13.3
Impact Factor
Theranostics 2019; 9(16):4567-4579. doi:10.7150/thno.31502 This issue Cite
Research Paper
A Comprehensive Study of Vesicular and Non-Vesicular miRNAs from a Volume of Cerebrospinal Fluid Compatible with Clinical Practice
1. Department of Genetics, Physical Anthropology and Animal Physiology, Faculty of Medicine and Nursing, University of The Basque Country (UPV/EHU), Leioa, Bizkaia, 48940, Spain.
2. Genome Analysis Platform, CIC bioGUNE, Derio, Bizkaia, 48980, Spain.
3. Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas (CIBERehd), Instituto de Salud Carlos III, Madrid, 28029, Spain.
4. Exosomes Lab, CIC bioGUNE, CIBERehd, Derio, Bizkaia, 48980, Spain.
5. Bioinformatics Unit, Centre Esther Koplovitz (CEK), CIBERehd, Barcelona, 08036, Spain.
6. Biochemistry Service, Cruces University Hospital, Barakaldo, Bizkaia, 48903, Spain.
7. Department of Pediatric Emergency, Cruces University Hospital, Barakaldo, Bizkaia, 48903, Spain.
8. Department of Pediatrics, University of The Basque Country (UPV/EHU), Leioa, Bizkaia, 48940, Spain.
9. BioCruces Health Research Institute, Barakaldo, Bizkaia, 48903, Spain.
10. IKERBASQUE, Basque Foundation for Science, Bilbao, Bizkaia, 48015, Spain.
Abstract
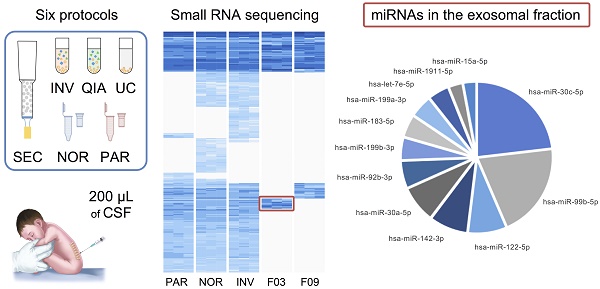
Cerebrospinal fluid (CSF) microRNAs (miRNAs) have emerged as potential biomarkers for minimally invasive diagnosis of central nervous system malignancies. However, despite significant advances in recent years, this field still suffers from poor data reproducibility. This is especially true in cases of infants, considered a new subject group. Implementing efficient methods to study miRNAs from clinically realistic CSF volumes is necessary for the identification of new biomarkers.
Methods: We compared six protocols for characterizing miRNAs, using 200-µL CSF from infants (aged 0-7). Four of the methods employed extracellular vesicle (EV) enrichment step and the other two obtained the miRNAs directly from cleared CSF. The efficiency of each method was assessed using real-time PCR and small RNA sequencing. We also determined the distribution of miRNAs among different CSF shuttles, using size-exclusion chromatography.
Results: We identified 281 CSF miRNAs from infants. We demonstrated that the miRNAs could be efficiently detected using only 200 µL of biofluid in case of at least two of the six methods. In the exosomal fraction, we found 12 miRNAs that might be involved in neurodevelopment.
Conclusion: The Norgen and Invitrogen protocols appear suitable for the analysis of a large number of miRNAs using small CSF samples.
Keywords: CSF miRNAs, CSF exosomes, microRNA profiling, infants, clinical samples
 Global reach, higher impact
Global reach, higher impact