13.3
Impact Factor
Theranostics 2018; 8(2):328-340. doi:10.7150/thno.21917 This issue Cite
Research Paper
Development and Comparison of Two Immuno-disaggregation Based Bioassays for Cell Secretome Analysis
1. Department of Biomedical Engineering, University of Akron, Akron, Ohio 44325, United States;
2. Department of Mechanical Engineering, University of Akron, Akron, Ohio 44325, United States.
Received 2017-7-14; Accepted 2017-10-17; Published 2018-1-1
Abstract
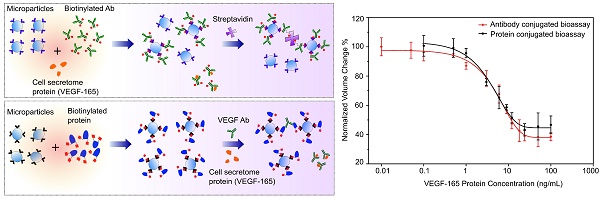
Cell secretome analysis has gained increasing attention towards the development of effective strategies for disease treatment. Analysis of cell secretome enables the platform to monitor the status of disease progression, facilitating therapeutic outcomes. However, cell secretome analysis is very challenging due to its versatile and dynamic composition. Here, we report the development of two immuno-disaggregation bioassays using functionalized microparticles for the quantitative analysis of the cell secretome.
Methods: We evaluated the feasibility of our developed immuno-disaggregation bioassays using antibody-conjugated MPs and protein-conjugated MPs for the detection of target cell secretome protein. The vascular endothelial growth factor (VEGF)-165 protein was tested as a model cell secretome protein in the serum and serum-free conditions. The status of MP aggregates was examined with a light microscopy and AccuSizerTM 780 Optical Particle Sizer. The accuracy of our bioassays measurement was compared with standard ELISA method.
Results: The developed bioassays successfully detected target VEGF protein present in serum-free buffer and serum-containing complete cell culture medium with high sensitivity and specificity. Additionally, the immuno-disaggregation bioassays using antibody-conjugated MPs and protein-conjugated MPs have a wide detection range from 0.01 ng/mL to 100 ng/mL and 0.5 ng/mL to 100 ng/mL, respectively. The sensitivity of the bioassay using antibody-conjugated MPs was approximately one order of magnitude higher than the bioassay using protein-conjugated MPs.
Conclusion: Our promising results indicate the potential of the developed bioassays as powerful platforms for the quantitative analysis of cell secretome.
Keywords: Immuno-disaggregation, competitive immunoreaction, cell secretome, vascular endothelial growth factor, microparticle.
Introduction
The importance of cell secretome analysis has been recognized in recent years due to the great potential of using cell secretome in disease treatment. The cell secretome represents the collection of proteins that are secreted and released from living cells1-3. Examples of secreted proteins in the cell secretome include growth factors, immunoregulatory cytokines, extracellular matrix-degrading enzymes, and cell motility factors4-6. Many groups have explored the potential applications of using secretome as new therapeutics and have reported encouraging in vivo results. The secretome obtained from porcine bone marrow-derived mesenchymal stem cells (MSCs), when injected into the infarcted porcine heart, resulted in significantly increased heart function, as evidenced by decreased expression of cardiac injury biomarkers and improved echocardiographic parameters7. The cell secretome produced by human adipose derived stem cells (ASCs) under the stimulation of tumor necrosis factor-α (TNF-α), when applied to rat excisional wound model, demonstrated accelerated wound closure, angiogenesis, and increased level of macrophage infiltration into the cutaneous wound as compared to the control groups8. The secretome of human peripheral blood mononuclear cells (MNCs), when used to treat spinal cord injured rat, has been shown to reduce acute axonal injury, attenuate cavity and to significantly enhance functional recovery compared to control animals9. These findings demonstrate the functional benefit of cell secretome for therapeutic applications. The ability to identify and quantify the specific proteins in cell secretome that are responsible for the therapeutic outcomes after cell secretome treatment will lead to the development of more effective strategies for disease treatment.
Cell secretome analysis is very challenging due to its versatile and dynamic composition. It has been reported that cell secretome is a complex soluble mixture consisting of various types of proteins and protein isoforms10,11. For example, the cell secretome of human adult cardiac progenitor cells (CPCs), when analyzed by reverse phase liquid chromatography and mass spectroscopy (LC-MS), have been shown to express more than 800 types of proteins12. In addition, the composition of cell secretome has been proven to be sensitive to environmental stimulations received by the cells. For instance, when grown as a monolayer under normoxic culture conditions, human mesenchymal stem cells (hMSCs) have been reported to secrete paracrine factors including vascular endothelial growth factor (VEGF; ~3.8 ng/mL), basic fibroblast growth factor (bFGF; ~30 pg/mL), interleukin 6 (IL-6; ~7.8 ng/mL), cathepsin B (~4.9 ng/mL), bone morphogenic protein 2 (BMP-2; ~12 pg/mL), and angiogenin (~0.9 ng/mL). However, when they were grown as 3D cellular spheroids under the same culture conditions, hMSCs have been demonstrated to secrete increased concentrations of VEGF (~28 ng/mL), bFGF (~46 pg/mL), IL-6 (~15 ng/mL), cathepsin B (~61 ng/mL), BMP-2 (~99 pg/mL), and angiogenin (~17 ng/mL)13. The protein concentration of cell secretome has been reported to be in a wide range covering from pg/mL to ng/mL level. Last but not least, the cell culture medium that contains the secreted proteins released from cells also possesses a high abundance of serum proteins, salts, and non-secreted proteins from dead cells, which would affect the accurate detection and analysis of the cell secretome14-16. To avoid the interference from serum proteins, serum-free culture medium has been used in many cell secretome analyses. However, the use of serum-deprived culture medium has been shown to affect cell functions (e.g., viability, proliferation and metabolism), which could potentially alter the composition of the cell secretome17,18. Other strategies have also been used to try and eliminate contaminants from the cell secretome prior to cell secretome analysis including centrifuge filtration and protein dialysis. Nevertheless, these methods result in protein loss during the procedures and could potentially decrease the sensitivity and accuracy of the downstream cell secretome analysis19.
Currently, the main technologies that have been successfully used in cell secretome analysis are enzyme-linked immunosorbent assay (ELISA) and liquid chromatography-mass spectroscopy (LC-MS). ELISA employs the immobilized specific antibody to capture the targeted protein from a mixture via high affinity antibody and antigen binding. It can capture and quantify the targeted protein with high specificity and sensitivity. However, ELISA is expensive, time-consuming and has a narrow protein concentration detection range (up to 1000 pg/mL)20,21. LC-MS utilizes a LC column to separate the protein mixture and MS to identify and measure the target protein. It can analyze multiple proteins at the same time to achieve high throughput. But LC-MS requires complicated instruments, well trained personnel and has been reported to have issues with low specificity and reproducibility22-24.
In this study, we aim to develop new bioassays using functionalized microparticles to facilitate analysis of the cell secretome. Our design principle uses the concept of competitive immunoreaction to create an inverse relationship between the average volume of microparticle (MP) aggregates and the concentration of the targeted protein. The higher the concentration of the targeted protein, the smaller the average volume of the MP aggregates. The lower the concentration of the targeted protein, the larger the average volume of the MP aggregates. Therefore, the protein concentration can be quantified from the volume of the MP aggregates. Based on this central design principle, we developed two distinct immuno-disaggregation based bioassays using different functionalized MPs and immunoreaction procedures. Vascular endothelial growth factor (VEGF)-165 was chosen to be used as a model protein of the cell secretome to validate our developed bioassays. VEGF-165 plays critical roles in angiogenesis and has been found in many cell secretomes. Here, we used the two developed bioassays to measure VEGF-165 samples with known concentrations in both serum-free and serum-containing solutions to prove the expected high sensitivity and specificity.
Experimental Section
Materials
Cell culture grade anti-biotin coated MACSiBeadTM microparticles and MACS BSA stock solution were purchased from Miltenyi Biotec (San Francisco, CA, USA). VEGF polyclonal antibody and biotinylated VEGF polyclonal antibody were purchased from ThermoFisher Scientific (Florence, KY, USA). Natural streptavidin protein was obtained from Abcam (Cambridge, MA, USA). Recombinant human VEGF-165 protein and biotinylated recombinant human VEGF protein were purchased from R&D Systems (Minneapolis, MN, USA). Cell culture grade 1X Phosphate Buffered Saline (PBS) was obtained from Corning (Manassas, VA, USA). MSCGM bullet kit was purchased from Lonza (Walkerville, MD, USA).
Immuno-disaggregation Bioassay using Antibody-conjugated Microparticles
Prior to the experiment, anti-biotin functionalized MPs (~3.5 µm nominal diameter) were mixed thoroughly with gentle agitation at room temperature to obtain a homogeneous suspension. The conjugation of biotinylated antibody to MPs was performed according to the manufacturer's recommendation. Briefly, the MPs were diluted to the concentration of 2x105 particles/mL and reacted with 800 ng/mL of biotinylated VEGF polyclonal antibody in 1X PBS containing 0.1% bovine serum albumin (BSA) buffer solution to form antibody-conjugated MPs. The conjugation reaction was carried out at 2-8ºC for 2 h with a constant rotation of 6 rpm by using a mini tube rotator (Fisher Scientific, Florence, KY, USA). After incubation, the solution mixture was centrifuged at 300 × g for 5 min at 4ºC and the supernatant containing unbound antibody was discarded. Next, the antibody-conjugated MPs were washed and resuspended in the desired volume of buffer solution. To induce the formation of MP aggregates through biotin-streptavidin binding, antibody-conjugated MPs (~5x104 counts) were reacted with various concentrations of natural streptavidin (SA) protein ranging from 250 ng/mL to 16000 ng/mL. The reaction mixture was incubated at 25ºC by rotating at a speed of 6 rpm for 45 min. The final reaction volume was maintained at 50 µL. An antibody-conjugated MPs control group (non-aggregates) was prepared by incubating antibody-conjugated MPs in a buffer solution under the same experimental conditions without SA protein.
For the immuno-disaggregation procedure, anti-biotin functionalized MPs at a concentration of 2x105 particles/mL were first mixed with 800 ng/mL of biotinylated VEGF polyclonal antibody in buffer solution. Subsequently, target cell secretome protein (recombinant human VEGF-165 protein) was added to the reaction mixture at the same time in concentrations varying from 0.01 ng/mL to 100 ng/mL. The mixture solution was then incubated at 2-8ºC for 2 h by maintaining a constant rotation of 6 rpm. After the incubation was complete, the solution was centrifuged and the supernatant containing unbound antibody and antibody-protein complexes was discarded. The obtained MPs were washed and resuspended in the desired volume of buffer solution. The aggregation of MPs was performed by mixing 5x104 of obtained antibody-conjugated MPs with 4000 ng/mL of SA protein for 45 min at 25ºC with a constant rotation of 6 rpm. The final reaction volume was maintained to 50 µL in buffer solution during the procedure.
Immuno-disaggregation Bioassay using Protein-conjugated Microparticles
The conjugation of biotinylated VEGF protein to MPs was carried out according to the recommendations from the manufacturer. Briefly, anti-biotin functionalized MPs prepared at a concentration of 2x105 particles/mL were conjugated with 800 ng/mL of biotinylated human VEGF protein in buffer solution containing 1X PBS/0.1% BSA. The conjugation reaction was performed at 2-8ºC for 2 h by rotating the sample mixture at a speed of 6 rpm. After conjugation, the unbound biotinylated VEGF protein was discarded from protein-conjugated MPs by centrifuging at 300 × g for 5 min at 4ºC. The obtained protein-conjugated MPs were then washed and resuspended in fresh buffer solution. Aggregation of MPs was performed by mixing 5x104 of protein-conjugated MPs with different concentrations of VEGF polyclonal antibody ranging from 50 ng/mL to 1600 ng/mL. The conjugation procedure was carried out at 25ºC for 45 min and 6 rpm rotation. The sample volume was maintained at 250 µL during the reaction step. Following the same protocol, the protein-conjugated MPs control group (non-aggregates) was prepared by incubating protein-conjugated MPs in buffer solution without incorporating the VEGF polyclonal antibody.
Immuno-disaggregation of MPs was performed by mixing 5x104 of protein-conjugated MPs with 400 ng/mL of the VEGF polyclonal antibody and various concentrations of cell secretome protein (recombinant human VEGF-165 protein) from 0.1 ng/mL to 100 ng/mL in 250 µL buffer solution. The reaction mixture was incubated at 25ºC for 45 min by rotating the samples at 6 rpm. The size and number of MP aggregates were then measured immediately after the incubation was complete to avoid non-specific interaction.
Preparation and Measurement of VEGF-165 Protein in Cell Culture Medium using Two Bioassays
To mimic the real cell secretome sample, known concentrations of target VEGF-165 protein was added to complete MSCGM Bullet kit supplemented with 1% antibiotic-antimycotic solution. The MP samples were prepared in 10% cell growth medium and 90% buffer solution containing target VEGF-165 protein at different concentrations of 0.01 ng/mL, 0.1 ng/mL, 1 ng/mL, 9 ng/mL, and 50 ng/mL. The prepared cell secretome samples were measured with both immuno-disaggregation bioassays using the respective experimental conditions mentioned above. For each experiment group, a negative control group that contained 10% cell culture medium was tested without target VEGF-165 protein. The normalized volume change percentage of MP samples obtained from cell culture medium were then compared with the samples prepared in 1X PBS buffer solution containing 0.1% BSA.
Measuring MP Aggregates
Microscope images of MP samples were captured with an inverted AxioVision A1 microscope (Carl Zeiss, Oberkochen, Germany). Following the manufacturer's recommendation, the counts and size distribution of MP samples were quantitatively measured with an AccuSizerTM 780 Optical Particle Sizer (Particle Sizing Systems, Port Richey, FL, USA) that has a detection range of 0.5-500 µm. Briefly, MP samples were diluted to ~3333 particles/mL in DI water (18.2 MΩ·cm; AQUA Solutions, Jasper, GA, USA) to a total volume of 15 mL. Once diluted, the samples were immediately measured using AccuSizer V software (Version 1.0; Particle Sizing Systems, Port Richey, FL, USA) at a flow rate of 10 mL/min. For analysis, the average aggregation volume (avg AV) and normalized volume change % were determined using the measured MPs whose size ranged from 1.5 µm to 10 µm for all the negative control and experimental groups.
Statistical Analyses
All experimental data are presented as mean ± standard error (SE) with three to eight replicates. Statistical analyses were performed using GraphPad Prism 5 software (GraphPad Software Inc., La Jolla, CA, USA). Multiple group comparisons were performed using one-way analysis of variance (ANOVA) and Tukey's post hoc test with a 95% confidence interval. Student's t-test was used for comparing the significant difference between two experimental groups. A p-value of less than 0.05 was considered to be statistically significant.
Results and Discussion
Design and Feasibility of the Immuno-disaggregation Bioassay using Antibody-conjugated MPs
A schematic design of the immuno-disaggregation bioassay using antibody-conjugated MPs is shown in Figure 1. For this design, the final measured MP aggregates are formed from MPs that are conjugated with biotinylated antibodies that specifically bound to target proteins in the cell secretome. The competitive reaction happens during the antibody conjugation procedure. As shown in Figure 1A, without the existence of target protein (VEGF-165), MPs conjugated with biotinylated VEGF antibodies form aggregates through specific biotin-streptavidin binding. However, if cell secretome containing the target protein is added during the antibody conjugation step, the available number of antibodies that are conjugated to MPs tend to decrease because of competitive binding of antibodies by the target protein. Therefore, under the same experimental conditions, the number or volume of the MP aggregates is reduced, as shown in Figure 1B. To test the feasibility of our design, we performed the following parallel experiments. For the negative control group, we conjugated MPs with biotinylated VEGF antibody in 0.1% BSA/1X PBS buffer solution containing no VEGF-165 protein and added streptavidin to form MP aggregates. For the experimental group, we conjugated MPs with biotinylated VEGF antibody in 0.1% BSA/1X PBS buffer solution containing 100 ng/mL VEGF-165 protein and added streptavidin to form MP aggregates. The MP aggregates formation was checked using a light microscope and the size and number of the formed aggregates were quantified with an AccuSizerTM 780 Optical Particle Sizing Systems (Particle Sizing Systems, Port Richey, FL, USA). In the negative control group, we observed the formation of MP aggregates (Figure 1C). We found that ~91.81% of MPs formed aggregates, of which, 58.50% consisted of 3 or 4 MPs per aggregate. In the experimental group, fewer MP aggregates were formed (Figure 1D). The percentage of MP aggregates consisting of more than 2 MPs were significantly reduced to 26.72%. Data from the particle size measurement also confirmed this trend. Average aggregation volume (avg AV) obtained from AccuSizerTM, which equals the total volume of MPs divided by the total particle counts, was used as the index to quantify the status of MP aggregates. The avg AV calculated from the negative control group (no VEGF-165) was 41.6 µm3, while the avg AV of the experimental group (100 ng/mL VEGF-165) was 27.4 µm3. Statistical analysis demonstrated a significant difference between the avg AVs derived from the negative control group and experiment group (Figure 1F). The decreased MP aggregation with the addition of VEGF proved the feasibility of our design.
Immuno-disaggregation bioassay using antibody-conjugated microparticles. (A, B) Schematic design of the immuno-disaggregation bioassay using VEGF as the example of targeted cell secretome protein. (C, D) Representative microscopy images of MP aggregates formed in the negative control group and the experiment group. (E) Representative microscopy image of MPs after antibody conjugation. (F) Comparison of average MP aggregate volumes between the negative group and experiment group (100 ng/mL of VEGF). Student's t-test showed statistically significant difference between the two groups with p-value < 0.01. Scale bar: 20 µm.
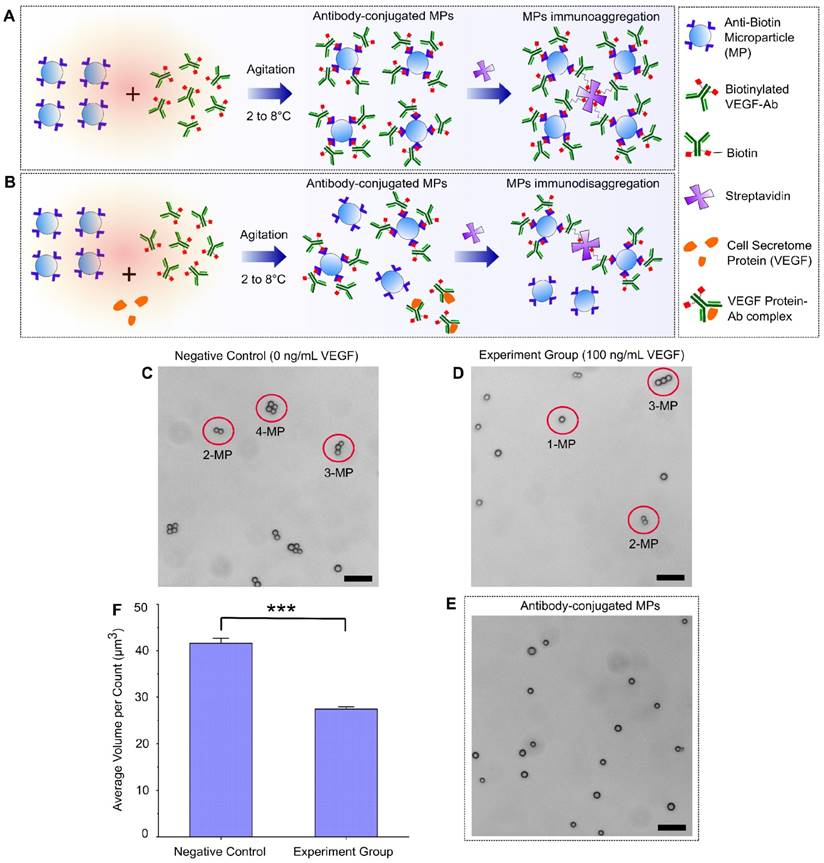
Several factors were identified via our feasibility testing that could affect the accuracy and sensitivity of this bioassay, which include antibody conjugation efficiency, MP self-aggregation and MP size distribution. The antibody conjugation procedure needs to be optimized to achieve maximal conjugation efficiency. In this experiment, we tested the effect of antibody concentration on conjugation efficiency. We found that the conjugation efficiency increased with increasing concentration of antibody until it reached a maximal plateau. The minimal concentration of antibody (800 ng/mL biotinylated VEGF antibody) that lead to maximal conjugation efficiency was chosen to be used in our bioassay. This allows maximal MP aggregation formation when used to test negative samples while avoiding the existence of excess unbound antibody that could quench the target protein during the process of competitive immunoreaction. MP self-aggregation could also affect the accuracy of our bioassay measurement. Therefore, we checked the biotinylated VEGF antibody-conjugated MPs using a microscope and size measurement. We did not observe any self-aggregation of our antibody-conjugated MPs (Figure 1E). The avg AV of MPs has no statistically significant difference before and after antibody conjugation. From the AccuSizerTM measurement, we did notice that the MPs we used have a relatively wide size distribution (nominal diameter range between 1.5 µm to 10 µm). We anticipate that the sensitivity of our immuno-disaggregation bioassay can be further enhanced by choosing MPs with a more uniform size distribution as reported by previous studies using MP-based bioassays25.
Development of Immuno-disaggregation Bioassay using Antibody-conjugated MPs
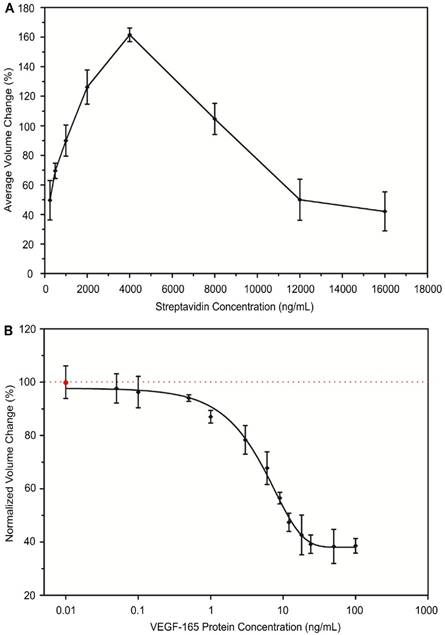
The optimal streptavidin (SA) concentration to achieve maximal aggregation of antibody-conjugated MPs was confirmed. SA, as the linker protein, binds the biotin molecules of the conjugated antibody to form aggregates. The concentration of SA affects the degree of MP aggregates formation. It has been reported that each SA molecule has four specific binding sites for biotin molecules26,27. Therefore, we expected that the optimal concentration of SA to achieve maximal aggregation of antibody-conjugated MPs is 4000 ng/mL using the 1:4 molar ratio of SA to biotin molecules. To confirm our estimation, we tested the effect of SA protein concentration on the aggregation of antibody-conjugated MPs. Eight different concentrations of SA ranging from 250 ng/mL to 16000 ng/mL were individually introduced to the same amount of biotinylated VEGF antibody-conjugated MPs to form aggregates under the same experimental conditions. The sizes of formed aggregates were measured and used to calculate the avg AV for comparison between groups. As shown in Figure 2A, the optimal SA concentration was 4000 ng/mL, as expected. The degree of aggregation increased with increasing concentration of SA until the optimal concentration was reached. After that, further increasing SA concentration negatively affected the aggregate formation. Similar phenomena have been reported by many other studies using specific biotin-streptavidin reaction28,29.
The inverse correlation between the degree of aggregation by antibody-conjugated MPs and the concentration of target protein in the cell secretome was examined. During the process of MP antibody conjugation, samples containing various concentrations of VEGF-165 were introduced. Then, VEGF antibody-conjugated MPs were triggered to form aggregates using the optimal concentration of SA (4000 ng/mL) and the degree of aggregation was measured. For each tested VEGF-165 concentration, a negative control was performed in parallel using sample containing no VEGF-165. The ratio between avg AV derived from the experimental group and the negative control group under the same experimental conditions was used to calculate the normalized volume change percentage to be compared between groups. Our results demonstrated an inverse relationship between the degree of aggregation by VEGF antibody-conjugated MPs and the concentration of VEGF-165 (Figure 2B). When the concentration of VEGF-165 increased from 0.01 ng/mL to 100 ng/mL, the normalized volume change percentage decreased from 99.8% to 39.4%. Using the inverse correlation, a standard curve was generated and sigmoidal dose-response curve fitting was performed, which produced a coefficient of determination (R2) of the fitting curve of ~0.86. Because of the specific biotin-streptavidin interaction, the bioassay was very selective over BSA and components of complete cell culture medium including serum during the conjugation reaction.
Design and Feasibility of the Immuno-disaggregation Bioassay using Protein-conjugated MPs
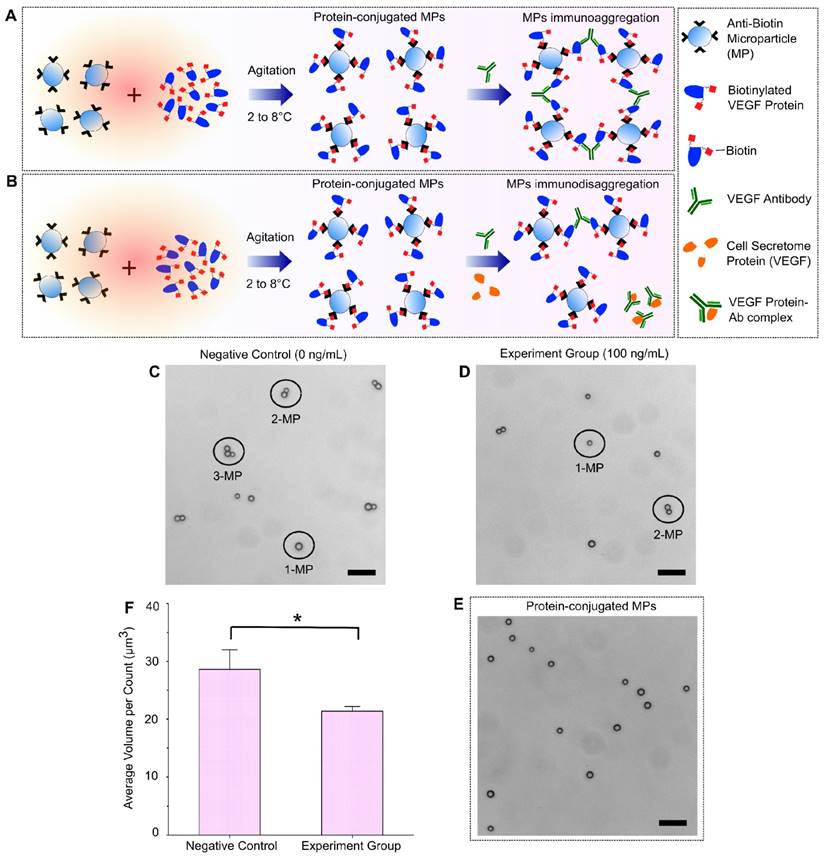
A schematic illustration of the immuno-disaggregation bioassay design using protein-conjugated MPs is demonstrated in Figure 3. In this bioassay, the measured MP aggregates were formed through high affinity antigen-antibody binding using MPs that were conjugated with biotinylated proteins. The competitive immunoreaction happens during the step of MP aggregates formation. As shown in Figure 3A, in the absence of target cell secretome protein (VEGF-165), aggregates are formed by the specific antibody-antigen binding between the added VEGF antibody and the VEGF proteins conjugated to the MPs. If cell secretome containing VEGF-165 is added during the aggregates formation, it competes with the VEGF conjugated with the MPs to bind VEGF antibody and therefore reduce the degree of MP aggregation (Figure 3B). We evaluated the feasibility of our immuno-disaggregation bioassay using protein-conjugated MPs by performing the following experiments. For the negative control group, the MPs were first conjugated with biotinylated VEGF protein following the protocol described in the Materials and Methods section. MP aggregates were then formed by adding VEGF antibody without cell secretome protein (VEGF-165). For the experiment group, under the same experimental conditions, we conjugated MPs with biotinylated VEGF protein and reacted them with VEGF antibody and target VEGF-165 cell secretome protein (100 ng/mL) at the same time to form MP aggregates. The MP aggregates formed in the negative control group and the experiment group were observed under a light microscope. As shown in Figure 3C, ~77.49% of MPs formed aggregates in the negative control group. Of all the formed aggregates, ~34.01% of aggregates contained more than 2 MPs. Less formation of MP aggregates was found in the experiment group (57.48%) and the number of MP aggregates with more than 2 MPs was significantly decreased to 28.96% (Figure 3D). The avg AV of experiment group (~21.38 µm3) was significantly reduced as compared to the negative control group (~28.63 µm3).
Development of immuno-disaggregation bioassay using antibody-conjugated MPs. (A) Confirmation of the optimal linker protein concentration to enable maximal aggregates formation. (B) Examining the inverse relationship between degree of antibody-conjugated MPs aggregation and target protein concentration. Red dotted line denotes 100% volume change.
We examined the factors that could potentially affect the sensitivity and efficacy of the immuno-disaggregation bioassay using protein-conjugated MPs. The conjugation of biotinylated protein to MPs is a critical step and needs to be optimized to obtain maximum conjugation efficiency while avoiding non-specific interaction. To optimize the conjugation of biotinylated protein to MPs, we performed the following experiments. First, the effect of biotinylated protein concentration on conjugation efficiency was examined. We found that the conjugation efficiency was directly related to the concentration of biotinylated protein. After the conjugation efficiency reached a plateau, it remained constant as the maximal conjugation efficiency. We chose the minimum concentration of protein (800 ng/mL biotinylated VEGF protein) that resulted in maximum conjugation efficiency for the development of this bioassay. Secondly, after conjugating biotinylated protein to MPs, samples were checked to see if the biotinylated proteins detach from MPs during the conjugation procedure, which would mix with the protein in the cell secretome and affect the accuracy of the bioassay. To check for detachment, VEGF-conjugated MPs were incubated in PBS buffer solution under the same experimental conditions as the negative control group (without VEGF antibody) to go through the whole aggregate formation procedure. After that, the supernatant was collected and the protein amount in the PBS buffer was measured using micro BCA protein assay kit (Thermo Scientific, Rockford, IL, USA). Our results confirmed that there was no significant difference in total protein content in the PBS buffer before and after the process, which proved that the conjugated biotinylated proteins would not detach from MPs to affect the bioassay measurement. Additionally, we checked the self-aggregation of protein-conjugated MPs using a light microscope and particle size measurement. Microscopy images showed no self-aggregation of our protein-conjugated MPs (Figure 3E). The MP size measurement from AccuSizerTM also indicated no statistical difference in avg AV of MPs before or after protein conjugation. Lastly, the same MPs were used in this bioassay and the previous bioassay to eliminate any differences caused by the MPs when comparing the two bioassays.
Immuno-disaggregation bioassay using protein-conjugated MPs. (A, B) Schematic illustration of immuno-disaggregation bioassay designed for the detection of target VEGF-165 cell secretome protein. (C, D) Representative microscopy images of MP aggregation in the negative control group and the experiment group. (E) Microscopy image of protein-conjugated MPs. (F) Measurement of average MP aggregate volumes in the negative control group and the experiment group (100 ng/mL of VEGF). Student's t-test showed a statistically significant difference between experimental groups with p-value < 0.05. Scale bar: 20 µm.
Development of Immuno-disaggregation Bioassay using Protein-conjugated MPs
The optimal concentration of VEGF antibody was examined to achieve the maximum aggregation of protein-conjugated MPs. In this design, VEGF antibody was used as a linker that binds to VEGF proteins that are conjugated to MPs with high affinity and facilitates MP aggregation. The degree of MP aggregation is dependent upon the concentration of VEGF antibody during the conjugation reaction. The optimum ratio of VEGF antibody to biotinylated VEGF protein that is conjugated to MPs leads to maximum aggregation of MPs through specific antigen-antibody binding. To achieve the optimal ratio, we tested the effect of VEGF antibody concentration on the formation of MP aggregates. We examined seven different concentrations of VEGF antibody ranging from 50 ng/mL to 1600 ng/mL. Under the same experimental conditions, each VEGF antibody concentration was mixed with the same amount of biotinylated VEGF protein-conjugated MPs to form aggregates. Once the aggregates formed, the size of MP aggregates was quantitatively measured with an AccuSizerTM. We achieved maximum aggregation of protein-conjugated MPs (92.71%) when the concentration of VEGF antibody was 400 ng/mL. As shown in Figure 4A, the aggregation of protein-conjugated MPs was directly related to the concentration of VEGF antibody until it reached the maximum conjugation efficiency. Afterwards, if the linker concentration was increased, the amount of VEGF antibody would be excessive, resulting in reduction of MP aggregation. Similar trends have been previously reported by many groups using MP-based immuno-aggregation assays30,31.
Development of immuno-disaggregation bioassay using protein-conjugated MPs. (A) Optimization of VEGF antibody concentration to achieve maximum MP aggregation. (B) Correlation of protein-conjugated MP aggregation and target cell secretome protein concentration (VEGF-165) showing an inverse relationship. Red dotted line denotes 100% volume change.
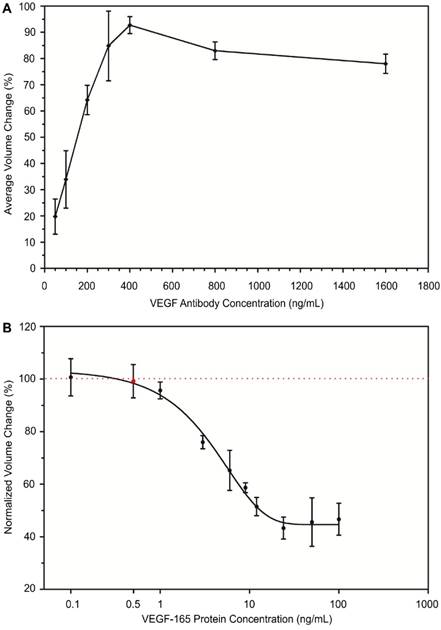
The relationship between the normalized volume change percentage of the formed MP aggregates using protein-conjugated MPs and concentration of target cell secretome protein (VEGF-165) was assessed. The VEGF protein-conjugated MPs were mixed with VEGF antibody (400 ng/mL) and samples containing various concentrations of VEGF-165 to form aggregates. For each experiment group, a negative control group was performed in parallel with MP samples containing no VEGF-165. The size and number of MP aggregates were measured from both groups. Then the normalized volume change percentage was determined by dividing avg AV obtained from the experiment group with that of the negative control group. Our results from the quantitative measurement of MP aggregates showed an inverse relationship between the normalized volume of formed aggregates and target VEGF-165 protein concentration. The normalized volume change percentage decreased from 100.5% to 46.6% when the concentration of VEGF-165 was increased from 0.1 ng/mL to 100 ng/mL (Figure 4B). The measurement range of this bioassay was 0.5 ng/mL to 100 ng/mL. A standard curve showing the correlation between the normalized volume change percentage and cell secretome protein concentration was generated using sigmoidal dose-response curve fitting (R2 ~ 0.80). The presence of BSA and cell culture medium components did not interfere with the target cell secretome protein detection, suggesting the selectivity of our developed bioassay due to specific antibody-antigen interaction.
Parallel Comparisons of Cell Secretome Detection using Two Immuno-disaggregation Bioassays
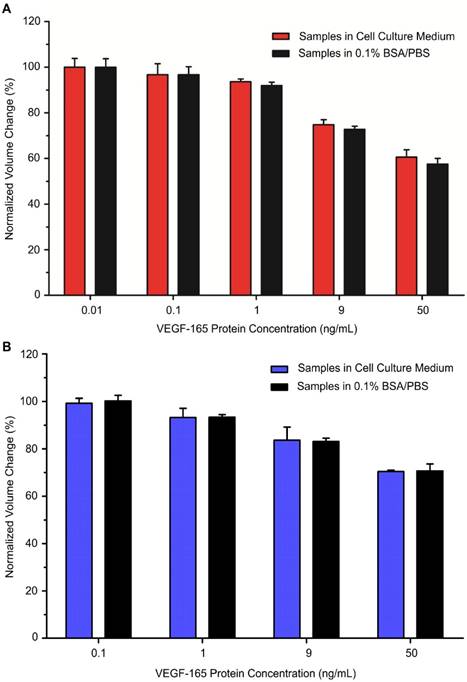
Using our developed bioassays, we tested the MP samples with various concentrations of model cell secretome protein (VEGF-165) that were prepared in serum-containing cell culture medium. Out of the total sample volume, each cell secretome sample contained 10% of complete cell culture medium. The cell secretome samples were quantitatively measured with an AccuSizerTM and the results were compared with corresponding data from the standard curves (Figures 2B and 4B). For each tested cell secretome protein concentration, a negative control group was prepared in parallel using cell culture medium containing no VEGF-165. For comparison between groups, the ratio between avg AV derived from the experimental group and the negative control group under the same experimental conditions was used to determine the normalized volume change percentage. Our data confirmed that target cell secretome protein (VEGF-165) prepared in serum-containing cell culture medium was detected with high specificity and sensitivity using our developed bioassays. For the immuno-disaggregation bioassay using antibody-conjugated MPs, the degree of MP aggregation obtained from cell culture medium samples had no statistically significant difference as compared to the standard curve (100% vs. 99.9% for 0.01 ng/mL; 96.7% vs. 96.7% for 0.1 ng/mL; 93.6% vs. 91.9% for 1 ng/mL; 74.8% vs. 72.7% for 9 ng/mL; 60.5% vs. 57.5% for 50 ng/mL) (Figure 5A). For the immuno-disaggregation bioassay using protein-conjugated MPs, compared to the standard curve, we did not find a significant difference in MP aggregation measured from cell culture medium samples (100% vs. 99.2% for 0.1 ng/mL; 93.3% vs. 93.2% for 1 ng/mL; 83.1% vs. 83.6% for 9 ng/mL; 70.7% vs. 70.4% for 50 ng/mL) (Figure 5B). To ensure the reproducibility of our bioassays, the coefficient of variation (CV) was measured from 4-8 independent samples. CV% of prepared cell culture samples were 10% or less from both immuno-disaggregation bioassays, indicating the high reproducibility of our detection methods.
Our developed immuno-disaggregation bioassays have the ability to measure a wide concentration range of cell secretome proteins in a serum environment. The cell secretome usually contains low concentrations of target secreted proteins and a high abundance of serum proteins (e.g., fetal bovine serum). For example, human monocytic cells, in response to inflammatory stimuli, have been shown to secrete cytokines including interleukin 1 beta (IL-1β), interleukin 8 (IL-8), and tumor necrosis factor (TNF) in the cell secretome at various concentrations ranging from ~500 pg/mL to 20,000 pg/mL6,32. In addition, to achieve real profiles of cell secretome, analysis of secreted proteins obtained from serum-containing growth medium is important. It provides quantitative information that reflects the true status of cells undergoing dynamic changes during in vitro culture18. In the present work, we demonstrated the development of two parallel immuno-disaggregation bioassays that are able to quantify secreted proteins present in serum-containing cell culture medium with high specificity and sensitivity. In the immuno-disaggregation bioassay using antibody-conjugated MPs, the MPs were conjugated with biotinylated VEGF antibody. MP aggregates were formed by reacting antibody-conjugated MPs with an optimum concentration of natural SA protein, which bound the biotin molecules of the conjugated antibodies. The target cell secretome protein was added during the biotinylated antibody conjugation step to induce competitive immunoreaction. In the immuno-disaggregation bioassay using protein-conjugated MPs, the MPs were conjugated with biotinylated VEGF protein and added VEGF antibody to form MP aggregates through specific antibody-antigen interaction. The competitive immunoreaction was induced by adding the target cell secretome protein during the MP aggregation step. The sensitivity of the immuno-disaggregation bioassay using antibody-conjugated MPs was approximately one order of magnitude higher than the immuno-disaggregation bioassay using protein-conjugated MPs, resulting in the former bioassay being a suitable candidate for detection of low concentrations of molecular targets. Moreover, our developed bioassays have a common detection range covering 0.5 ng/mL to 100 ng/mL, which can measure cell secretome proteins as low as 500 pg/mL. Compared to the immuno-disaggregation bioassay using protein-conjugated MPs, the antibody-conjugated MPs-based immuno-disaggregation approach can measure cell secretome proteins down to 10 pg/mL, a level of sensitivity that is comparable to conventional ELISA. In this work, we have successfully demonstrated the feasibility of detecting target cell secretome protein in serum-containing cell culture medium using our developed bioassays. Although cell lines could be used to mimic clinical specimens, they cannot address the complex variability present in clinical samples. Therefore, the next critical step will be measuring the real clinical samples that aim to improve therapeutic outcomes. In the future study, we plan to investigate applications in detecting patients' samples that could facilitate disease diagnosis and monitoring.
Detection of cell secretome protein using developed immuno-disaggregation bioassays. (A) Immuno-disaggregation bioassay using antibody-conjugated MPs. (B) Immuno-disaggregation bioassay using protein-conjugated MPs. Student's t-test showed no significant difference between samples prepared in buffer solution and cell culture medium at each corresponding concentration.
MP-based immuno-aggregation strategies have been extensively used to achieve high sensitivity protein analysis33-40. The MPs are functionalized with recognition biomolecules to capture and detect target analytes. For example, Yu et al. had developed colorimetric metal-linked immunosorbent assay (MeLISA) for the detection of biomarkers in the serum samples41. MeLISA utilizes the sandwich-type immunoreaction to capture target proteins between the capture antibodies that are coated on polystyrene plate and the detection antibodies conjugated with silver nanoparticles (AgNPs). Upon the addition of gold nanoparticles (AuNPs) solution containing hydrogen peroxide (H2O2), the silver ions are produced leading to the formation of AuNPs aggregation that yields a distinct color change depending upon the concentration of AgNPs bound to target protein. Similarly, Zhao et al. demonstrated an immunofluorescence assay using functionalized AgNPs for the detection of α-fetoprotein (AFP), C-reactive protein (CRP), and human serum samples34. Compared with other immunoassays including conventional ELISA, which focuses on a two-dimensional approach to protein targeting, MP-based immuno-aggregation methods offer several advantages such as high surface to volume ratio and enhanced assay kinetics for biomolecular binding42,43. Additionally, they require simple sample preparation in order to not compromise reproducibility and are highly specific. However, a MP-based immuno-aggregation approach employs non-competitive molecular binding between recognition biomolecules and target proteins that usually requires a large amount of targets to form MP aggregates, which could compromise the sensitivity of the assay. For instance, Thanh et al. reported the detection of Protein A antibody in serum through aggregation-based immunoassay that utilizes Protein A-functionalized gold nanoparticles. After mixing the functionalized gold nanoparticles with different concentrations of target Protein A antibody, the formed aggregates showed distinct absorption value differences depending upon the rate of aggregation. The limit of detection of the assay was ~1 µg/mL31. Compared to a MP-based immuno-aggregation method, the measurement of target proteins using an immuno-disaggregation approach could enable higher sensitivity, facilitating the detection of low concentrations of molecular targets24. For example, Chen et al. used an immuno-disaggregation approach to detect Kanamycin in milk and was able to achieve a limit of detection of 0.1 ng/mL33. Xiang et al. also reported an immuno-disaggregation method to develop personal glucose meters with a detection limit of 0.4 ng/mL37. The immuno-disaggregation strategy involves a competitive immunoreaction induced by the target protein, which reduces the volume of MP aggregates with increasing concentration of the target. Because the assay is designed for an inverse relationship between target protein and volume of MP aggregates, it requires a small amount of target protein to induce a volume change in MP aggregates, which results in improved sensitivity. To the best of our knowledge, this is the first study using immuno-disaggregation approaches for cell secretome analysis. In addition to using antibody-conjugated MPs to achieve immuno-disaggregation as reported by other groups, we also developed a protein-conjugated MPs-based approach. The side by side comparison of both immuno-disaggregation approaches will provide valuable information for future bioassay development.
Conclusions
In the present study, two innovative immuno-disaggregation based bioassays were developed for the quantitative analysis of the cell secretome. Using VEGF-165 protein as our model cell secretome protein, we demonstrated the concept of competitive immunoreaction in our conjugation procedure that resulted in an inverse relationship between the volume of MP aggregates and target protein concentration. The formation of MP aggregates was dependent upon the concentration of target protein. Based on the degree of MP aggregation, the amount of target proteins present in serum-containing complete cell culture medium could be accurately detected with high specificity. The developed bioassays offer a wide common detection range between 0.5 ng/mL to 100 ng/mL. Compared to our bioassay using protein-conjugated MPs, the antibody-conjugated MPs based bioassay can measure target protein concentration as low as 0.01 ng/mL, a sensitivity that is comparable to conventional ELISA. Moreover, our developed bioassays can be used to detect and measure any secreted proteins present in the cell secretome as long as biotinylated probes against the target protein of interest are available. These attractive results indicate the potential of our detection strategies as suitable candidates in disease treatment and monitoring.
Abbreviations
MSCs: Mesenchymal stem cells; MSCGM: Mesenchymal stem cell growth medium; ASCs: Adipose derived stem cells; TNF-α: Tumor necrosis factor-α; CPCs: Cardiac progenitor cells; LC-MS: Liquid chromatography and mass spectroscopy; VEGF: Vascular endothelial growth factor; bFGF: Basic fibroblast growth factor; IL-6: Interleukin 6; BMP-2: Bone morphogenic protein 2; ELISA: Enzyme-linked immunosorbent assay; MP: Microparticle; SA: Streptavidin; avg AV: Average aggregation volume; BSA: Bovine serum albumin; PBS: Phosphate buffered saline; Ab: Antibody.
Acknowledgements
This work was supported by the National Science Foundation (DBI-IDBR-1353720 and ECCS 1625544).
Supplementary Material
Supplementary figures and tables.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mukherjee P, Mani S. Methodologies to Decipher the Cell Secretome. Biochim Biophys Acta. 2013. 1834(11):2226-2232
2. Chaudhuri S, Maurya P, Kaur M, Tiwari A, Borth N, Bhatnagar S, Kumar N. Investigation of CHO Secretome: Potential Way to Improve Recombinant Protein Production from Bioprocess. J Bioprocess Biotech. 2015:5 (7)
3. Stastna M, Van Eyk JE. Secreted Proteins as a Fundamental Source for Biomarker Discovery. Proteomics. 2012;12(4-5):722-735
4. Xue H, Lu B, Lai M. The Cancer Secretome: A Reservoir of Biomarkers. J Transl Med. 2008;6(1):52
5. Blaber SP, Webster RA, Hill CJ, Breen EJ, Kuah D, Vesey G, Herbert BR. Analysis of in Vitro Secretion Profiles from Adipose-Derived Cell Populations. J Transl Med. 2012;10(1):172
6. Skalnikova KH. Proteomic Techniques for Characterisation of Mesenchymal Stem Cell Secretome. Biochimie. 2013;95(12):2196-2211
7. Nguyen BK, Maltais S, Perrault LP, Tanguay JF, Tardif JC, Stevens LM, Borie M, Harel F, Mansour S, Noiseux N. Improved Function and Myocardial Repair of Infarcted Heart by Intracoronary Injection of Mesenchymal Stem Cell-Derived Growth Factors. J Cardiovasc Transl Res. 2010;3(5):547-558
8. Heo SC, Jeon ES, Lee IH, Kim HS, Kim MB, Kim JH. Tumor Necrosis Factor-α-Activated Human Adipose Tissue-Derived Mesenchymal Stem Cells Accelerate Cutaneous Wound Healing through Paracrine Mechanisms. J Invest Dermatol. 2011;131(7):1559-1567
9. Haider T, Höftberger R, Rüger B, Mildner M, Blumer R, Mitterbauer A, Buchacher T, Sherif C, Altmann P, Redl H, Gabriel C, Gyöngyösi M, Fischer MB, Lubec G, Ankersmit HJ. The Secretome of Apoptotic Human Peripheral Blood Mononuclear Cells Attenuates Secondary Damage Following Spinal Cord Injury in Rats. Exp Neurol. 2015;267:230-242
10. Polisetty RV, Gupta MK, Nair SC, Ramamoorthy K, Tiwary S, Shiras A, Chandak GR, Sirdeshmukh R. Glioblastoma Cell Secretome: Analysis of Three Glioblastoma Cell Lines Reveal 148 Non-Redundant Proteins. J Proteomics. 2011;74(10):1918-1925
11. Drago D, Cossetti C, Iraci N, Gaude E, Musco G, Bachi A, Pluchino S. The Stem Cell Secretome and Its Role in Brain Repair. Biochimie. 2013;95(12):2271-2285
12. Sharma S, Mishra R, Bigham GE, Wehman B, Khan MM, Xu H, Saha P, Goo YA, Datla SR, Chen L, Tulapurkar ME, Taylor BS, Yang P, Karathanasis S, Goodlett DR, Kaushal S. A Deep Proteome Analysis Identifies the Complete Secretome as the Functional Unit of Human Cardiac Progenitor Cells Novelty and Significance. Circ Res. 2017;120(5):816-834
13. Potapova IA, Gaudette GR, Brink PR, Robinson RB, Rosen MR, Cohen IS, Doronin SV. Mesenchymal Stem Cells Support Migration, Extracellular Matrix Invasion, Proliferation, and Survival of Endothelial Cells In Vitro. Stem Cells. 2007;25(7):1761-1768
14. Parѐ B, Deschѐnes LT, Pouliot R, Duprѐ N, Gros-Louis F. An Optimized Approach to Recover Secreted Proteins from Fibroblast Conditioned-Media for Secretomic Analysis. Front Cell Neurosci. 2016;10:70
15. Chevallet M, Diemer H, Van Dorssealer A, Villiers C, Rabilloud T. Toward a Better Analysis of Secreted Proteins: The Example of the Myeloid Cells Secretome. Proteomics. 2007;7(11):1757-1770
16. Makridakis M, Vlahou A. Secretome Proteomics for Discovery of Cancer Biomarkers. J Proteomics. 2010;73(12):2291-2305
17. Brown KJ, Formolo CA, Seol H, Marathi RL, Duguez S, An E, Pillai D, Nazarian J, Rood BR, Hathout Y. Advances in the Proteomic Investigation of the Cell Secretome. Expert Rev Proteomics. 2012;9(3):337-345
18. Weng Y, Sui Z, Shan Y, Jiang H, Zhou Y, Zhu X, Liang Z, Zhang L, Zhang Y. In-Depth Proteomic Quantification of Cell Secretome in Serum-Containing Conditioned Medium. Anal Chem. 2016;88(9):4971-4978
19. Stastna M, Van Eyk JE. Investigating the Secretome: Lessons About the Cells That Comprise the Heart. Circ Cardiovasc Genet. 2012;5(1):o8-o18
20. Chen Y, Xianyu Y, Sun J, Niu Y, Wang Y, Jiang X. One-Step Detection of Pathogens and Cancer Biomarkers by the Naked Eye Based on Aggregation of Immunomagnetic Beads. Nanoscale. 2016;8(2):1100-1107
21. Leng SX, McElhaney JE, Walston JD, Xie D, Fedarko NS, Kuchel GA. ELISA and Multiplex Technologies for Cytokine Measurement in Inflammation and Aging Research. J Gerontol Ser A Biol Sci Med Sci. 2008;63(8):879-884
22. Wiener MC, Sachs JR, Deyanova EG, Yates NA. Differential Mass Spectrometry : A Label-Free LC - MS Method for Finding Significant Differences in Complex Peptide and Protein Mixtures. Anal Chem. 2004;76(20):6085-6096
23. Grebe SKG, Singh RJ. LC-MS/MS in the Clinical Laboratory - Where to from Here? Clin Biochem Rev. 2011;32(1):5-31
24. Wang S, Zhang Y, Pang G, Zhang Y, Guo S. Tuning the Aggregation/Disaggregation Behavior of Graphene Quantum Dots by Structure-Switching Aptamer for High-Sensitivity Fluorescent Ochratoxin A Sensor. Anal Chem. 2017;89(3):1704-1709
25. Liu Y, Liu Y, Mernaugh RL, Zeng X. Single Chain Fragment Variable Recombinant Antibody Functionalized Gold Nanoparticles for a Highly Sensitive Colorimetric Immunoassay. Biosens Bioelectron. 2009;24(9):2853-2857
26. Ren C, Carvajal D, Shull KR, Szleifer I. Streptavidin-Biotin Binding in the Presence of a Polymer Spacer. A Theoretical Description. Langmuir. 2009;25(20):12283-12292
27. Sano T, Cantor CR. Cooperative Biotin Binding by Streptavidin. J Biol Chem. 1990;265(6):3369-3373
28. Aslan K, Luhrs CC, Pérez-Luna VH. Controlled and Reversible Aggregation of Biotinylated Gold Nanoparticles with Streptavidin. J Phys Chem B. 2004;108(40):15631-15639
29. Shi X, Dong S, Li M, Liu X, Zhang Q, Zhao W, Zong C, Zhang Y, Gai H. Counting Quantum Dot Aggregates for the Detection of Biotinylated Proteins. Chem Commun. 2015;51(12):2353-2356
30. Wu H, Han Y, Yang X, Chase GG, Tang Q, Lee CJ, Cao B, Zhe J, Cheng G. A Versatile Microparticle-Based Immunoaggregation Assay for Macromolecular Biomarker Detection and Quantification. PLoS One. 2015;10(2):e0115046
31. Thanh NTK, Rosenzweig Z. Development of an Aggregation-Based Immunoassay for Anti-Protein A Using Gold Nanoparticles. Anal Chem. 2002;74(7):1624-1628
32. Groessl M, Luksch H, Rösen-Wolff A, Shevchenko A, Gentzel M. Profiling of the Human Monocytic Cell Secretome by Quantitative Label-Free Mass Spectrometry Identifies Stimulus-Specific Cytokines and Proinflammatory Proteins. Proteomics. 2012;12(18):2833-2842
33. Chen YP, Zou MQ, Qi C, Xie MX, Wang DN, Wang YF, Xue Q, Li JF, Chen Y. Immunosensor Based on Magnetic Relaxation Switch and Biotin-streptavidin System for the Detection of Kanamycin in Milk. Biosens Bioelectron. 2013;39(1):112-117
34. Zhao LJ, Yu RJ, Ma W, Han HX, Tian H, Qian RC, Long YT. Sensitive Detection of Protein Biomarkers Using Silver Nanoparticles Enhanced Immunofluorescence Assay. Theranostics. 2017;7(4):876-883
35. Rajan NK, Rajauria S, Ray T, Pennathur S, Cleland AN. A Simple Microfluidic Aggregation Analyzer for the Specific, Sensitive and Multiplexed Quantification of Proteins in a Serum Environment. Biosens Bioelectron. 2016;77:1062-1069
36. Molina-Bolívar JA, Galisteo-González F. Latex Immunoagglutination Assays. J Macromol Sci Part C Polym Rev. 2005;45(1):59-98
37. Xiang Y, Lu Y. Portable and Quantitative Detection of Protein Biomarkers and Small Molecular Toxins Using Antibodies and Ubiquitous Personal Glucose Meters. Anal Chem. 2012;84(9):4174-4178
38. Han Y, Wu H, Liu F, Cheng G, Zhe J. Label-Free Biomarker Assay in a Microresistive Pulse Sensor via Immunoaggregation. Anal Chem. 2014;86(19):9717-9722
39. Woolley CF, Hayes MA. Sensitive Detection of Cardiac Biomarkers Using a Magnetic Microbead Immunoassay. Anal Methods. 2015;7(20):8632-8639
40. Reverté L, Garibo D, Flores C, Diogène J, Caixach J, Campàs M. Magnetic Particle-Based Enzyme Assays and Immunoassays for Microcystins: From Colorimetric to Electrochemical Detection. Environ Sci Technol. 2013;47(1):471-478
41. Yu RJ, Ma W, Liu XY, Jin HY, Han HX, Wang HY, Tian H, Long YT. Metal-Linked Immunosorbent Assay (MeLISA): The Enzyme-Free Alternative to ELISA for Biomarker Detection in Serum. Theranostics. 2016;6(10):1732-1739
42. Lee JT, Sudheendra L, Kennedy IM. Accelerated Immunoassays Based on Magnetic Particle Dynamics in a Rotating Capillary Tube with Stationary Magnetic Field. Anal Chem. 2012;84(19):8317-8322
43. Kim D, Herr AE. Protein Immobilization Techniques for Microfluidic Assays. Biomicrofluidics. 2013;7(4):41501
Author contact
![]() Corresponding authors: Dr. Ge Zhang, Department of Biomedical Engineering, University of Akron, Olson Research Center 301L, Akron, OH 44325-0302; Phone: (330) 972-5237; E-mail: gezhangedu and Dr. Jiang Zhe, Department of Mechanical Engineering, University of Akron, Auburn Science and Engineering Center 435A, Akron, OH 44325-0302; Phone: (330) 972-7737; E-mail: jzheedu
Corresponding authors: Dr. Ge Zhang, Department of Biomedical Engineering, University of Akron, Olson Research Center 301L, Akron, OH 44325-0302; Phone: (330) 972-5237; E-mail: gezhangedu and Dr. Jiang Zhe, Department of Mechanical Engineering, University of Akron, Auburn Science and Engineering Center 435A, Akron, OH 44325-0302; Phone: (330) 972-7737; E-mail: jzheedu
 Global reach, higher impact
Global reach, higher impact