13.3
Impact Factor
Theranostics 2018; 8(14):3766-3780. doi:10.7150/thno.24333 This issue Cite
Research Paper
Molecular signatures of liver dysfunction are distinct in fungal and bacterial infections in mice
1. Department for Anesthesiology and Intensive Care Medicine, AG Nanophysiology, Jena University Hospital, Jena, Germany
2. Center for Sepsis Control and Care, Jena University Hospital, Jena, Germany
3. Department of General, Visceral and Vascular Surgery, Experimental Transplantation Surgery, Jena University Hospital, Jena, Germany
4. Research Group Systems Biology/Bioinformatics, Leibniz Institute for Natural Product Research and Infection Biology (Hans Knöll Institute), Jena, Germany
5. Department of Bioinformatics, Friedrich-Schiller-University, Jena, Germany
6. Research Group Applied Systems Biology, Leibniz Institute for Natural Product Research and Infection Biology (Hans Knöll Institute), Jena, Germany
7. Department of Clinical Chemistry and Laboratory Medicine, Jena University Hospital, Jena, Germany
8. Electron Microscopy Center, Jena University Hospital, Jena, Germany
9. Institute of Medical Microbiology, Jena University Hospital, Jena, Germany
10. Bloomsbury Institute of Intensive Care Medicine, University College London, London, UK
11. Research Group Microbial Immunology, Leibniz Institute for Natural Product Research and Infection Biology (Hans Knöll Institute), Jena, Germany
12. Friedrich-Schiller-University, Jena, Germany
Abstract
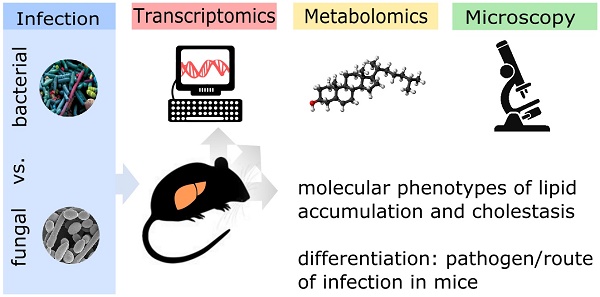
Rationale: The liver is a central organ not only for metabolism but also immune function. Life-threatening infections of both bacterial and fungal origin can affect liver function but it is yet unknown whether molecular changes differ depending on the pathogen. We aimed to determine whether the hepatic host response to bacterial and fungal infections differs in terms of hepatic metabolism and liver function.
Methods: We compared murine models of infection, including bacterial peritoneal contamination and infection (PCI), intraperitoneal and systemic C. albicans infection, at 6 and 24 h post-infection, to sham controls. The molecular hepatic host response was investigated by the detection of regulatory modules based on large-scale protein-protein interaction networks and expression data. Topological analysis of these regulatory modules was used to reveal infection-specific biological processes and molecular mechanisms. Intravital microscopy and immunofluorescence microscopy were used to further analyze specific aspects of pathophysiology such as cholestasis.
Results: Down-regulation of lipid catabolism and bile acid synthesis was observed after 6 h in all infection groups. Alterations in lipid catabolism were characterized by accumulation of long chain acylcarnitines and defective beta-oxidation, which affected metabolism by 6 h. While PCI led to an accumulation of unconjugated bile acids (BA), C. albicans infection caused accumulation of conjugated BA independent of the route of infection. Hepatic dye clearance and transporter expression revealed reduced hepatic uptake in fungal infections vs. defects in secretion following polybacterial infection.
Conclusion: Molecular phenotypes of lipid accumulation and cholestasis allow differentiation between pathogens as well as routes of infection at early stages in mice. Targeted metabolomics could be a useful tool for the profiling of infected/septic patients and the type of pathogen, with subsequent customization and targeting of therapy.
Keywords: sepsis, cholestasis, Candida albicans, metabolism, host response
 Global reach, higher impact
Global reach, higher impact