13.3
Impact Factor
Theranostics 2019; 9(19):5532-5541. doi:10.7150/thno.34070 This issue Cite
Research Paper
Tumor-derived DNA from pleural effusion supernatant as a promising alternative to tumor tissue in genomic profiling of advanced lung cancer
1. Department of Pulmonary Medicine, Zhongshan Hospital, Fudan University, Shanghai, 200032, China
2. Shanghai Respiratory Research Institute, Shanghai, 200032, China
3. Translational Medicine Research Institute, Geneseeq Technology Inc., Toronto, Ontario, M5G 1L7, Canada
4. Medical Department, Nanjing Geneseeq Technology Inc., Nanjing, Jiangsu, 210032, China
5. School of Public Health, Nanjing Medical University, Nanjing, Jiangsu, 210029, China
*These authors contributed equally to this work.
Abstract
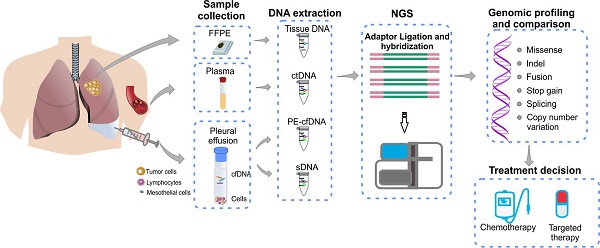
Pleural effusion (PE) is commonly observed in advanced lung cancer and was suggested to contain both cell-free tumor DNA and tumor cells. Molecular profiling of PE represents a minimally invasive approach of detecting tumor driver mutations for clinical decision making, especially when tumor tissues are not available. The objective of this study is to investigate the efficacy and precision of detecting gene alterations in PE samples to address the feasibility in clinical use.
Methods: Sixty-three metastatic lung cancer patients with (n=30, cohort 1) or without (n=33, cohort 2) matched tumor tissues were enrolled in this study. PE and plasma samples of each patient were collected simultaneously. Supernatant and cell precipitate of PE were processed separately to extract cfDNA (PE-cfDNA) and sediment DNA (sDNA). All samples were subjected to targeted next-generation sequencing (NGS) of 416 cancer-related genes.
Results: PE supernatants contain more abundant tumor DNA than PE sediments and plasma samples, suggested by higher mutant allele frequencies (MAF) and elevated mutation detection rate in PE-cfDNA (98.4% vs. 90.5% in PE sDNA vs. 87% in plasma cfDNA). In Cohort 1 with matched tumor tissue, tumor mutational burden (TMB) of PE-cfDNA was similar as tumor tissues (6.4 vs. 5.6), but significantly higher than PE sDNA (median TMB: 3.3) and plasma cfDNA (median TMB: 3.4). Ninety-three percent (27 out of 29) of tissue-determined driver mutations were detected in PE-cfDNA, including alterations in ALK, BRAF, EGFR, ERBB2, KRAS, NF1, PIK3CA, and RET, while only 62% were captured in plasma cfDNA. PE-cfDNA also has the highest detection rate of EGFR driver mutations in the full cohort (71% vs. 68% in PE sDNA vs. 59% in plasma cfDNA). Mutation detection from cytological negative and hemorrhagic PE is challenging. Comparatively, PE-cfDNA demonstrated absolute superiority than PE sDNA in such a scenario, suggesting that it is an independent source of tumor DNA and therefore less influenced by the abundance of tumor cells.
Conclusion: Genomic profiling of PE-cfDNA offers an alternative, and potentially more meticulous approach in assessing tumor genomics in advanced lung cancer when tumor tissue is not available. Our data further demonstrate that in hemorrhagic or cytologically negative PE samples, PE-cfDNA has higher mutation detection sensitivity than sDNA and plasma cfDNA, and therefore is a more reliable source for genetic testing.
Keywords: lung cancer, pleural effusion, cell free DNA, genomic profiling, next-generation sequencing
 Global reach, higher impact
Global reach, higher impact