13.3
Impact Factor
Theranostics 2019; 9(2):449-465. doi:10.7150/thno.29055 This issue Cite
Research Paper
MiR-454-3p-Mediated Wnt/β-catenin Signaling Antagonists Suppression Promotes Breast Cancer Metastasis
1. Affiliated Cancer Hospital & Institute of Guangzhou Medical University; Key Laboratory of Protein Modification and Degradation; State Key Laboratory of Respiratory Disease, School of Basic Medical Sciences, Guangzhou Medical University, Guangzhou 511436, China;
2. Guangdong Key Laboratory for Genome Stability and Human Disease Prevention, Department of Biochemistry and Molecular Biology, Health Science Center, Shenzhen University, Shenzhen, 518060, China;
3. Key Laboratory of Molecular Target & Clinical Pharmacology, School of Pharmaceutical Sciences & the Fifth Affiliated Hospital, Guangzhou Medical University, Guangzhou 511436, China;
4. Department of Pathogen Biology and Immunology, School of Basic Courses, Guangdong Pharmaceutical University, Guangzhou, 510006, China;
5. Department of Ultrasonography, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangzhou 510623, China;
6. Department of Urologic Oncosurgery, Affiliated Cancer Hospital & Institute of Guangzhou Medical University, Guangzhou 510095, China;
7. Department of Radiation Oncology, Affiliated Cancer Hospital & Institute of Guangzhou Medical University, Guangzhou 510095, China.
*These authors contributed equally to this work.
Received 2018-8-7; Accepted 2018-11-8; Published 2019-1-1
Abstract
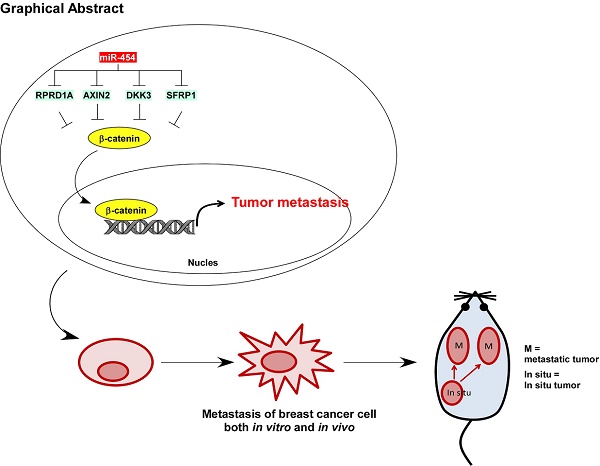
The Wnt/β-catenin pathway is constitutively active and promotes multiple tumor processes, including breast cancer metastasis. However, the underlying mechanism by which the Wnt/β-catenin pathway is constitutively activated in breast cancer metastasis remains unclear. Inhibition of Wnt antagonists is important for Wnt/β-catenin signaling activation, and post-transcriptional regulation of these antagonists by microRNAs (miRNAs) might be a possible mechanism underlying signaling activation. Regulation of nuclear pre-mRNA domain-containing 1A (RPRD1A) is a known inhibitor of cell growth and Wnt/β-catenin signaling activity, but the function and regulatory mechanism of RPRD1A in breast cancer have not been clarified. The aim of this study was to understand how regulators of the Wnt/β-catenin pathway may play a role in the metastasis of this cancer.
Methods: RPRD1A expression and its association with multiple clinicopathological characteristics was analyzed immunohistochemically in human breast cancer specimens. miR-454-3p expression was analyzed using real-time PCR. RPRD1A or miR-454-3p knockdown and overexpression were used to determine the underlying mechanism of their functions in breast cancer cells. Xenografted tumor model, 3D invasive culture, cell migration and invasion assays and sphere formation assay were used to determine the biofunction of RPRD1A and miR-454-3p in breast cancer. Electrophoretic mobility shift assay (EMSA), luciferase reporter assay, and RNA immunoprecipitation (RIP) were performed to study the regulation and underlying mechanisms of RPRD1A and miR-454-3p expression and their correlation with the Wnt/β-catenin pathway in breast cancer.
Results: The Wnt/β-catenin signaling antagonist RPRD1A was downregulated and its upstream regulator miR-454-3p was amplified and overexpressed in metastatic breast cancer, and both were correlated with overall and relapse-free survival in breast cancer patients. The suppression by miR-454-3p on RPRD1A was found to activate Wnt/β-catenin signaling, thereby promoting metastasis. Simultaneously, three other negative regulators of the Wnt/β-catenin pathway, namely, AXIN2, dickkopf WNT signaling pathway inhibitor (DKK) 3 and secreted frizzled related protein (SFRP) 1, were also found to be targets of miR-454-3p and were involved in the signaling activation. miR-454-3p was found to be involved in early metastatic processes and to promote the stemness of breast cancer cells and early relapse under both in vitro and in vivo conditions.
Conclusions: The findings indicate that miR-454-3p-mediated suppression of Wnt/β-catenin antagonist RPRD1A, as well as AXIN2, DKK3 and SFRP1, sustains the constitutive activation of Wnt/β-catenin signaling; thus, miR-454-3p and RPRD1A might be potential diagnostic and therapeutic targets for breast cancer metastasis.
Keywords: miR-454-3p, RPRD1A, Wnt/β-catenin signaling, breast cancer, metastasis
Introduction
Recent advances in the technologies used for the early diagnosis and treatment of breast cancer have greatly improved the survival and prognosis of breast cancer patients. The 5-year survival rate is now 90%; it is 99% in early in situ cancer patients; and, it is 85% even in patients with local progression. However, patients who have metastases at the time of diagnosis have been reported to have a 5-year survival rate of only 26% [1]. Indeed, metastasis is the main cause of mortality in breast cancer patients. For improving survival in such patients, it is important to understand the molecular and genetic mechanisms underlying the metastasis of this cancer and eventually identify targets for therapeutic strategies.
Dysregulation of Wnt/β-catenin signaling, which is necessary for many vital biological processes, such as embryonic development, organogenesis, tissue regeneration, hematopoiesis, cell survival, cellular proliferation and differentiation and stem cell renewal [2, 3], is associated with many diseases, including osteoporosis, neurodegenerative diseases, and cardiovascular diseases, and numerous human malignancies [3, 4]. It has been demonstrated that atypical activation of the Wnt/β-catenin signaling pathway drives tumor initiation and progression, including promotion of cell proliferation, migration, invasion, angiogenesis and resistance to chemotherapy [5-7]. With regard to breast cancer metastasis, aberrant activation of Wnt/β-catenin has been observed, but the molecular basis for the deregulation remains puzzling.
RPRD1A is a known inhibitor of cell growth that has been reported to exert its effects via inhibition of Wnt/β-catenin signaling activity. Overexpression of RPRD1A was found to suppress cell growth by decreasing the expression of cyclin D1 and c-Myc—two Wnt-targeted genes that are critical for cell growth—and attenuating canonical Wnt signaling by disrupting β-catenin/TCF4 interaction [8]. Further, RPRD1A was found to inhibit chicken DF-1 cell proliferation by downregulating the expression of downstream regulatory genes of the Wnt/β-catenin pathway, including β-catenin, TCF4, and cyclin D1 [9]. Moreover, RPRD1A was found to interact with HDAC2 and reduce the amount of histone H3 in the TCF4-binding region, and thus, act as an intrinsic transcriptional repressor of Wnt/β-catenin-mediated gene transcription [10]. At present, the biological function, clinical relevance and regulatory mechanism of RPRD1A in breast cancer have not been clarified.
The present study demonstrated that RPRD1A, as a negative regulator of Wnt/β-catenin signaling, is downregulated in metastatic breast cancer, and that miR-454-3p plays an essential role in promoting breast cancer metastasis by inhibiting RPRD1A and thereby sustaining Wnt/β-catenin signaling. The findings also indicated that RPRD1A was indeed downregulated in metastatic breast cancer, and that its expression was correlated with patient survival and prognosis. The results further showed that RPRD1A suppression; as well suppression of the Wnt antagonists AXIN2, DKK3 and SFRP1, is post-transcriptionally mediated by miR-454-3p via sustaining Wnt/β-catenin signaling activity. Furthermore, a detailed analysis of the role of miR-454-3p in breast cancer metastasis revealed that it is involved in early metastatic events, promotes the stemness of breast cancer cells, and promotes early distant relapse. In conclusion, RPRD1A and miR-454-3p might both be essential regulators of the Wnt/β-catenin pathway and may, therefore, also have potential as prognostic indicators as well as diagnostic and therapeutic targets for breast cancer.
Methods
Cell culture
Primary normal mammary epithelial cells (NMEC) were established according to a previous report [11]. The breast cancer cell lines ZR-75-30, MCF-7, ZR-75-1, BT-549, BT-474, SKBR3, T47D, MDA-MB-415, MDA-MB-435, MDA-MB-468, MDA-MB-231 and MDA-MB-453 were purchased from the ATCC and maintained in DMEM (Gibco, Grand Island, NY) or RPMI-1640 (Gibco) supplemented with 10% FBS (HyClone, Logan, UT) and 1% penicillin/ streptomycin (Gibco). The breast cancer cell line MCF-7, which has low invasive capability, was used to study the metastasis-promoting effects of RPRD1A downregulation or miR-454 overexpression, and MDA-MB-231, a highly metastatic breast cell line, was chosen to study the metastasis-inhibiting effects. The breast cell line ZR-75-1, which has moderate invasive ability, was used to study both promotive and inhibitory effects [12-15].
Tissue specimens
This study was conducted on a total of 232 paraffin-embedded breast cancer samples, which were histopathologically and clinically diagnosed at the Sun Yat-sen University Cancer Center between 1999 and 2007. Eight breast cancer tissues and their corresponding paired adjacent noncancerous breast tissue specimens were stored by freezing them in liquid nitrogen until further use. The investigation was conducted in accordance with ethical standards and according to the tenets of the Declaration of Helsinki and national and international guidelines. Clinical and clinicopathological classification and stage were determined according to the American Joint Committee on Cancer criteria. For the clinical materials used in our study, prior patient consent and approval from the Institutional Research Ethics Committee of Sun Yat-sen University were obtained. Clinical information about the samples is shown in Table S1.
Immunohistochemistry analysis
For immunohistochemistry (IHC) analysis, paraffin-embedded specimens were cut into 4-µm sections, deparaffinized with xylene, rehydrated, and then submerged in EDTA-containing antigen retrieval buffer and microwaved for antigenic retrieval, as previously described [16, 17]. A 1% bovine serum albumin (BSA) solution was used to block nonspecific binding. The sections were then incubated with anti-RPRD1A, anti-AXIN2, anti-DDK3, anti-SFRP1 (Abcam, Cambridge, MA) and anti-β-catenin (Cell Signaling, Danvers, MA) antibodies overnight at 4 °C. The tissue sections were treated with biotinylated anti-rabbit secondary antibody (Thermo Fisher Scientific, MA), and this was followed by further incubation with the streptavidin-horseradish peroxidase complex (Thermo Fisher Scientific), immersion in 3,3ʹ-diaminobenzidine, counterstaining with 10% Mayer's hematoxylin, dehydration and mounting.
The stained tumor sections were examined and scored independently by two observers for positively stained tumor cells and the intensity of immunohistochemical signals. According to the proportion of positively stained tumor cells, the sections were scored as follows: 0, no positive tumor cells; 1, <10% positive tumor cells; 2, 10-50% positive tumor cells; and 3, >50% positive tumor cells. The intensity of staining was graded according to the following criteria: 0, no staining; 1, weak staining (light yellow); 2, moderate staining (yellow brown); and 3, strong staining (brown). The staining index (SI) was calculated as the staining intensity score multiplied by the proportion of positive tumor cells. We assessed expression of the indicated protein in IHC-stained tumor sections based on the SI scores as 0, 1, 2, 3, 4, 6 and 9. Cut-off values were chosen on the basis of a measure of heterogeneity with the log-rank test, with respect to survival analysis.
Immunofluorescence staining
Cells were plated on coverslips, harvested at 24 h, washed with ice-cold phosphate-buffered saline (PBS), and fixed with 2% paraformaldehyde for 10 min. Following this, the coverslips were blocked with 1% BSA for 30 min and incubated with primary antibody against β-catenin (Cell Signaling) for 1 h at room temperature. After washing with PBS, the coverslips were incubated with a fluorescein isothiocyanate-conjugated goat anti-mouse secondary antibody (Jackson Immuno Research, West Grove, PA) for 30 min. Cell nuclei were counterstained with 4', 6 - diamidino -2- phenylindole (DAPI, 5 ng/mL) for 10 min. The coverslips were washed with PBS, mounted with anti-fading regent (Invitrogen, Carlsbad, CA) and stored in the dark until evaluation. Gray-level images were acquired under a laser scanning microscope (Carl Zeiss Co. Ltd., Jena, Germany).
RNA extraction and real-time quantitative PCR
Total miRNA from cultured cells was extracted using the mirVana miRNA Isolation Kit (Ambion, Austin, TX), and total miRNA from paraffin-embedded tissue was extracted using the RecoverAllTM Total Nucleic Acid Isolation Kit (Ambion). cDNAs were synthesized and real-time PCR was performed using the GoTaq® 2-Step RT-qPCR System (Promega, Madison, WI) in an ABI Prism 7500 Sequence Detection System (Applied Biosystems, Foster City, CA). The expression of miRNA was defined based on the threshold cycle (Ct), and relative expression levels were calculated as 2-[(Ct of miR-454-3p) - (Ct of U6)] after normalization with U6 small nuclear RNA expression. Total RNA extraction and quantitation of mRNA were performed as described previously [18]. The median of its relative expression value was chosen as the cut-off value to categorize the samples as high and low miR-454-3p expression.
Western blotting
Western blotting analysis was performed according to a standard method previously described, using anti-RPRD1A, anti-Axin2, anti-Dkk3, anti-SFRP1 (Abcam) or anti-E-cadherin, anti-α-catenin, anti-N-cadherin, anti-vimentin and anti-β-catenin (Cell Signaling) antibodies. The blotting membranes were stripped and re-probed with an anti-α-tubulin antibody (Sigma, Saint Louis, MO) as the loading control.
Plasmids and transfection
The human miR-454-3p gene was PCR-amplified from genomic DNA and cloned into a pMSCV-puro retroviral vector. The miR-454-3p anti-sense strand was cloned into the miRZip plasmid purchased from System Biosciences (San Francisco, CA), as described in a previous report [11]. The 3ʹ-UTR regions of human RPRD1A, AXIN2, DKK3 and SFRP1, generated by PCR amplification of genomic DNA, were cloned into the pGL3-luciferase reporter plasmid (Promega). The TOP Flash and FOP Flash reporters containing the wild-type and mutated TCF/LEF DNA-binding sites, respectively, were purchased from Upstate Biotechnology (Lake Placid, NY). Transfection of siRNAs (Ribo Biotech, Guangzhou) or plasmids was performed using the Lipofectamine 2000 reagent (Invitrogen). Stable cell lines expressing miR-454-3p or miRZip-454-3p were generated via retroviral infection using HEK293T cells and selected with 0.5 μg/mL puromycin for 10 days.
Xenografted tumor model and tumor tissue staining
BALB/c-nude mice (female, 4-5 weeks of age, weighing 18-20 g) were purchased from the Center of Experimental Animals of Guangzhou University of Chinese Medicine. All the experimental procedures were approved by the Institutional Animal Care and Use Committee of Guangzhou Medical University. The BALB/c nude mice were randomly divided into the indicated groups. The indicated cells were inoculated subcutaneously into the mammary fat pad of the mice. Tumor volume was calculated using the equation L × W2 / 2 (L = length, W = width). Thirty days after tumor implantation, the mice were sacrificed. The mammary tumors were then moved and weighed. For analysis of metastasis status, nude mice were intravenously injected with miR-454-3p-transduced or miR-454-3p-silenced cells or control cells via the lateral tail vein. The lungs were collected to count surface metastases under a dissecting microscope at 40 days after tumor implantation. The tumors and lungs were fixed in formalin and embedded in paraffin using the routine method. Serial 6.0-μm sections were cut and subjected to H&E staining with Mayer's hematoxylin solution. IHC assay was performed using anti-RPRD1A, anti-AXIN2, anti-DKK3, anti-SFRP1 (Abcam) and anti-β-catenin antibodies (Cell Signaling).
3D invasive culture
The Matrigel matrix (BD Biosciences, San Jose, CA) was used for the 3D spheroid invasion assay. The indicated cells (1×104) were trypsinized, mixed with 5% Matrigel and seeded on 2% Matrigel-coated 24-well plates. The culture medium was refreshed every other day, and microscopic images were obtained at 2-day intervals for 2-3 weeks.
Cell migration and invasion assays
The Transwell migration (without Matrigel) assay and Matrigel invasion assay were performed as previously described [11, 18]. Cells (1×104) to be tested were plated on the top surface of the polycarbonate Transwell filter without (for cell migration assay) or with Matrigel coating (for cell invasion assay) in the upper chamber of the BioCoatTM Invasion Chambers (BD, Bedford, MA) and incubated at 37 °C for 22 h; this was followed by removal of cells inside the upper chamber with cotton swabs. Cells that had migrated and invaded the bottom surface of the membrane were fixed in 1% paraformaldehyde, stained with hematoxylin, photographed and quantified by counting the number of cells in five random 200× magnification fields.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) was performed using the LightShift™ Chemiluminescent EMSA Kit from Thermo Scientific™ (Thermo Fisher Scientific) according to the manufacturer's instruction. Briefly, the binding reaction solutions contained 10 μg nuclear extract, 1 μg of double-stranded poly (dI-dC)-poly (dI-dC), and 20 fmol of biotin-labeled probe in 20 μL of 1× EMSA buffer. The incubation was carried out for 30 min at room temperature. The EMSA binding reaction mixture was resolved by electrophoresis in a 6% non-denaturing polyacrylamide gel in 0.5×Tris-borate-EDTA (TBE) buffer. After electrophoresis, the DNA was transferred to a Nylon membrane using 0.5 × TBE buffer. The DNA was then cross-linked to the nylon membrane by UV (120 mJ/cm2). The biotinylated oligonucleotides were detected on the membrane according to the manufacturer's instructions.
Luciferase reporter assay
Cells (1×104) were seeded in triplicate in 48-well plates and allowed to settle for 24 h. Then, 100 ng of the indicated plasmids 100 ng (e.g., luciferase reporter plasmids or the control plasmid) plus 1 ng of the pRL-TK Renilla plasmid was transfected into cells using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instruction. Forty-eight hours after transfection, luciferase and Renilla signals were measured using the Dual Luciferase Reporter Assay Kit according to the manufacturer's instructions (Promega).
RNA immunoprecipitation
Cells were co-transfected with HA-Ago1 together with 100 nM miR-454, and this was followed by HA-Ago1 immunoprecipitation with the HA-antibody. Real-time PCR analysis of the immunoprecipitate was used to determine the association of the mRNA of AMER1, AXIN2, CBY1, DKK3, RPRD1A, SFRP1, TCF7L1, 5S rRNA and GAPDH with the RISC complex. 5S rRNA and GAPDH were both used as negative controls.
Sphere formation assay
Five hundred cells were seeded in ultra-low attachment 6-well plates (Corning, Painted Post, NY) for 10-14 days. Spheres that were established were maintained in DMEM/F12 serum-free medium (Invitrogen) supplemented with 2% B27 (BD PharMingen, Carlsbad, CA), 20 ng/mL epidermal growth factor (EGF), 20 ng/mL basic fibroblast growth factor (bFGF), 0.4% BSA, and 5 μg/mL insulin (Sigma). The spheres that were generated were photographed and counted under a light microscope (Carl Zeiss Co. Ltd.).
Statistical analysis
Statistical tests for the present data analysis included Fisher's exact test, log-rank test, chi-square test, and Student's two-tailed t-test. Bivariate correlations between study variables were determined using Spearman's rank correlation analysis. Survival curves were plotted by the Kaplan-Meier method and compared with the log-rank test. The significance of various variables associated with survival was analyzed by univariate and multivariate Cox regression analyses. Statistical analyses were performed using the SPSS 11.0 statistical software package. Data are shown as the mean ± SD values, and P-values ≤ 0.05 were considered to indicate statistical significance.
Results
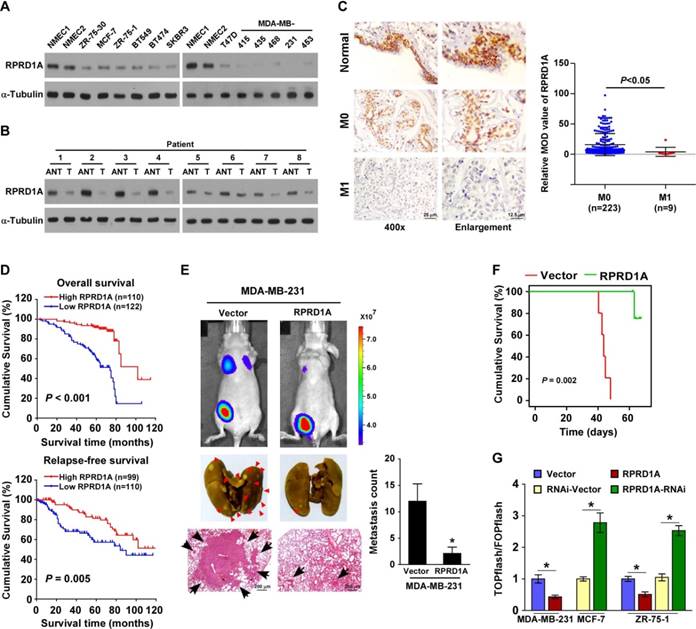
RPRD1A downregulation is correlated with breast cancer metastasis
As expected, the expression of the RPRD1A protein was clearly reduced in 12 breast cancer cell lines compared with 2 normal mammary epithelial cells, and in 8 clinical breast cancer samples compared with their paired adjacent non-tumor tissues (Figure 1A-B). Interestingly, IHC staining showed that RPRD1A expression was evident in normal breast tissues; further, its expression was reduced in breast cancer tissues without distance metastasis (M0) and was markedly repressed in breast cancer samples with distance metastasis (M1) (M0 vs. M1, P < 0.05; Figure 1C). Thus, RPRD1A downregulation might be involved in breast cancer metastasis. Concordantly, statistical analysis revealed that RPRD1A levels were inversely correlated with N/M classification (N: P < 0.001; M: P = 0.026) in patients with breast cancer (n = 232; Table S1 and Table S2), and patients with lower RPRD1A expression not only had shorter overall survival but also showed poorer relapse-free survival (P < 0.001 and P = 0.005 respectively; Figure 1D). This further confirmed the correlation between RPRD1A downregulation and breast cancer metastasis.
Upregulation of RPRD1A inhibits breast cancer metastasis and Wnt/β-catenin signaling
To further investigate the biological effect of RPRD1A on breast cancer metastasis, the MDA-MB-231 cell line, which is a highly metastatic breast cancer cell line, was engineered to stably overexpress RPRD1A and injected into the mammary gland fat pads of nude mice. Bioluminescence imaging and histological analyses showed that mice injected with MDA-MB-231/vector control cells displayed prominent lung metastasis, while the volume of RPRD1A- overexpressing tumors was similar to that of the control tumors. According to the results of growth curve and bioluminescence analyses, only slight metastasis was found in mice injected with RPRD1A- overexpressing MDA-MB-231 cells (Figure 1E and Figure S1A-B). This indicates that the capability of MDA-MB-231 cells to induce lung metastasis was significantly impaired by RPRD1A upregulation. Similarly, the mice injected with RPRD1A-overexpressing MDA-MB-231 cells survived significantly longer than those injected with vector control cells (Figure 1F).
Since previous reports have demonstrated that RPRD1A acts as a negative regulator of Wnt/β-catenin signaling, we investigated whether Wnt/β-catenin signaling was involved in the molecular mechanism and found that the transactivity of β-catenin and the expression of well-known β-catenin downstream target genes were significantly decreased in the RPRD1A-overexpressing breast cancer cells but increased in the RPRD1A-silenced cells (Figure 1G and Figure S1C). Thus, these results further supported the inhibitory effects of RPRD1A on breast cancer progression and Wnt/β-catenin signaling.
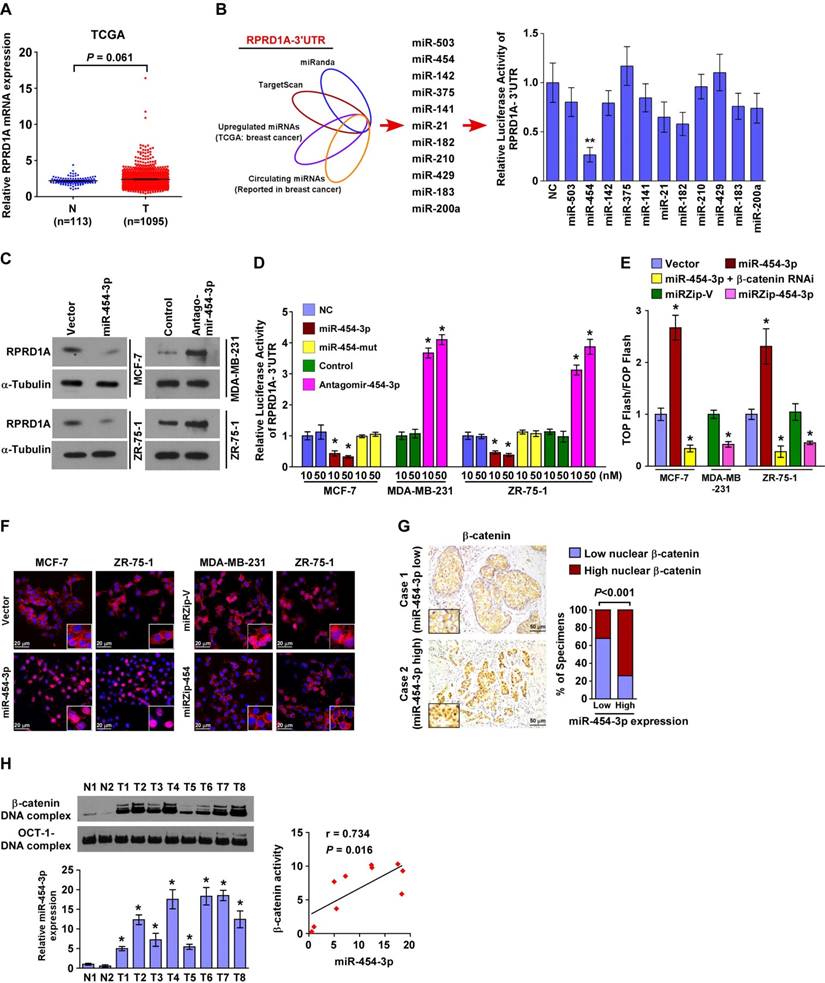
miR-454-3p directly suppresses RPRD1A and activates Wnt/β-catenin signaling in breast cancer cells
Consistent with the analyses of multiple published microarray datasets (TCGA, NCBI/GEO/ GSE10797, 9574, 5764; P > 0.05; Figure 2A and Figure S2A), our real-time PCR analysis also showed no significant alteration in RPRD1A mRNA levels in clinical breast cancer samples when they were compared to their adjacent non-tumor tissues (n = 8, including 3 samples with N/M metastasis; Figure S2B). This indicates that RPRD1A downregulation may occur at the post-transcriptional level in breast cancer.
When the miRNAs that potentially target the RPRD1A 3ʹUTR based on public algorithms (TargetScan and miRANDA) were integrated with the upregulated miRNAs reported in TCGA breast cancer samples and the circulating miRNAs reported in breast cancer [19-27], the results showed that miR-454-3p might be involved in the regulation of RPRD1A protein in breast cancer (Figure 2B). Indeed, western blotting analysis revealed that overexpression of miR-454-3p significantly repressed the expression of RPRD1A protein, but inhibition of endogenous miR-454-3p had the opposite effect (Figure 2C and Figure S2C). Furthermore, the luciferase reporter assay showed that the luciferase activity of RPRD1A-3ʹUTR was drastically decreased in miR-454-3p -overexpressing breast cancer cells but increased in miR-454-silenced cells, whereas forced expression of a mutant miR-454-3p (miR-454-3p-mu: UUCAGCAAUAUUGCUUAUAGGGU) had no inhibitory effect on RPRD1A-3ʹUTR luciferase activity (Figure 2D). Collectively, these results demonstrate that RPRD1A is a bona fide target of miR-454-3p.
Since RPRD1A is known to be a Wnt/β-catenin antagonist, we next examined the effect of miR-454-3p on Wnt/β-catenin signaling. Gene set enrichment analysis (GSEA) revealed that the miR-454 levels were positively correlated with β-catenin-activated gene signatures; this indicates that miR-454-3p activates Wnt/β-catenin signaling (Figure S3A). Moreover, overexpression of miR-454-3p significantly increased β-catenin transactivity and the expression of multiple well-known β-catenin downstream target genes, but silencing of miR-454-3p had the opposite effect (Figure 2E and Figure S3B). Nuclear β-catenin was increased in miR-454-3p-overexpressing cells and decreased when miR-454-3p was suppressed (Figure 2F). The correlation studies in 232 breast cancer specimens showed that the expression of nuclear β-catenin was strongly associated with miR-454-3p levels (P < 0.001; Figure 2G). Importantly, the DNA-binding activity of β-catenin was significantly upregulated in breast cancer tissues that have high miR-454-3p expression (r = 0.734, P = 0.016; Figure 2H). This further provides evidence that miR-454-3p contributes to the activation of Wnt/β-catenin signaling in breast cancer.
RPRD1A is downregulated in breast cancer and correlates with breast cancer metastasis. (A-B) Western blotting analysis of RPRD1A in 12 breast cancer cells (A) and 8 clinical breast cancer samples paired with their adjacent non-tumor tissues (B). α-tubulin served as the loading control. ANT: adjacent non-tumor; T: tumor. (C) IHC staining and statistical analyses of the average MOD of RPRD1A expression in normal breast tissues and breast cancer tissues without (M0; n=223) or with (M1; n=9) distant metastasis. Magnification, ×400 (left); enlarged images (right). The MOD value was determined with the Image-Pro Plus software. MOD: mean optical density. (D) Kaplan-Meier curves for breast cancer patients with low and high expression of RPRD1A. (E) Bioluminescence images of subcutaneous tumors showing distant metastasis signals (left, upper). Representative bright field images (left, middle) and quantification (right) of metastases in the lungs (arrows indicate surface metastatic nodules). Lung metastases in the mice were confirmed by H&E staining (left, lower). (F) Kaplan-Meier curves for RPRD1A-overexpressing mice (P = 0.002) compared with control mice (log-rank test). (G) Luciferase assay of TCF/LEF transcriptional activity in the indicated cells. Each bar represents the mean ± SD value from three independent experiments. *P < 0.05.
miR-454-3p directly suppresses RPRD1A in breast cancer cells. (A) Expression of RPRD1A in human breast cancer clinical specimens from the TCGA mRNA expression array data. (B) Illustration of miR-454-3p selection. Luciferase activity of reporters containing the 3ʹ-UTR of RPRD1A in the indicated miRNA-transfected cells was assessed for the selection of target genes (right). (C) Western blotting analysis of RPRD1A in the indicated cells. (D) Luciferase activities of reporters containing the 3ʹ-UTR of RPRD1A in miR-454-transduced cells, miR-454-3p-silenced cells, control cells or miR-454-3p-mutant-transfected cells. (E) Luciferase assay of TCF/LEF transcriptional activity in the indicated cells. (F) Subcellular β-catenin localization in the indicated cells was assessed by immunofluorescence staining. Magnification, ×200. (G) IHC staining of β-catenin in tissues with low and high miR-454-3p expression. Percentage of specimens (right) showing low or high miR-454-3p expression in relation to subcellular β-catenin localization. Magnification, ×200. (H) The DNA-binding activity of β-catenin as determined by the EMSA assay and the correlation between the DNA-binding activity of β-catenin and miR-454-3p expression in clinical breast cancer samples (n = 8) in comparison with normal breast tissues (n = 2). Each bar represents the mean ± SD value from three independent experiments. *P < 0.05.
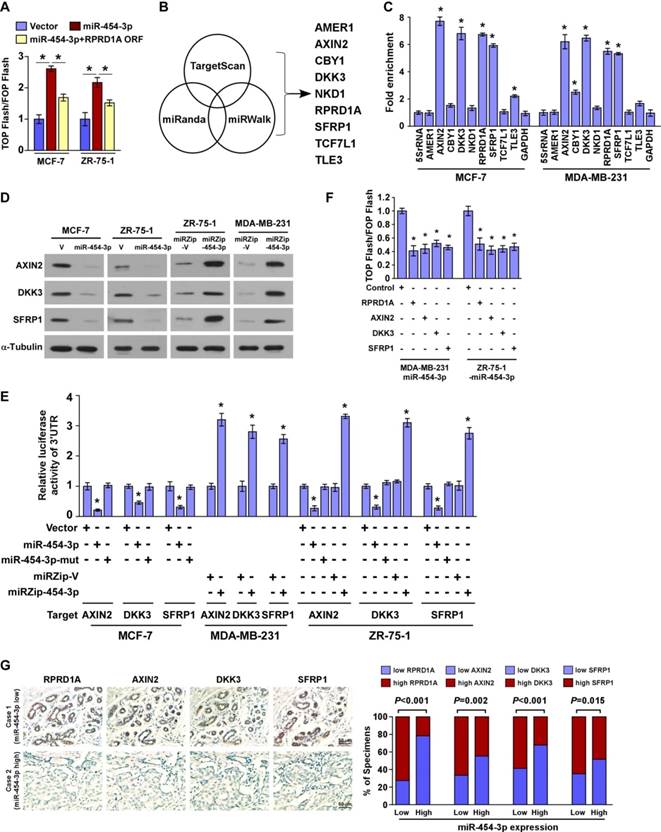
miR-454-3p directly suppresses multiple key components in the Wnt/β-catenin cascade
It has been reported that RPRD1A attenuates Wnt/β-catenin signaling by disrupting the interaction of β-catenin with TCF4 in the nucleus. We therefore tried to investigate whether miR-454-3p might be involved in this mechanism. We found that restoration of RPRD1A expression in miR-454-3p-transduced cells to a level that is comparable to that in control cells via transfection with the RPRD1A ORF (without the 3ʹUTR) only partially abrogated miR-454-3p-induced β-catenin activity (Figure 3A and Figure S3C). This indicates that, in addition to RPRD1A, other targets of miR-454-3p may also contribute to Wnt/β-catenin signaling activation. Indeed, analysis using publicly available algorithms predicted that multiple negative regulators of the Wnt/β-catenin pathway, including AMER1, AXIN2, CBY1, DKK3, NKD1, SFRP1, TCF7L1 and TLE3, might also be potential targets of miR-454-3p (Figure 3B). However, miRNP immunoprecipitation assay revealed that miR-454-3p only specifically associated with the 3′-UTRs of AXIN2, DKK3 and SFRP1 in both MCF-7 and MDA-MB-231 breast cancer cells, but did not associate with the 3′-UTRs of AMER1, NKD1 and TCF7L1 (Figure 3C). Further, western blotting analysis revealed that the expressions of AXIN2, DKK3 and SFRP1 were dramatically decreased in miR-454-3p-transduced breast cancer cells but were increased in miR-454-suppressed cells (Figure 3D). We also found that overexpression of miR-454-3p reduced the reporter activities driven by the 3′-UTRs of the AXIN2, DKK3 and SFRP1 transcripts, while silencing of miR-454-3p elevated these activities (Figure 3E). Further, individual overexpressing of RPRD1A, AXIN2, DKK3 and SFRP1 potently inhibited Wnt/β-catenin signaling activity in miR-454-transduced cells (Figure 3F). Negative correlations between miR-454-3p levels and protein expression of AXIN2, DKK3 and SFRP1 were also observed in clinical breast cancer specimens (Figure 3G). These findings further demonstrate that these Wnt/ β-catenin antagonists are suppressed by miR-454-3p and, as a result, Wnt/β-catenin activation is sustained in breast cancer metastasis.
miR-454-3p overexpression is correlated with shorter relapse-free survival in breast cancer patients and induces in vivo breast cancer metastasis
In agreement with the public breast cancer miRNA array data (TCGA, n normal = 103, n tumor = 1077; Figure S4A), real-time PCR analysis revealed that miR-454-3p was markedly elevated in breast cancer cell lines, especially the cell lines with high metastatic capability, and in the primary tumors of breast cancer patients with metastasis (Figure 4A-B). Furthermore, statistical analysis revealed that the miR-454-3p levels were strongly correlated with the metastatic status and N/M classification (N: P < 0.001, M: P = 0.017) in breast cancer patients (n = 232; Table 1). Importantly, patients with higher miR-454-3p expression had poorer overall survival and shorter relapse-free survival (P < 0.001 and P = 0.018 respectively; Figure 4C-D); this was further confirmed by TCGA data analysis (P < 0.001 and P = 0.037 respectively; Figure S4B). Moreover, the miR-454-3p level was recognized as an independent prognostic factor in breast cancer, as was the RPRD1A level (P < 0.001 and P = 0.002 respectively; Table S3). Consistent with the clinical correlation observed between miR-454-3p expression and breast cancer metastasis, GSEA of published breast cancer datasets also showed that miR-454 expression was significantly correlated with metastatic gene signatures (Figure S4C). Collectively, these results indicate that miR-454-3p plays vital roles in breast cancer metastasis.
Correlation between clinicopathological features and expression of miR-454-3p.
| Patient characteristics | miR-454-3p expression | P-value | ||
|---|---|---|---|---|
| Low | High | |||
| Age (years) | ≤ 53 | 88 | 74 | 0.045 |
| > 53 | 28 | 42 | ||
| Clinical stage | I | 35 | 4 | < 0.001 |
| II | 71 | 36 | ||
| III | 9 | 67 | ||
| IV | 1 | 9 | ||
| T classification | T1 | 49 | 18 | < 0.001 |
| T2 | 61 | 58 | ||
| T3 | 5 | 27 | ||
| T4 | 1 | 13 | ||
| N classification | N0 | 73 | 21 | < 0.001 |
| N1 | 31 | 39 | ||
| N2 | 6 | 36 | ||
| N3 | 6 | 20 | ||
| M classification | N0 | 115 | 108 | 0.017 |
| Yes | 1 | 8 | ||
| Vital status | Alive | 98 | 59 | < 0.001 |
| Dead | 18 | 57 | ||
To further examine the biological role of miR-454-3p in breast cancer metastasis, luciferase-expressing MCF-7 cells transduced with an miR-454-3p or control vector were injected into the mammary gland fat pads of nude mice. As expected, no visible metastasis was found in mice transplanted with the control vector-transduced MCF-7 cells, according to the bioluminescence imaging and histological analysis findings. Strikingly, mice bearing MCF-7/miR-454-3p tumors displayed prominent lung metastasis, whereas blockage of Wnt/β-catenin signaling by β-catenin silencing drastically inhibited the promotive effect of miR-454-3p on MCF-7 cell metastasis (Figure 4E and Figure S4D). Consistently, the lung metastatic ability of MDA-MB-231 cells was significantly impaired by miR-454-3p silencing (Figure 4F). Similarly, treatment with an miR-454-3p antagomir that was injected via the tail vein (2 times per week for 3 weeks) also extensively reduced the number of tumor nests formed in the lungs by MDA-MB-231 cells compared with the antagomir control group (Figure 4F). Further, mice injected with cells with high miR-454-3p expression had shorter survival than those injected with cells that had low miR-454-3p expression, while silencing of miR-454-3p significantly extended their survival (Figure 4G). These results provide further evidence for the role of miR-454-3p in human breast cancer metastasis.
miR-454-3p suppresses multiple key components in the Wnt/β-catenin cascade in breast cancer cells. (A) Luciferase assay of TCF/LEF transcriptional activity in the indicated cells. (B) Illustration of miR-454-3p downstream target gene selection. (C) miRNP immunoprecipitation assay revealed that AXIN2, DKK3, RPRD1A and SFRP1 mRNAs were recruited to miRNP complexes following immunoprecipitation with Ago2. IgG immunoprecipitation was used as a negative control. (D) Western blotting analysis of AXIN2, DKK3 and SFRP1 in the indicated cells. (E) Luciferase activities of reporters containing the 3ʹ-UTRs of AXIN2, DKK3 and SFRP1 in miR-454-3p-transduced cells, miR-454-3p-silenced cells, control cells or miR-454-3p-mutant-transfected cells. (F) Luciferase assay of TCF/LEF transcriptional activity in AXIN2-, RPRD1A-, DKK3- or SFRP1-expressing cells. (G) IHC staining of RPRD1A, AXIN2, DDK3 and SFRP1 in tissues with low and high miR-454-3p expression. Percentage of specimens (right) showing low or high miR-454-3p expression in relation to RPRD1A, AXIN2, DDK3 and SFRP1 expression. Each bar represents the mean ± SD value from three independent experiments. *P < 0.05.
miR-454-3p overexpression correlates with shorter survival in breast cancer patients and promotes in vivo metastasis of breast cancer. (A) Expression of miR-454-3p in breast cancer cell lines. (B) Expression of miR-454-3p in stage I-IV human breast cancer clinical specimens. (C-D) Kaplan-Meier curves for breast cancer patients with low and high expression of miR-454-3p. (E-F) Bioluminescence images of subcutaneous tumors showing distant metastasis signals (left). Representative bright field images (middle) and quantification of metastases (right) in the lungs (arrows indicate surface metastatic nodules). Lung metastases in the mice were confirmed by H&E staining (middle, lower). (G) Kaplan-Meier curves for the indicated mice (log-rank test). (H) Expression of miR-454-3p in the peripheral blood circulation of the indicated mice. (I) Expression of miR-454-3p in the serum of breast cancer patients with (M1) or without (M0) metastasis. Each bar represents the mean ± SD value from three independent experiments. *P < 0.05.
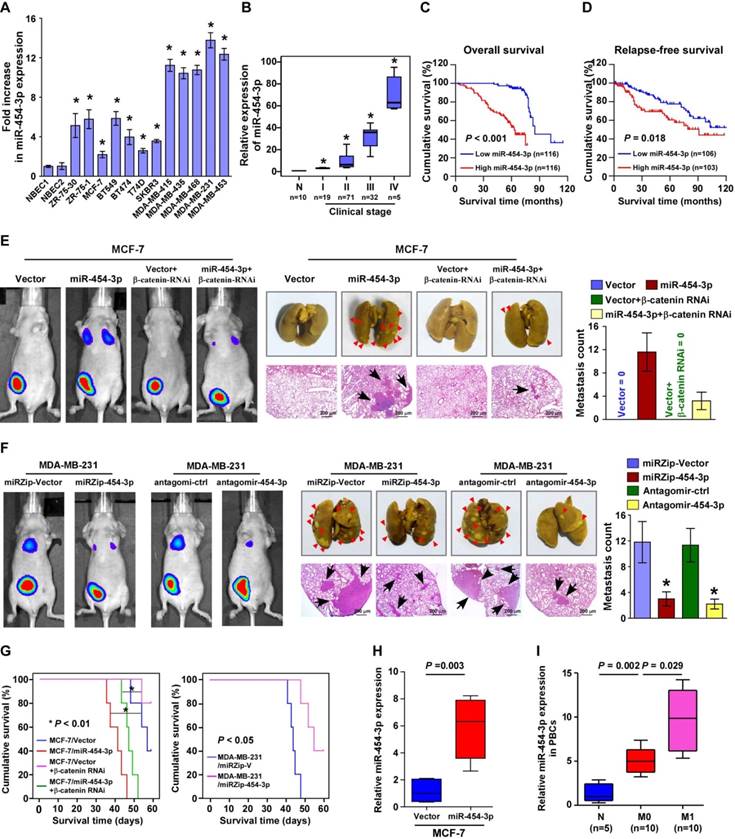
An important in vivo finding was that the miR-454-3p levels were dramatically elevated in the peripheral blood circulation of mice bearing MCF-7/ miR-454-3p cells compared with mice bearing control tumors; this finding was consistent with the results obtained for exosomes and circulating tumor cells in the blood (Figure 4H and Figure S4E-F). Consistent with these findings, we found that the expression of miR-454-3p was lower in the serum of breast cancer patients who did not have metastasis and that it was markedly increased in the serum of breast cancer patients with metastasis in comparison to healthy subjects (Figure 4I). This indicates that miR-454-3p might be a potential marker for the detection of breast cancer metastasis.
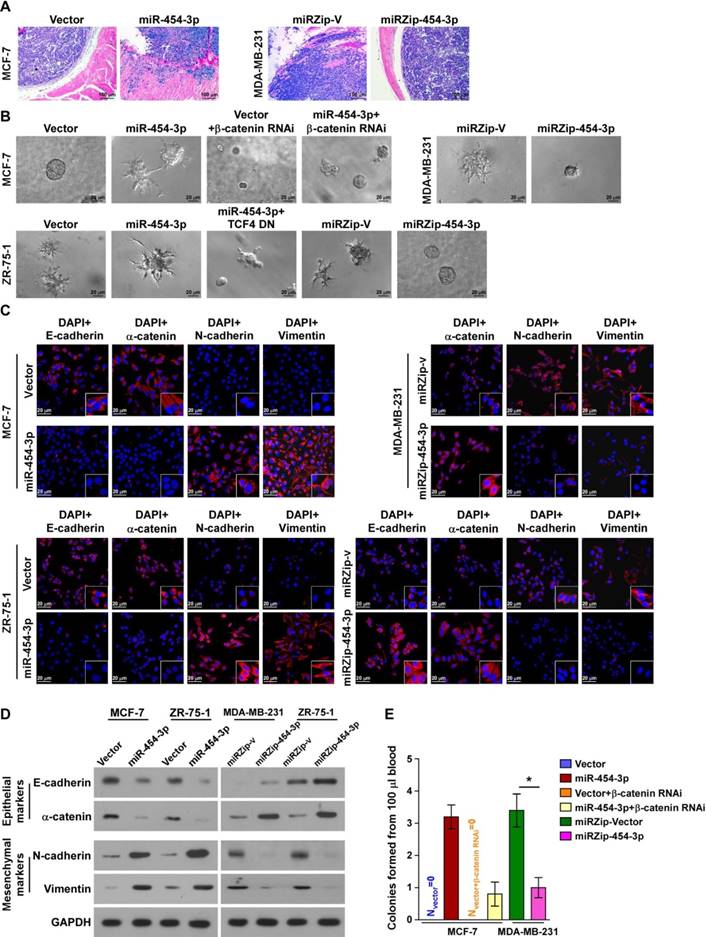
miR-454-3p contributes to early events in the breast cancer metastasis cascade. (A) H&E staining of the borders of primary tumors in the indicated mice. (B) 3D spheroid invasion assays of the indicated cells. (C-D) Immunofluorescence staining (C) and western blotting analysis (D) of EMT markers in the indicated cells. Magnification, ×400. (E) Colonies from peripheral blood samples of the indicated mice. Each bar represents the mean ± SD value from three independent experiments. *P < 0.05.
miR-454-3p contributes to early events in the breast cancer metastasis cascade
Next, we attempted to identify the specific steps of the metastatic process in which miR-454-3p is involved. Microscopic examination of tumor morphology showed that the borders of miR-454-3p- transduced MCF-7 primary tumors displayed spike-like structures that penetrated into the surrounding muscle, but the control tumors were well encapsulated (Figure 5A). In contrast, silencing of miR-454-3p drastically inhibited the local invasive capability of MDA-MB-231/vector tumors (Figure 5A). The pro-invasive role of miR-454-3p in breast cancer was further confirmed by 3D-spheroid invasion assays and Matrigel-coated Transwell assays (Figure 5B and Figure S5A). These results indicate that miR-454-3p promotes local invasion of breast cancer cells.
We also found that miR-454-3p-transduced MCF-7 and ZR-75-1 cells displayed a dramatic morphological change: the typical cobblestone-like appearance of the epithelium had been altered to a spindle-like, mesenchymal morphology (Figure S5B). Further, levels of epithelial markers, including E-cadherin and α-catenin, were drastically decreased while levels of the mesenchymal markers N-cadherin and vimentin were increased in miR-454-3p-transduced MCF-7 and ZR-75-1 cells; opposite findings were obtained in MDA-MB-231 and ZR-75-1 cells transfected with miRZip-454-3p (in comparison with the indicated control cells, as indicated by immunofluorescence staining and western blotting analysis; Figure 5C-D). These findings indicate that miR-454-3p might promote the transition from the epithelial to the mesenchymal phenotype.
Blood samples (100 μL) obtained from mice of the indicated groups were cultured in the presence of puromycin to select for circulating tumor cells. Strikingly, more than three colonies of tumor cells were recovered from the blood samples of the mice bearing MCF-7/miR-454-3p tumors, whereas no colonies were recovered from the blood samples of mice bearing MCF-7/vector control tumors. Further, the average number of colonies recovered from the blood samples of the tumor-bearing mice was markedly higher in the MDA-MB-231/miRZip-V group (3.4 colonies/100 μL blood) than in the MDA-MB-231/miRZip-454-3p group (1 colony/100 μL blood) (Figure 5E). This indicates that miR-454-3p promotes the dissemination of breast cancer cells in circulation and their survival. Taken together, these results indicate that miR-454-3p contributes to early events in the breast cancer metastasis cascade.
miR-454-3p promotes the stemness of breast cancer cells and early distant relapse of breast cancer
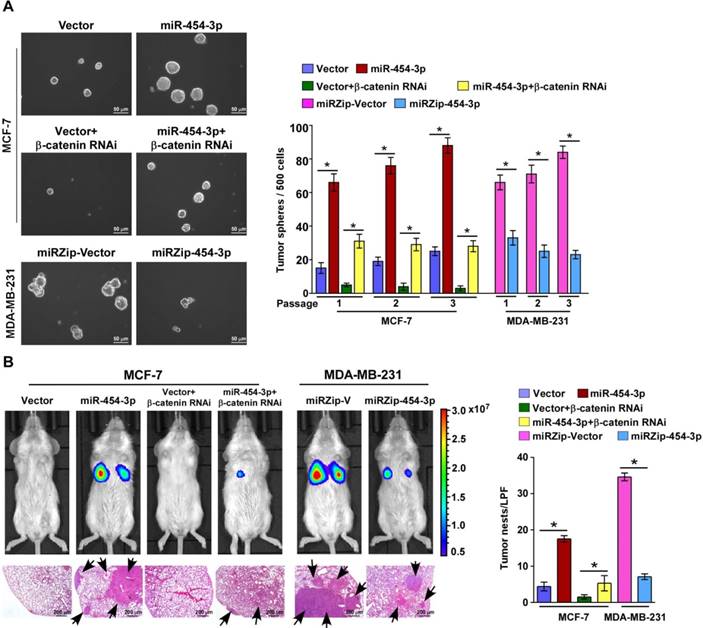
Recent reports have indicated that only a few metastatic cells with high tumor-initiating capacity can develop into overt metastatic foci showing distant relapse [28]. Therefore, we further examined the effect of miR-454-3p on the stem cell-like traits of breast cancer cells and early distant relapse of breast cancer. In agreement with the GSEA results, according to which miR-454 expression was positively correlated with the stemness signatures, the expressions of stemness-associated genes, including MYC, SOX2, OCT4, NANOG and SNAIL, were significantly increased in miR-454-3p-transduced cells and decreased in miR-454-3p-silenced cells (Figure S6A-B). Furthermore, the tumor sphere formation assay showed that MCF7/miR-454-3p breast cancer cells formed larger and more tumor spheres than the vector controls (Figure 6A). Moreover, overexpression of miR-454-3p markedly increased the proportion of side population (SP) breast cancer cells but silencing of miR-454-3p decreased their proportion (Figure S6C). Collectively, these results demonstrate that miR-454-3p enhances the stem-like traits of breast cancer cells.
Next, we examined the effect of miR-454-3p on early distant relapse of breast cancer in a lung colonization model. Nude mice were intravenously injected with control cells and miR-454-3p-dysregulated cells via their lateral tail veins, and lung metastatic foci were detected by bioluminescence imaging after 4 weeks. As shown in Figure 6B, the number of tumor nests in the lungs formed by the miR-454-3p-transduced MCF-7 cells was significantly higher than that formed by the control cells; further, silencing of miR-454-3p drastically inhibited the capability of MDA-MB-231/vector cells to form lung metastases. This finding indicates that miR-454-3p overexpression promotes formation of lung metastatic foci from breast cancer cells. Importantly, a limiting dilution assay revealed that MCF-7/miR-454-3p cells exhibited greater capability to reform tumors in the lungs compared with control cells, but the capability of lung relapse of MDA-MB-231 cells was drastically inhibited by miR-454-3p downregulation (Figure S6D). These findings together indicate that miR-454-3p promotes early distant relapse of breast cancer.
miR-454-3p is amplified in breast cancers
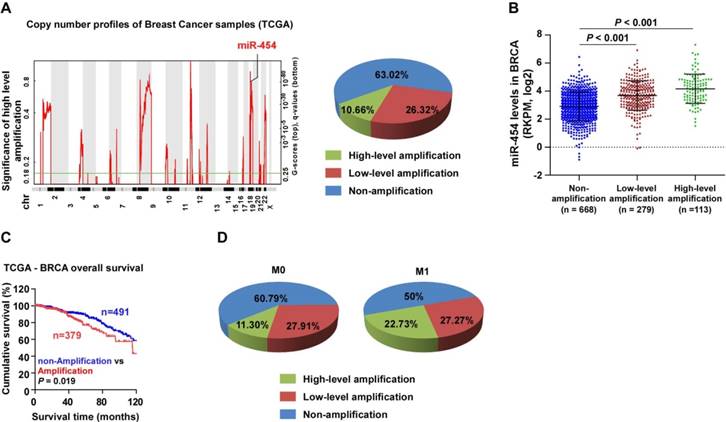
The miR-454 locus is located on chromosome 17q22, which is frequently amplified in various human cancers [29, 30]. Analysis of miR-454 copy number variation (CNV) in the TCGA dataset also showed that the miR-454 locus was amplified in 36.98% of breast cancer samples and that miR-454 expression was significantly associated with miR-454 CNV (Figure 7A-B). Furthermore, we found that patients with breast cancer who had miR-454 amplification had poorer survival outcome than patients with miR-454 non-amplification (P = 0.019; Figure 7C). In addition, analysis of miR-454 CNV in the TCGA data showed that the percentage of miR-454 gene amplification (including low- and high- level amplification) in metastatic cases (M1, totally 50%) was higher than that in non-metastatic cases (M0, totally 39.21%); in particular, high-level amplification was much higher in metastatic cases (22.73%) than in non-metastatic cases (11.3%) (Figure 7D).
Interestingly, analysis of the TCGA datasets revealed that the miR-454 CNV and miR-454 levels were also markedly amplified in subsets of primary tumors, including bladder urothelial carcinoma (BLCA), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), prostate adenocarcinoma (PRAD), stomach adenocarcinoma (STAD), and uterine corpus endometrial carcinoma (UCEC) (Figure S7A-B). Further, the miR-454 levels were significantly upregulated, and a positive correlation was found between the miR-454 levels and miR-454 CNV in LIHC and KIRC (Figure S7C). Consistent with this finding, higher miR-454 expression was associated with shorter overall survival in LIHC and KIRC patients (Figure S7D-E). These results indicate that miR-454 overexpression might contribute to tumor progression in multiple cancers.
The oncogenic function of miR-454-3p in other human cancers was further examined in LIHC and KIRC. Overexpression of miR-454-3p significantly increased β-catenin luciferase reporter activity in both the LIHC and KIRC cell lines, while silencing of miR-454-3p decreased this activity. Moreover, the tumor sphere formation assay showed that the number and size of tumor spheres formed were significantly higher in miR-454-3p-transduced LIHC and KIRC cells but was lower in miR-454-3p-silenced cells as compared with the respective control cells (Figure S7F). Taken together, these results demonstrate that aberrantly amplified miR-454-3p promotes cancer stem cell-like traits by activating the Wnt/β-catenin pathways, thus resulting in poor outcomes in cancer patients.
miR-454-3p promotes early distant relapse in breast cancer. (A) Images (left) and quantification (right) of tumor spheres formed from the indicated cells. (B) Bioluminescence images of distant metastasis signals in mice bearing the indicated cells injected through the tail vein (upper). Lung metastases in the mice were confirmed by H&E staining (lower image: arrows indicate surface metastatic nodules). The number of lung tumor nests in each group was counted under low-power fields (LPFs) (right). NOD/SCID mice were used in the study. Each bar represents the mean ± SD value from three independent experiments. *P < 0.05.
miR-454-3p is amplified in breast cancer. (A) Analysis of miR-454 copy number variation (CNV) in the TCGA dataset. (B) Expression of miR-454 corresponding to different amplification levels in the TCGA dataset. (C) Kaplan-Meier curves for miR-454 in miR-454 amplification and non-amplification groups (miR-454 amplification groups: CNV gain and amplification; miR-454 non-amplification groups: diploid CNV; P = 0.019, log-rank test). (D) Analysis of miR-454 CNV in non-metastatic (M0) and metastatic (M1) cases from the TCGA dataset.
Discussion
In the present study, we demonstrated that RPRD1A, as an inhibitor of breast cancer metastasis, displayed an inverse correlation with both poor prognosis and early recurrence in patients with breast cancer; thus, it shows potential as a prognostic indicator of breast cancer metastasis. With regard to the mechanism, RPRD1A was found to be post-transcriptionally suppressed by miR-454-3p in metastatic breast cancer tissue, as a result of which the antagonistic effect of RPRD1A on the Wnt/β-catenin signaling pathway was abrogated and the activity of this pathway was sustained. Further analyses into the mechanisms and functions of miR-454-3p showed that it was positively correlated with breast cancer metastasis and promoted the metastatic processes of breast cancer under both in vivo and in vitro conditions.
Wnt/β-catenin signaling has been confirmed to promote tumor metastasis by inducing epithelial-to-mesenchymal transition (EMT) and cancer cell stemness [31, 32]. EMT, a process by which cancer cells lose their epithelial properties and acquire a mesenchymal phenotype and become motile and invasive, is considered to be a crucial component of the metastatic process [33, 34]. Although the role of EMT in carcinoma metastasis is unclear [35, 36], it has been found that EMT promotes breast cancer metastasis [37-39]. Moreover, EMT has also been associated with the induction of cancer stem cells, drug resistance, and immune suppression [40, 41]. In our study, we found that downregulation of RPRD1A or ectopic expression of miR-454-3p is followed by breast cancer EMT, as indicated by upregulation of the mesenchymal markers N-cadherin and vimentin and downregulation of the epithelial markers E-cadherin and α-catenin; this process finally results in breast cancer metastasis. Thus, EMT may be one of the many biological processes underlying breast cancer development through which miR-454-3p exerts its promotive effects. Moreover, miR-454-3p expression was positively correlated with the expression of stemness-associated genes, including MYC, SOX2, OCT4, NANOG and SNAIL, and the stemness signatures; this demonstrates that miR-454- 3p can enhance the stem-like traits of breast cancer cells. In addition to EMT and stem cell processes, other processes that are required for tumor metastasis include angiogenesis, lymphangiogenesis and reverse transition from the mesenchymal to epithelial morphology to establish evident metastasis at a secondary site [42, 43]. Given these other processes that are involved, the comprehensive function of miR-454-3p in breast cancer metastasis needs to be further explored as it may participate in other metastasis-related processes.
Multiple potential cancer therapeutic strategies based on regulation of Wnt/β-catenin signaling have been studied. For example, prodigiosin, as a potent inhibitor of the Wnt/β-catenin pathway, was found to block Wnt/β-catenin signaling by targeting multiple regulators of the pathway, including low-density lipoprotein-receptor-related protein 6, Dishevelled, and glycogen synthase kinase-3-beta, and thereby exhibit potent cytotoxic activity against human cancer cells [44]. The DNA repair enzyme O6-methylguanine-DNA methyltransferase, which is commonly overexpressed in cancers and is involved in the progression of chemoresistance, was found to be downregulated in response to pharmacological or genetic inhibition of Wnt activity, and this in turn restored the chemosensitivity of DNA-alkylating drugs [45]. Moreover, dysregulation of multiple direct target genes of Wnt/β-catenin signaling in immune cells and tumor cells has been demonstrated to be responsible for the failure of cancer immunotherapy. It was proposed that targeting Wnt/β-catenin signaling potentially improved the clinical outcomes of cancer patients by overcoming primary, adaptive, and acquired resistance to immunotherapy [46]. All these findings strongly indicate that combination therapy that targets Wnt/β-catenin signaling would be highly beneficial, and that it is important to explore the detailed mechanism of this signaling pathway during tumor development. However, the effective therapeutic targets of Wnt/β-catenin signaling have not been exactly clarified. In addition, inhibition of Wnt antagonists is important for Wnt/β-catenin signaling activation [47, 48], and post-transcriptional regulation of these antagonists by miRNAs might be a possible mechanism of the signaling activation. Multiple studies have reported that miRNAs upregulate Wnt/β-catenin pathway activity by targeting several Wnt antagonists [49-51]. Based on these findings, it is possible that breast cancer metastasis is mediated via inhibition of Wnt antagonists by certain miRNAs. Our findings showed that suppression of the Wnt antagonists RPRD1A, as well as AXIN2, DKK3 and SFRP1, by miR-454-3p upregulation led to Wnt/β-catenin activation followed by aggressive breast cancer progression. All these findings indicate the role of a novel regulatory mechanism involving miR-454-3p and Wnt/β-catenin signaling pathway components in the metastasis of breast cancer and suggest a potential therapeutic strategy of breast cancer.
Although the functions of miRNAs and their regulatory mechanisms in Wnt/β-catenin signaling require further investigation, several miRNA-targeted therapeutics are under clinical investigation [52]. In one such study, inhibition of miR-122 was found to result in a substantial reduction in hepatitis C virus (HCV) infection load and reduction in liver damage. Recently, two companies (Roche/Santaris and Regulus Therapeutics) have been engaged in clinical trials with anti-miR-122 LNAs as a therapeutic agent against HCV infections, and the projects have reached phase II trials for the treatment of hepatitis. RG-125, an anti-miRNA against miR-103/107, has recently been investigated in the context of non-alcoholic steatohepatitis treatment. MRG-201, an miR-29 miRNA mimic (miRagen Therapeutics), is being tested for the treatment of scleroderma, and MRG-106, an LNA-based antimiR-155 (miRagen Therapeutics), is being tested for the treatment of cutaneous T cell lymphoma (mycosis fungoides subtype); both drugs are in phase I clinical trials. The success of these phase I and II trials has led to additional phase II studies with long-term follow-ups, more patients and multidrug combinations, which could have promising implications in clinical therapeutics. Here, we have shown that miR-454-3p could be a potential diagnostic marker for patients with breast cancer, and also act as a promoter of breast cancer aggressiveness. Inhibition of miR-454-3p resulted in inhibition of breast cancer metastasis in a mouse model, indicating its potential clinical applications as a therapeutic target.
Herein, we demonstrated that inhibition of miR-454-3p or ectopic expression of Wnt/β-catenin antagonists, including RPRD1A, AXIN2, DKK3 and SFRP1, led to Wnt/β-catenin signaling activity inhibition, which subsequently led to attenuation of breast cancer metastasis. Such studies on the role of Wnt/β-catenin signaling molecules in tumor therapy could shed light on promising strategies for cancer therapy.
Conclusion
In the current study, suppression of RPRD1A or upregulation of miR-454-3p was found to be significantly correlated with both poor prognosis and early recurrence in breast cancer via Wnt/β-catenin signaling activation. Thus, RPRD1A and other known Wnt/β-catenin antagonists might represent promising targets in the development of diagnosis and treatment strategies for metastatic breast cancer.
Abbreviations
BLCA: bladder urothelial carcinoma; Dkk: dickkopf WNT signaling pathway inhibitor; EMT: epithelial-to-mesenchymal transition; GSEA: gene set enrichment analysis; IHC: immunohistochemistry; KIRC: kidney renal clear cell carcinoma; KIRP: kidney renal papillary cell carcinoma; LIHC: liver hepatocellular carcinoma; LUAD: lung adenocarcinoma; LUSC: lung squamous cell carcinoma; miRNAs: microRNAs; NMEC: normal mammary epithelial cells; PRAD: prostate adenocarcinoma; RPRD1A: regulation of nuclear pre-mRNA domain containing 1A; SFRP: secreted frizzled related protein; STAD: stomach adenocarcinoma; UCEC: uterine corpus endometrial carcinoma.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by the Natural Science Foundation of China (No. 81830082, 91740119, 81672874, 81502194), Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (GDUPS), the Distinguished Young Scholar of Guangdong Province (No. 2015A030306033), the Natural Science Foundation of Guangdong Province (No. 2015A030313468), the Young Scholar of Science and Technology of Guangdong Province (2016TQ03R801), Innovative Academic Team of Guangzhou Education System (1201610014), the Science and Technology Program of Guangzhou (201604020001, 201803010098), Natural Science Foundation research team of Guangdong Province (2018B030312001, 2018B030311009), The National Funds for developing local colleges and universities (B16056001), the Research Team of Department of Education of Guangdong Province (2017KCXTD027), Guangzhou key medical discipline construction project fund; Guangdong traditional Chinese medicine bureau project (20161178), and Guangzhou traditional Chinese medicine and traditional Chinese and western medicine science and technology project (2016A011020; 20182A011025).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in china, 2015. CA Cancer J Clin. 2016;66:115-32
2. Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, Kemler R. Lack of beta-catenin affects mouse development at gastrulation. Development. 1995;121:3529-37
3. Nusse R, Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985-99
4. Hermans KC, Blankesteijn WM. Wnt signaling in cardiac disease. Compr Physiol. 2015;5:1183-209
5. Tammela T, Sanchez-Rivera FJ, Cetinbas NM, Wu K, Joshi NS, Helenius K. et al. A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature. 2017;545:355-9
6. Barker N, Clevers H. Mining the Wnt pathway for cancer therapeutics. Nat Rev Drug Discov. 2006;5:997-1014
7. Zhu J, Wu G, Li Q, Gong H, Song J, Cao L. et al. Overexpression of suprabasin is associated with proliferation and tumorigenicity of esophageal squamous cell carcinoma. Sci Rep. 2016;6:21549
8. Wu Y, Zhang Y, Zhang H, Yang X, Wang Y, Ren F. et al. P15RS attenuates Wnt/{beta}-catenin signaling by disrupting {beta}-catenin.TCF4 interaction. J Biol Chem. 2010;285:34621-31
9. Jin K, Chen H, Zuo Q, Huang C, Zhao R, Yu X. et al. CREPT and P15RS regulate cell proliferation and cycling in chicken DF-1 cells through the Wnt/beta-catenin pathway. J Cell Biochem. 2018;119:1083-92
10. Liu C, Zhang Y, Li J, Wang Y, Ren F, Zhou Y. et al. P15RS/RPRD1A (p15ink4b-related sequence/regulation of nuclear pre-mrna domain-containing protein 1a) interacts with HDAC2 in inhibition of the Wnt/beta-catenin signaling pathway. J Biol Chem. 2015;290:9701-13
11. Jiang L, Yu L, Zhang X, Lei F, Wang L, Liu X. et al. Mir-892b silencing activates NF-kappaB and promotes aggressiveness in breast cancer. Cancer Res. 2016;76:1101-11
12. Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T. et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515-27
13. Comsa S, Cimpean AM, Raica M. The story of MCF-7 breast cancer cell line: 40 years of experience in research. Anticancer Res. 2015;35:3147-54
14. Kao J, Salari K, Bocanegra M, Choi YL, Girard L, Gandhi J. et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS One. 2009;4:e6146
15. Holliday DL, Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011;13:215
16. Jiang L, Zhou J, Zhong D, Zhou Y, Zhang W, Wu W. et al. Overexpression of SMC4 activates TGFbeta/smad signaling and promotes aggressive phenotype in glioma cells. Oncogenesis. 2017;6:e301
17. Song J, Xie C, Jiang L, Wu G, Zhu J, Zhang S. et al. Transcription factor AP-4 promotes tumorigenic capability and activates the WNT/beta-catenin pathway in hepatocellular carcinoma. Theranostics. 2018;8:3571-83
18. Jiang L, Wu J, Yang Y, Liu L, Song L, Li J. et al. Bmi-1 promotes the aggressiveness of glioma via activating the NF-kappab/MMP-9 signaling pathway. BMC Cancer. 2012;12:406
19. Huo D, Clayton WM, Yoshimatsu TF, Chen J, Olopade OI. Identification of a circulating microrna signature to distinguish recurrence in breast cancer patients. Oncotarget. 2016;7:55231-48
20. Matamala N, Vargas MT, Gonzalez-Campora R, Minambres R, Arias JI, Menendez P. et al. Tumor microRNA expression profiling identifies circulating microRNAs for early breast cancer detection. Clin Chem. 2015;61:1098-106
21. Jacob NK, Cooley JV, Yee TN, Jacob J, Alder H, Wickramasinghe P. et al. Identification of sensitive serum microrna biomarkers for radiation biodosimetry. PLoS One. 2013;8:e57603
22. McGuire A, Brown JA, Kerin MJ. Metastatic breast cancer: The potential of mirna for diagnosis and treatment monitoring. Cancer Metastasis Rev. 2015;34:145-55
23. Jung EJ, Santarpia L, Kim J, Esteva FJ, Moretti E, Buzdar AU. et al. Plasma microrna 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer. 2012;118:2603-14
24. Lu L, Mao X, Shi P, He B, Xu K, Zhang S. et al. MicroRNAs in the prognosis of triple-negative breast cancer: A systematic review and meta-analysis. Medicine (Baltimore). 2017;96:e7085
25. Mishra S, Srivastava AK, Suman S, Kumar V, Shukla Y. Circulating miRNAs revealed as surrogate molecular signatures for the early detection of breast cancer. Cancer Lett. 2015;369:67-75
26. Wang PY, Gong HT, Li BF, Lv CL, Wang HT, Zhou HH. et al. Higher expression of circulating miR-182 as a novel biomarker for breast cancer. Oncol Lett. 2013;6:1681-6
27. Bovy N, Blomme B, Freres P, Dederen S, Nivelles O, Lion M. et al. Endothelial exosomes contribute to the antitumor response during breast cancer neoadjuvant chemotherapy via microrna transfer. Oncotarget. 2015;6:10253-66
28. Adhikari AS, Agarwal N, Iwakuma T. Metastatic potential of tumor-initiating cells in solid tumors. Front Biosci (Landmark Ed). 2011;16:1927-38
29. Barlund M, Tirkkonen M, Forozan F, Tanner MM, Kallioniemi O, Kallioniemi A. Increased copy number at 17q22-q24 by cgh in breast cancer is due to high-level amplification of two separate regions. Genes Chromosomes Cancer. 1997;20:372-6
30. Wu G, Sinclair C, Hinson S, Ingle JN, Roche PC, Couch FJ. Structural analysis of the 17q22-23 amplicon identifies several independent targets of amplification in breast cancer cell lines and tumors. Cancer Res. 2001;61:4951-5
31. Zhang Q, Bai X, Chen W, Ma T, Hu Q, Liang C. et al. Wnt/beta-catenin signaling enhances hypoxia-induced epithelial-mesenchymal transition in hepatocellular carcinoma via crosstalk with HIF-1alpha signaling. Carcinogenesis. 2013;34:962-73
32. Li G, Su Q, Liu H, Wang D, Zhang W, Lu Z. et al. Frizzled7 promotes epithelial-to-mesenchymal transition and stemness via activating canonical Wnt/beta-catenin pathway in gastric cancer. Int J Biol Sci. 2018;14:280-93
33. Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442-54
34. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871-90
35. Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST. et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472-6
36. Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H. et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525-30
37. Su S, Liu Q, Chen J, Chen J, Chen F, He C. et al. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell. 2014;25:605-20
38. Neelakantan D, Zhou H, Oliphant MUJ, Zhang X, Simon LM, Henke DM. et al. EMT cells increase breast cancer metastasis via paracrine gli activation in neighbouring tumour cells. Nat Commun. 2017;8:15773
39. Zhang K, Corsa CA, Ponik SM, Prior JL, Piwnica-Worms D, Eliceiri KW. et al. The collagen receptor discoidin domain receptor 2 stabilizes snail1 to facilitate breast cancer metastasis. Nat Cell Biol. 2013;15:677-87
40. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY. et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704-15
41. Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during snail-induced emt of cancer cells. Cancer Cell. 2009;15:195-206
42. Choi WW, Lewis MM, Lawson D, Yin-Goen Q, Birdsong GG, Cotsonis GA. et al. Angiogenic and lymphangiogenic microvessel density in breast carcinoma: Correlation with clinicopathologic parameters and vegf-family gene expression. Mod Pathol. 2005;18:143-52
43. Gunasinghe NP, Wells A, Thompson EW, Hugo HJ. Mesenchymal-epithelial transition (MET) as a mechanism for metastatic colonisation in breast cancer. Cancer Metastasis Rev. 2012;31:469-78
44. Wang Z, Li B, Zhou L, Yu S, Su Z, Song J. et al. Prodigiosin inhibits wnt/beta-catenin signaling and exerts anticancer activity in breast cancer cells. Proc Natl Acad Sci U S A. 2016;113:13150-5
45. Wickstrom M, Dyberg C, Milosevic J, Einvik C, Calero R, Sveinbjornsson B. et al. Wnt/beta-catenin pathway regulates MGMT gene expression in cancer and inhibition of Wnt signalling prevents chemoresistance. Nat Commun. 2015;6:8904
46. Wang B, Tian T, Kalland KH, Ke X, Qu Y. Targeting Wnt/beta-catenin signaling for cancer immunotherapy. Trends Pharmacol Sci. 2018;39:648-58
47. Valencia A, Roman-Gomez J, Cervera J, Such E, Barragan E, Bolufer P. et al. Wnt signaling pathway is epigenetically regulated by methylation of wnt antagonists in acute myeloid leukemia. Leukemia. 2009;23:1658-66
48. Chim CS, Pang R, Fung TK, Choi CL, Liang R. Epigenetic dysregulation of wnt signaling pathway in multiple myeloma. Leukemia. 2007;21:2527-36
49. Lv C, Li F, Li X, Tian Y, Zhang Y, Sheng X. et al. Mir-31 promotes mammary stem cell expansion and breast tumorigenesis by suppressing Wnt signaling antagonists. Nat Commun. 2017;8:1036
50. Cai J, Fang L, Huang Y, Li R, Xu X, Hu Z. et al. Simultaneous overactivation of Wnt/beta-catenin and tgfbeta signalling by mir-128-3p confers chemoresistance-associated metastasis in nsclc. Nat Commun. 2017;8:15870
51. Song XL, Huang B, Zhou BW, Wang C, Liao ZW, Yu Y. et al. Mir-1301-3p promotes prostate cancer stem cell expansion by targeting SFRP1 and GSK3beta. Biomed Pharmacother. 2018;99:369-74
52. Rupaimoole R, Slack FJ. Microrna therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203-22
Author contact
![]() Corresponding author: Lili Jiang, MD., Affiliated Cancer Hospital & Institute of Guangzhou Medical University; Key Laboratory of Protein Modification and Degradation; State Key Laboratory of Respiratory Disease, School of Basic Medical Sciences, Guangzhou Medical University, Guangzhou 511436, China; Phone: +86(20)37104123; E-mail: jiangliliedu.cn.
Corresponding author: Lili Jiang, MD., Affiliated Cancer Hospital & Institute of Guangzhou Medical University; Key Laboratory of Protein Modification and Degradation; State Key Laboratory of Respiratory Disease, School of Basic Medical Sciences, Guangzhou Medical University, Guangzhou 511436, China; Phone: +86(20)37104123; E-mail: jiangliliedu.cn.
 Global reach, higher impact
Global reach, higher impact