, Bayesian optimization, breast cancer, condition-specific regulation.
 Global reach, higher impact
Global reach, higher impact