13.3
Impact Factor
Theranostics 2019; 9(15):4542-4557. doi:10.7150/thno.35282 This issue Cite
Research Paper
Host-derived fecal microRNAs can indicate gut microbiota healthiness and ability to induce inflammation
1. Institute for Biomedical Sciences, Center for Inflammation, Immunity and Infection, Digestive Disease Research Group, Georgia State University, Atlanta, GA, USA.
2. Neuroscience Institute, Georgia State University, Atlanta, GA, USA
3. Veterans Affairs Medical Center, Decatur, GA, USA
Abstract
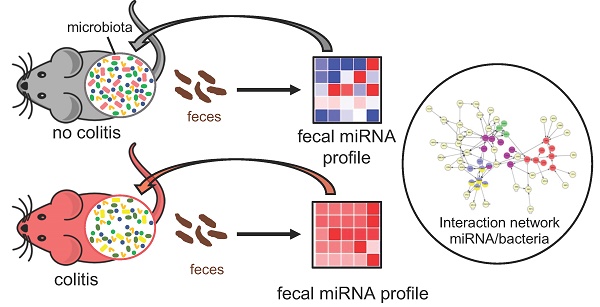
Disruption of intestine-microbiota symbiosis can result in chronic gut inflammation. We hypothesize that assessing the initial inflammatory potential of the microbiota in patients is essential and that host-derived miRNAs, which can be found in feces, could fulfill this function. We investigated whether the gut microbiota composition impacts the fecal miRNA profile and thereby indicates its ability to influence intestinal inflammation.
Methods: We used high-throughput qPCR to compare fecal miRNA profile between germ-free and conventional mice. Conventionalization of germfree mice by various colitogenic and non-colitogenic microbiotas (IL10-/- and TLR5-/- associated microbiota) was performed.
Results: We identified 12 fecal miRNAs impacted by the presence of a microbiota. Conventionalization of germfree mice by various colitogenic and non-colitogenic microbiotas associated with the development of intestinal inflammation (IL10-/- and TLR5-/- associated microbiota) yielded distinctively altered fecal miRNA profiles compared to that of mice receiving a “healthy” microbiota. Correlation analysis revealed the existence of interactions between the 12 abovementioned miRNAs and specific microbiota members.
Conclusion: These results showed that fecal miRNA profile can be differentially and specifically impacted by microbiota composition, and that miRNA could importantly serve as markers of the colitogenic potential of the microbiota. This is particularly relevant to assess individual state of the microbiota in patients with dysbiosis-related disorders, such as IBD and potentially determine their ability to respond to therapeutics.
Keywords: miRNA, microbiota, inflammation, feces, colitis
 Global reach, higher impact
Global reach, higher impact