13.3
Impact Factor
Theranostics 2019; 9(22):6601-6617. doi:10.7150/thno.37549 This issue Cite
Research Paper
Dual-platform affinity proteomics identifies links between the recurrence of ovarian carcinoma and proteins released into the tumor microenvironment
1. Institute of Molecular Biology and Tumor Research (IMT), Center for Tumor Biology and Immunology (ZTI), Philipps University, Marburg, Germany
2. Clinic for Gynecology, Gynecological Oncology and Gynecological Endocrinology, Center for Tumor Biology and Immunology (ZTI), Philipps University, Marburg, Germany
3. Experimental Tumor Biology, Clinic for Hematology, Oncology and Immunology, Center for Tumor Biology and Immunology (ZTI), Philipps University, Marburg, Germany
4. Clinic for Gynecology, Gynecological Oncology and Gynecological Endocrinology, University Hospital of Giessen and Marburg (UKGM), Marburg, Germany
5. Institute of Pharmacology, Biochemical-Pharmacological Center (BPC), Philipps University, Marburg, Germany
6. Department of Pharmacology, Max-Planck-Institute for Heart and Lung Research, Bad Nauheim, Germany
7. Biomolecular Mass Spectrometry, Max-Planck-Institute for Heart and Lung Research, Bad Nauheim, Germany
8. German Centre for Cardiovascular Research (DZHK), Partner Site Rhine-Main, Max-Planck-Institute for Heart and Lung Research, Bad Nauheim, Germany
Abstract
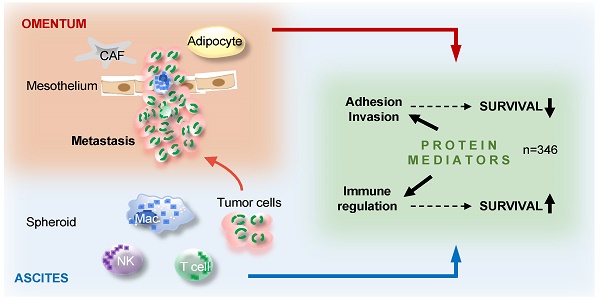
The peritoneal fluid (ascites), replete with abundant tumor-promoting factors and extracellular vesicles (EVs) reflecting the tumor secretome, plays an essential role in ovarian high-grade serous carcinoma (HGSC) metastasis and immune suppression. A comprehensive picture of mediators impacting HGSC progression is, however, not available.
Methods: Proteins in ascites from HGSC patients were quantified by the aptamer-based SOMAscan affinity proteomic platform. SOMAscan data were analyzed by bioinformatic methods to reveal clinically relevant links and functional connections, and were validated using the antibody-based proximity extension assay (PEA) Olink platform. Mass spectrometry was used to identify proteins in extracellular microvesicles released by HGSC cells.
Results: Consistent with the clinical features of HGSC, 779 proteins in ascites identified by SOMAscan clustered into groups associated either with metastasis and a short relapse-free survival (RFS), or with immune regulation and a favorable RFS. In total, 346 proteins were linked to OC recurrence in either direction. Reanalysis of 214 of these proteins by PEA revealed an excellent median Spearman inter-platform correlation of ρ=0.82 for the 46 positively RFS-associated proteins in both datasets. Intriguingly, many proteins strongly associated with clinical outcome were constituents of extracellular vesicles. These include proteins either linked to a poor RFS, such as HSPA1A, BCAM and DKK1, or associated with a favorable outcome, such as the protein kinase LCK. Finally, based on these data we defined two protein signatures that clearly classify short-term and long-term relapse-free survivors.
Conclusion: The ascites secretome points to metastasis-promoting events and an anti-tumor response as the major determinants of the clinical outcome of HGSC. Relevant proteins include both bone fide secreted and vesicle-encapsulated polypeptides, many of which have previously not been linked to HGSC recurrence. Besides a deeper understanding of the HGSC microenvironment our data provide novel potential tools for HGSC patient stratification. Furthermore, the first large-scale inter-platform validation of SOMAscan and PEA will be invaluable for other studies using these affinity proteomics platforms.
 Global reach, higher impact
Global reach, higher impact