13.3
Impact Factor
Theranostics 2020; 10(4):1479-1499. doi:10.7150/thno.40659 This issue Cite
Review
Re-recognition of pseudogenes: From molecular to clinical applications
1. State Key Laboratory of Cancer Biology, Department of Immunology, Fourth Military Medical University, Xi'an, Shaanxi, 710032, P.R. China.
2. Department of Hematology and Oncology, Children's Hospital of Soochow University, Suzhou, Jiangsu, 215025, P.R. China.
3. Department of Medical Genetics and Developmental Biology, Fourth Military Medical University, Xi'an, Shaanxi, 710032, P.R. China.
Received 2019-9-25; Accepted 2019-11-12; Published 2020-1-1
Abstract
Pseudogenes were initially regarded as “nonfunctional” genomic elements that did not have protein-coding abilities due to several endogenous inactivating mutations. Although pseudogenes are widely expressed in prokaryotes and eukaryotes, for decades, they have been largely ignored and classified as gene “junk” or “relics”. With the widespread availability of high-throughput sequencing analysis, especially omics technologies, knowledge concerning pseudogenes has substantially increased. Pseudogenes are evolutionarily conserved and derive primarily from a mutation or retrotransposon, conferring the pseudogene with a “gene repository” role to store and expand genetic information. In contrast to previous notions, pseudogenes have a variety of functions at the DNA, RNA and protein levels for broadly participating in gene regulation to influence the development and progression of certain diseases, especially cancer. Indeed, some pseudogenes have been proven to encode proteins, strongly contradicting their “trash” identification, and have been confirmed to have tissue-specific and disease subtype-specific expression, indicating their own value in disease diagnosis. Moreover, pseudogenes have been correlated with the life expectancy of patients and exhibit great potential for future use in disease treatment, suggesting that they are promising biomarkers and therapeutic targets for clinical applications. In this review, we summarize the natural properties, functions, disease involvement and clinical value of pseudogenes. Although our knowledge of pseudogenes remains nascent, this field deserves more attention and deeper exploration.
Keywords: pseudogene, classification, function, diagnosis, prognosis, therapeutics
Introduction
Since the completion of the Human Genome Project, multiple genomic sequencing analyses have been successively accomplished in different organisms, providing numerous clues for the thorough identification of the genome, transcriptome and proteome. According to sequencing data, the entire human genome possesses approximately more than three billion bases; however, only 2% of DNA sequences encode “functional” proteins [1], and the other 98% are regarded as “trash” elements that evolved from neutral selection without coding ability. In that period, pseudogenes, along with other noncoding factors, were all categorized as “trash” sequences.
The first pseudogene was identified in 1977 when several mutations were simultaneously discovered in its DNA sequence. Due to internal mutations, the pseudogene lost its coding ability and served as a homologous gene copy as its counterpart in the genome [2]. Since then, pseudogenes have been broadly identified in a series of organisms ranging from prokaryotes to eukaryotes [3]. Nevertheless, because of the previous “nonfunctional” label, pseudogenes have for decades been considered as “junk DNA”, “genomic fossils” and “gene relics”; a number of strategies were even developed to focus on eliminating a pseudogene when attempting to determine its parental gene [4, 5]. With the wide application of next-generation sequencing technology, pseudogenes have gradually been found to exert parental gene-dependent and parental gene-independent functions at the DNA, RNA and protein levels; these sequences are thus involved in transcriptional and posttranscriptional modulation, participating in the physiological maintenance of endogenous homeostasis and in the pathological process of disease. Notably, a small fraction of pseudogenes reportedly maintain or have regained protein-coding capacity [6], suggesting that pseudogenes also act as conspicuous elements that contributes to the transcriptome and proteome of different species.
Currently, discovered “functional” pseudogenes comprise only a small fraction of the total, whereas the majority of pseudogenes have an “unknown” status with no established identification or function. Moreover, a number of nonfunctional “dying” pseudogenes indeed are present in the genome, e.g., the ancient O-acyltransferase-like pseudogene (ACYL3), increasing the complexity of pseudogene distribution [7]. Pseudogenes are evolutionally conserved [8], with properties of both predisposition in unique disease subtypes [9] and tissue-specificity [10], further highlighting the potential correlation between pseudogenes and certain diseases and the necessity to study their functions and mechanisms in these diseases.
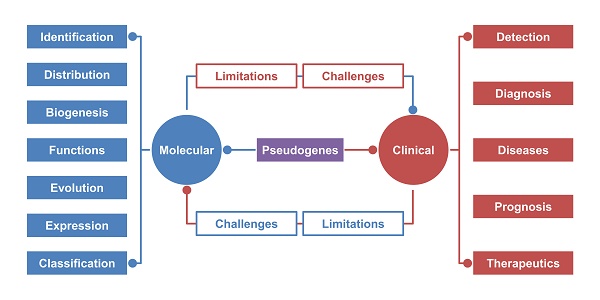
In this review, we summarize the identification, expression, evolution, biogenesis and function of pseudogenes. We also present evidence of pseudogene involvement in different diseases and the promising correlation between pseudogene and diagnosis, prognosis and therapeutics in the clinic. Finally, we discuss the current detection methods, limitations and challenges of pseudogene exploration to optimize existing protocols to increase their efficiency for further pseudogene research.
Identification: A Long-Term Investigation
The first identification and naming of a pseudogene in human history was in 1977, when Jacq et al. [2] found a gene copy that was homologous to 5S rRNA in Xenopus laevis. By comparing its DNA sequence with that of 5S rRNA, they discovered a 16 base pair (bp)-deficiency and a 14 bp-mismatch condition within the 5'-terminal of this copy. In addition, its mRNA expression could barely be detected, suggesting that this gene possessed no coding capability and was considered “nonfunctional”. It was presumed that this type of aberrant gene, displaying high sequence homology to functional genes, lost its coding ability due to different mutations, such as a frameshift mutation or premature stop codon in the genome, and was termed a “pseudogene” [11].
Since then, a large number of pseudogenes have been gradually discovered from monocellular organisms to multicellular organisms and from prokaryotes to eukaryotes [12] with the aid of next-generation sequencing technologies. However, because of the high homology of pseudogene sequences to those of parental genes (termed “ancestral gene”, “cognate gene” or “counterpart”), an emerging issue faced by pseudogene analysis is how to distinguish them from their counterparts. There have long been many attempts to identify pseudogenes more accurately. First, the Ka/Ks index (rate of the nucleotide nonsynonymous to synonymous substitution) was applied as a criterion to identify pseudogenes [13] because during evolution, these sequences are under neutral selection, as opposed to positive or purifying selection. Therefore, the Ka/Ks index should be close to or equal to one [14]. In fact, under the guidance of the Ka/Ks index, over 8,000 processed pseudogenes have been identified in various species [15]. In this case, the Ka/Ks index serves as an initial step during pseudogene identification.
In addition to the Ka/Ks index, features of pseudogenes, such as a special category [16] and transcriptional capacity [17], became new proof for pseudogene identification, e.g., processed pseudogenes should be found in the same genome that contains their paralogs, whereas unitary pseudogenes exist alone without paralogs. Several pseudogenes were later confirmed to be transcribed, which is easily identified through RNA transcripts. Although this method may help to increase accuracy, the approach is time-consuming and not efficient because a large amount of manual work needs to be performed and pseudogenes without transcripts are difficult to identify. Therefore, this strategy is better used together with other methods. Further trials are needed to expand the search scope without decreasing accuracy.
With the rapid development of next-generation sequencing technology, strategies focusing on pseudogene identification were changed to depend on bioinformatics, which significantly promoted our recognition of pseudogenes in the whole genomes of different species. By conducting established pipelines with public databases, a large amount of comprehensive information can be acquired in a short time. However, the efficiency of this method has several limitations. 1) Because pipelines require information related to the genome, transcriptome and proteome, they are not suitable for pseudogene detection in atypical organisms. 2) Pseudogenes that are not transcribed are outside the testing range for RNA sequencing analysis. 3) Some pseudogenes only have a few nucleotides that differ from their parental genes in sequence, and an objective evaluation is needed to determine whether these differences derive from genomic mutations or sequencing errors. 4) Low expression levels and small coverage of RNA sequencing analysis are likely to result in negative results for a specific pseudogene [17, 18]. Notably, despite these drawbacks, bioinformatics has become the most effective and accurate strategy for pseudogene identification.
In conclusion, pseudogene identification is a long-term investigative process from more manual works to more intelligent innovations, bringing our recognition of pseudogenes into a new era. However, more efforts should still be made to improve the breadth and precision of our current methods to help better understand pseudogenes.
Distribution: Extremely Wide and Uneven
The distribution of pseudogenes can be classified into two perspectives, macroscopic and microscopic. From a macroscopic perspective, the distribution of pseudogenes relies on species that are different and range from monocellular organisms to multicellular organisms: monocellular organisms have few or no pseudogenes with exclusive effects, whereas multicellular organisms, including prokaryotes and eukaryotes, possess many more pseudogenes [3]. Almost 11,000 pseudogenes have been identified in the complete human genome [19], and more than two-thirds (over 8,000) have been verified as processed pseudogenes [15].
From a microscopic perspective, only 10% of the genes in the entire human genome can be detected with at least one pseudogene counterpart. Moreover, the distribution of pseudogenes per coding gene is markedly uneven [20, 21]. Notably, pseudogenes are frequently located in regions undergoing DNA duplication, deletion or chromosomal rearrangement [22], which may give rise to more mutations in those sequences. In addition to the global distribution of pseudogenes, the total amount of transcribed pseudogenes varies widely, ranging from 6% [23] to 20% [24]. Compared with their parental genes, the RNA transcripts of pseudogenes also change significantly in abundance because decreased levels are found for the majority of pseudogene transcripts [25]. Nonetheless, for some examples, such as pseudogenes of POU class 5 homeobox 1 (OCT4), the levels are almost equal to or even increased [26], further indicating the uneven distribution of pseudogenes at the molecular level.
Taken together, these details highlight the extremely wide and uneven distribution of pseudogenes within the genome at macroscopic and microscopic levels, suggesting their intrinsic diversity and complexity in genomes.
Expression: A Spatiotemporal and Unique Pattern
The expression pattern of a pseudogene shows a strongly spatiotemporal property compared with that of its parental gene, and these expression patterns appear to occur in two completely opposite phases. In fact, most pseudogenes are expressed in parallel with their parental genes, e.g., loss of phosphatase and tensin homolog pseudogene 1 (PTENP1), a processed pseudogene of PTEN at chromosome 9p13.3, can lead to a remarkable reduction in the level of PTEN [27]. Both the PTEN and PTENP1 loci may be deleted in melanoma [28], suggesting a positive spatiotemporal correlation between the parental gene and its pseudogene. However, several pseudogenes exhibit an expression pattern that is entirely different from that of their parental genes, e.g., the 5-hydroxytryptamine receptor 7 (HTR7) pseudogene can be detected in the liver and kidney, whereas its counterpart HTR7 is exclusively present in the central nervous system (CNS). Additionally, RNA transcripts of secretory blood group 1, pseudogene (SEC1P) have been found in all tumor cell lines detected, but those of its parental gene fucosyltransferase 2 (FUT2) were not found in six leukemia cell lines despite the same chromosomal location and almost 70% homology, as supported by evidence from Koda et al. [29]. Therefore, spatiotemporal expression specificity is probably the reason that pseudogenes can function in a parental gene-dependent or parental gene-independent manner.
In addition to a spatiotemporal expression pattern that is different from that of its parental gene, a pseudogene also shows a unique expression profile in different specimens and under various conditions. First, pseudogenes frequently display a tissue-specific expression profile in different organs, tissues, and even blood; for example, SUMO1P, a pseudogene of small ubiquitin like modifier 1 (SUMO1), is upregulated in gastric cancer (GC) tissues compared with benign gastric disease tissues [10], and expression of integrator complex subunit 6 pseudogene 1 (INTS6P1) in the plasma of hepatocellular carcinoma (HCC) patients is significantly decreased compared with the plasma of non-HCC patients [30]. Pseudogenes also appear to be expressed in a specific disease subtype. For instance, Kalyana-Sundaram et al. performed an RNA-seq analysis on samples from 13 cancers and their corresponding normal tissues and found 218 pseudogenes and 40 pseudogenes that were only present in the cancer samples and a single cancer subtype, respectively [31]. Similarly, the pseudogene Nanog homeobox retrogene P8 (NANOGP8) is aberrantly expressed in cancer cell lines, though its counterpart NANOG is not [32]. Furthermore, different physiological or pathological conditions may lead to alterations in pseudogene expression, such as cell differentiation [33], diabetes [34], asthma [35] and cancer [36, 37]. Moreover, single-nucleotide polymorphisms (SNPs) can occur in pseudogene sequences to induce variants, such as alleles of poly(ADP-ribose) polymerase (PADPRP)-processed pseudogene [38], E2F transcription factor 3 pseudogene 1 (E2F3P1) [39] and OCT4-pg1 [40]. Finally, epigenetic modifications, such as DNA methylation, are involved in modulating pseudogene expression, e.g., the promoter region of PTENP1 in GC cells is dramatically enriched with DNA methylation, leading to an epigenetic silencing effect [41]. In conclusion, a pseudogene has its own expression pattern, which is different from that of the parental gene, in some disease conditions, serving as a potential biomarker in clinical applications.
Evolution: “Molecular Fossil” and “Gene Repository”
The identification of pseudogenes has revealed an interesting phenomenon in which pseudogenes are highly homologous to their parental genes because of their origin, strongly indicating their evolutionary conservation. In addition, the ratio of nucleotide nonsynonymous to synonymous (Ka/Ks) mutations of a pseudogene is close to or equal to one, which is relatively high, suggesting that despite the mutations involved, the pseudogene is under an evolutionary constraint [8]. Moreover, with the preservation of a specific sequence, a pseudogene has its own identity when evaluating genetic relationships and evolutionary distances between species, acting as a “molecular fossil” or “gene relic” in the genome [42]. For instance, Marques et al. [43] found that a total of 48 pseudogenes are conserved in various specimens, including humans, mice, rats and dogs. Another recent study identified 68 pseudogenes that are conserved in humans and two other mammals [44], indicating high evolutionary conservation of the pseudogene in primates.
In fact, pseudogenes are under neutral selection pressure to be maintained in the human genome [15], and they gradually develop functions that are similar to or even greater than those of their counterparts [45], functioning as a “gene repository” to store and expand genetic information. Furthermore, the number of pseudogenes in the genomes of multicellular organisms is much higher than that in the genomes of monocellular organisms, and monocellular organisms are capable of excluding genes that have become pseudogenes, further indicating the “gene repository” role of pseudogenes in higher organisms [3].
Nevertheless, despite some current evidence proving the conserved evolution of pseudogenes, more efforts should be made to increase the proof and to elucidate the underlying mechanisms.
Biogenesis and Classification
Biogenesis: A pseudogene is regarded as a product and a reservoir of gene mutations
Due to the duplication and transcriptional properties of the human genome, more than one product of a gene is produced, which significantly promotes genetic information heritance but lays a foundation for pseudogene biogenesis. Pseudogenes are primarily derived from two events. 1) Mutation: a gene that is newly generated during DNA duplication or multiple mutations (such as insertion, deletion, frameshift, premature stop codon, and splicing error in the coding or regulatory regions) can give rise to loss of its function, especially the protein-coding property, and can transfer it to a pseudogene [46]. Similarly, for the original functional gene, the accumulation of mutations in certain domains can also convert it to a “nonfunctional” pseudogene [25]. 2) Retrotransposon: reversely transcribed cDNA may randomly reintegrate into the genome by forming an inappropriate locus or mutation, leading to the biogenesis of a functionally insufficient pseudogene [17]. Notably, based on the abovementioned evidence, the biogenesis of pseudogenes is more likely to proceed during high-synthesis and high-metabolism DNA events, which provide more opportunities for mutations; this is supported by evidence that pseudogenes may be present at a higher rate in reproductive cells than in somatic cells [47]. Therefore, pseudogenes serve as an outcome and the simultaneous storage of gene mutations in the human genome.
In theory, any sequence in the genome can give rise to a pseudogene because the key trigger is a mutation that frequently and inevitably occurs. However, some elements are likely to affect pseudogene biogenesis. 1) Type of nucleotide: Pseudogene biogenesis is less common in regions enriched with GC nucleotides, which is probably due to their negative effects on the accumulation of mutations [48]. 2) Length of the gene: A different coding gene length tends to produce a different subtype of pseudogene, e.g., a processed pseudogene is generally found in a short coding gene [49]. 3) Gene condition: As mentioned above, certain genes that are frequently involved in a high duplication status can increase the possibility for mutations, such as highly expressed genes in cell division and metabolism [50]. 4) Pseudogene: Evidence shows that the parental gene is not the only source of a pseudogene, which can also derive from another pseudogene [51]. These findings not only reveal the diversity of pseudogene biogenesis but also provide novel approaches for pseudogene identification.
Classification: A pseudogene is categorized via a distinct biogenesis process
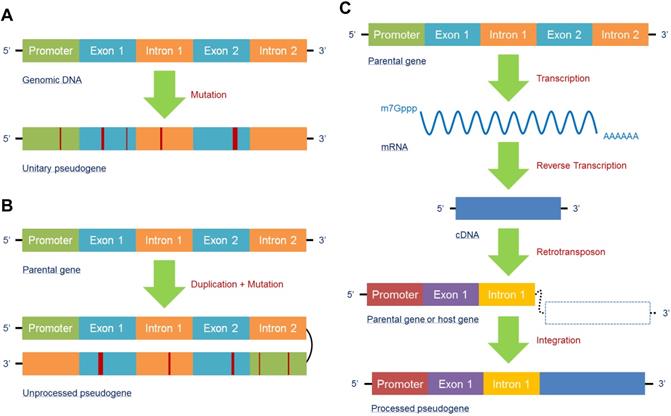
In accordance with the unique biogenesis mechanism, pseudogenes can generally be classified into three types: unitary pseudogenes, unprocessed pseudogenes, and processed pseudogenes. A unitary pseudogene, as its name suggests, is derived from a single coding gene copy with a few mutations that restrain transcription or translation. Hence, there is no fully functional genomic counterpart for the unitary pseudogene in the same genome, though orthologs can be found in related species [16] (Figure 1A). An unprocessed pseudogene, also known as a duplicated pseudogene, is the product of aberrant DNA duplication with mutation. Although it is located in the same region of a chromosome and contains the introns with flanking sequences of its counterpart, its ability to be transcribed or encode proteins is lost due to mutations in coding or regulatory sequences; in contrast, its counterpart retains its original functions [52] (Figure 1B). A processed pseudogene is different from the above two types because its main mechanism of formation is the retrotransposon of mRNA transcripts. As a result, processed pseudogenes may contain poly(A) tails without introns and regulatory sequences and integrate randomly into the genome; thus, they are more likely to be found in a new location far from their counterparts or on different chromosomes [53]. Additionally, as retrotransposon is not a high-fidelity process, mutations may occur within the processed pseudogene to suppress its function [54]. Moreover, transcription of a processed pseudogene relies on the regulatory elements of its host gene because it lacks a promoter, which is different from its parental gene [24] (Figure 1C). In fact, because of the specific biogenesis mechanisms and structures, the abovementioned three subtypes of pseudogenes can display a variety of functions that help them participate in the regulation of gene networks and diseases.
Pseudogenes are mainly generated in three forms. (A) The unitary pseudogene is derived from a coding gene with several mutations involved, leading to loss of its transcription and translation capacities, with have no fully functional counterpart in the same genome. (B) Due to unfaithful duplication, the duplicated gene generates a mutated gene copy that eventually becomes an unprocessed pseudogene; the original gene copy is fully functional. (C) A processed pseudogene derives from an mRNA that has been reverse transcribed into a cDNA and then synthesized into a host gene or parental gene via retrotransposon. Processed pseudogenes can be found far from their counterparts or on different chromosomes.
Functions
At different levels, a pseudogene serves a variety of functions other than that of a “nonfunctional” gene “trash” or “relic”. For example, at the DNA level, pseudogenes can impact their parental gene or host gene sequences by random insertion or DNA sequence exchange, thus further influencing their structures and functions. At the RNA level, RNA transcripts of pseudogenes can function as antisense RNAs, small interfering RNAs (siRNAs) and competing endogenous RNAs (ceRNAs) to regulate target gene expression at the posttranscriptional level. At the protein level, pseudogenes may be able to encode a protein or peptide to act as a “functional” gene involved in a gene regulation network. Therefore, pseudogenes are important because they thoroughly influence the human genome under different conditions, especially in diseases.
Function of pseudogenes at the DNA level
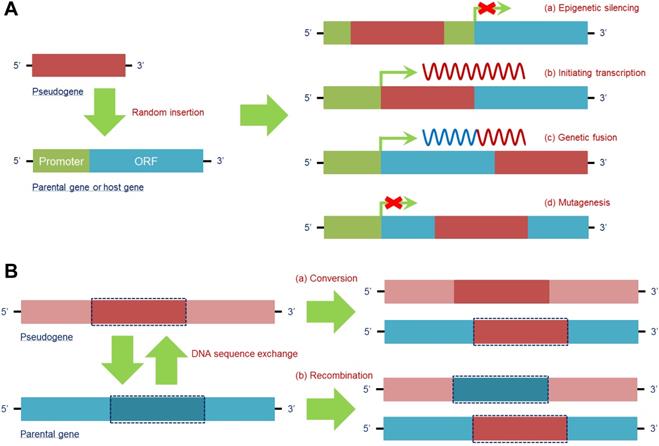
Random insertion
The DNA sequence of a pseudogene is able to randomly insert into the host gene to exert different effects, which mainly depend on the specific region of insertion. The insertion forms and effects are as follows.
1) Epigenetic silencing: By inserting into the upstream regulatory regions, particularly a promoter, a pseudogene may destroy its “landing site” and prevent host gene transcription, e.g., pseudogene protein tyrosine phosphatase non-receptor type 12 (PTPN12) inserts into the promoter region of MAX dimerization protein MGA (MGA), which acts as a potential lung cancer suppressor, to inactivate its expression and promote a malignant phenotype in NCI-H2009 cells [31]. Given that mutation occurring in antitumor genes is a key trigger of tumorigenesis, pseudogene-induced epigenetic silencing of antitumor genes provides a new form of mutation during tumor development.
2) Initiating transcription: When a pseudogene insertion site occurs in the intron or 3'-untranslated region (3'-UTR) of a host gene, the pseudogene is capable of using the transcriptional launch of the host gene to help trigger its own expression; in contrast, a pseudogene is unlikely to be expressed when inserting into an intergenic region. Notably, when a pseudogene insertion site is in the 3'-UTR of a host gene, all 3'-UTR-induced posttranscriptional regulation is lost [55]. In fact, this feature may promote the detection of some pseudogenes that cannot be transcribed in their original gene loci.
3) Genetic fusion: A processed pseudogene, which is inserted into a more downstream intron site of a host gene, tends to be cotranscribed with its own host gene, giving rise to a fusion RNA transcript that is partially derived from the pseudogene and partially derived from the host gene. For example, Koda et al. discovered a fusion gene of FUT2 and its pseudogene SEC1P, which is formed by an unequal crossing-over. The fusion gene contains the 5' region of SEC1P, indicating that it shares the same promoter region with SEC1P [29]. Because SEC1P can be detected in multiple cancer cell lines, expression of the FUT2 and SEC1P fusion is expected in tumors in vivo.
4) Mutagenesis: Transcription of the host gene can be abrogated when the insertion site of a pseudogene is within an exon, similar to an insertion mutation in a coding sequence [56]. This situation leads to functional loss of normal genes, especially those that are significant for homeostasis, which would result in microenvironment disruption and/or disease.
Therefore, pseudogene insertion can be regarded as a new area for recognizing disease mechanisms, especially for tumors because insertion frequently occurs (Figure 2A).
DNA sequence exchange
In addition to random insertion into a host gene, a pseudogene can perform a parental gene-dependent function by exchanging DNA sequences with the gene. The main pattern of this function exhibits two types. 1) Conversion: The DNA sequence in the parental gene can be substituted by the homologous sequence in the pseudogene. After this replacement, the newly substituted section and the rest of the sequence in the host gene become identical. For instance, the hybrid alleles PMS1 homolog 2, mismatch repair system component (PMS2) can carry sequences from its pseudogene, PMS2 C-terminal like pseudogene (PMS2CL), in exons 13-15, tracing back to an intrachromosomal recombination that possibly modulated cancer susceptibility in carriers [57]. Furthermore, conversion between pseudogenes and their parental genes provides a great opportunity for activation of oncogenes or inactivation of oncosuppressor genes [58]. 2) Recombination: Homologous sequences between a pseudogene and its counterpart can be exchanged, disrupting the original function of the parental gene. For instance, intron 2 of BRCA1 DNA repair associated (BRCA1) and intron 2 of its pseudogene PsiBRCA1 can be exchanged by recombination, transferring BRCA1 into a “nonfunctional” gene without a tumor suppressive effect [59], which constitutes a new mechanism for inactivating antitumor genes. In this case, DNA sequence exchange between the pseudogene and its counterpart provides a chance for disease occurrence (Figure 2B).
Pseudogenes have a series of regulatory effects at the DNA level. (A) DNA of the pseudogene can be randomly inserted into the parental gene or other host genes to regulate their transcription. By insertion into the promoter region of a target gene, a pseudogene is able to epigenetically silence its expression (a). In addition, pseudogenes can utilize the transcriptional mechanism of host genes to achieve their own transcription (b). Moreover, when the insertion site of a pseudogene is in a more downstream intron site of the host gene, a fusion gene is formed, and a chimeric RNA transcript is then produced (c). In fact, pseudogene insertion occurring in the coding region of a target gene can lead to mutagenesis and simultaneously loss of its own function (d). (B) DNA of the parental gene can also be influenced by pseudogenes via sequence exchange due to the high similarity of the sequences. Parental gene DNA can be substituted in a conversion with (a) or exchanged in a homologous recombination with (b) specific pseudogene DNA, thus finally affecting the function of the parental gene at the DNA level. Abbreviations: ORF: open reading frame.
Function of pseudogenes at the RNA level
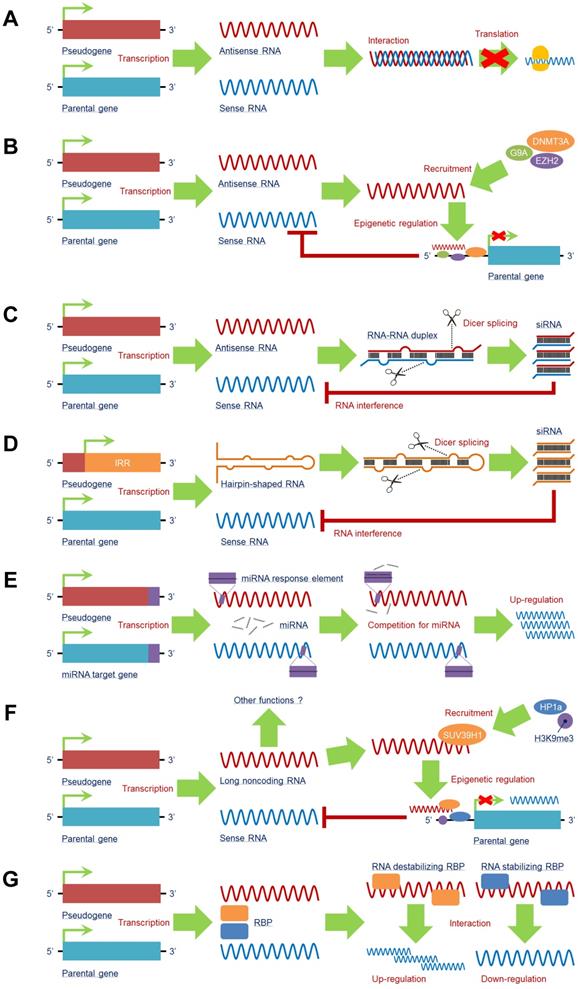
Transcription as an antisense transcript
A pseudogene can produce antisense RNA transcripts that directly interact with the mRNA of its counterpart, generating a double-stranded RNA-RNA duplex to restrain translation of the counterpart. A canonical example of this function is the pseudogene of neuronal nitric oxide synthase (nNOS), which is a natural antisense regulator and transcribed with a significant antisense region to nNOS, resulting in the formation of a double-stranded RNA-RNA duplex and a decline in nNOS protein synthesis. Both the nNOS and pseudoNOS transcripts are present in the same neuron, and the activity of the nNOS enzyme is substantially suppressed [60], suggesting that the pseudogene-mediated antisense mechanism can regulate the translation of some neuron-dependent genes and that simultaneously transcribed pseudogenes are a potential source of a new class of regulatory genes in the central nervous system (Figure 3A).
Intriguingly, potential crosstalk exists between the pseudogene antisense RNA transcript and epigenetic regulation, e.g., the antisense RNA alpha isoform of PTENP1 can recruit DNA methyltransferase 3 alpha (DNMT3A) and enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) to the promoter region of PTEN to suppress its transcription, whereas the beta isoform of PTENP1 antisense RNA still performs the traditional function [61]. OCT4-pg5 is able to produce to an antisense RNA that enhances enrichment of histone trimethylated at lysine 27 (H3K27me3) to repress OCT4 transcription by recruiting EZH2 and euchromatic histone lysine methyltransferase 2 (G9A) to its promoter region [62] (Figure 3B), indicating that pseudogenes may have both transcriptional and postregulatory effects on parental genes.
Processing into siRNAs
Several pseudogenes are capable of generating endogenous siRNAs. There are two major mechanisms that pseudogenes rely on for processing into siRNAs. One is from RNA-RNA duplexes formed by sense mRNA and antisense RNA transcript from counterpart pseudogenes (Figure 3C); the other is from hairpin-shaped RNA generated by the inverted repeat region of the pseudogene (Figure 3D). Both products can be sliced into siRNAs by Dicer. The single-stranded siRNAs are incorporated into an RNA-induced silencing complex to initiate the RNA interference (RNAi) process to regulate the counterpart gene [63].
Pseudogene RNA exists in various forms and plays a vital role in target gene expression. A pseudogene can be transcribed into antisense RNA that (A) interacts with or (B) recruits multiple negative epigenetic regulators, such as G9A, DNMT3A, and EZH2, to the promoter region of the parental gene to induce an inhibitory effect on its transcription. Some pseudogenes can produce endogenous siRNAs by (C) interacting with sense RNA to form a double-stranded RNA-RNA duplex or (D) being transcribed from the inverted repeat region. Both of these products may undergo Dicer splicing to produce siRNAs, which mediate an RNA interference effect to reduce sense RNA. (E) By containing similar miRNA response elements (MREs) with the miRNA target gene, including the parental gene, pseudogene RNA is capable of competing for miRNAs by serving as a miRNA decoy to enhance expression of miRNA target genes at the posttranscriptional level. (F) Pseudogenes can also generate long noncoding RNAs (lncRNAs) to trigger epigenetic regulations of parental genes; other functions of pseudogene-derived lncRNAs still need to be investigated. (G) Pseudogene RNA is able to compete for RNA-binding proteins (RBPs) with the RNA transcripts of the parental gene. The comprehensive effect on the transcription of the parental gene mainly relies on the subtype of RBP: RNA-stabilizing RBPs and RNA-destabilizing RBPs. Abbreviations: DNMT3A: DNA methyltransferase 3 alpha; EZH2: enhancer of zeste 2 polycomb repressive complex 2 subunit; G9A: euchromatic histone lysine methyltransferase 2; H3K9me3: histone trimethylated at lysine 9; HP1a: Heterochromatin Protein 1A; IRR: inverted repeat region; RBP: RNA binding protein; SUV39H1: suppressor of variegation 3-9 homolog 1.
Protein phosphatase, Mg2+/Mn2+-dependent 1K pseudogene (PPM1KP), a partially retro-transcribed pseudogene with an inverted repeat region, can fold into a hairpin-shaped structure to produce two endogenous specific siRNAs that negatively regulate its cognate genes PPM1K and NIMA-related kinase 8 (NEK8) [64], leading to alterations in hepatocellular cell mitochondrial activation and proliferation. This further illustrates double pseudogene functions as both parental gene dependent and independent.
In fact, multiple pseudogenes have been found to create siRNAs to inhibit the functions of their counterpart via the RNAi mechanism, in humans [65] and also in mice [66], insects [67], and plants [68, 69], suggesting that siRNAs derived from pseudogenes are not exclusive to humans and may be discovered in other species not yet been reported.
Competition for miRNAs
MicroRNAs (miRNAs) comprise a cluster of small noncoding RNAs that function as negative regulators of target genes by interacting with miRNA response elements (MREs) in the mRNA 3'-UTRs at the posttranscriptional level. Theoretically, any RNA possessing MREs can sponge miRNAs, which are also categorized as ceRNAs [70], including pseudogene RNA transcripts. In fact, pseudogene RNA transcripts may contain several MREs that are the same as those in their counterparts, leading to direct competition for miRNAs and allowing pseudogenes to modulate their counterparts. By sharing similar MREs, target genes other than their counterparts can also be regulated by pseudogenes in a ceRNA manner [18, 71] (Figure 3E).
Processed pseudogenes HMGA1P6 and HMGA1P7, located at 13q12.12 and 6q23.2, share high sequence homology with their parental gene high mobility group AT-hook 1 (HMGA1) in the 3'-UTR. Within this region is a perfect match with the conserved seed sequences of HMGA1-targeting miRNAs, e.g., miR-15, miR-16, miR-214 and miR-761. In addition, HMGA1P6 and HMGA1P7 show a similar expression pattern with HMGA1 due to their competition as ceRNAs for the endogenous binding sites of HMGA1-targeting miRNAs; they therefore function as oncogenic regulators of their parental gene HMGA1 in human anaplastic thyroid carcinomas [72].
Similarly, BRAFP1, a pseudogene of B-Raf proto-oncogene, serine/threonine kinase (BRAF), is a genomic gain and aberrantly expressed in various human cancers. BRAFP1 serves as a miRNA decoy that sponges miR-30a, miR-182, miR-590 and miR-876 to regulate expression of BRAF and to activate the mitogen-activated protein kinase (MAPK) signaling pathway, inducing lymphoma in vivo [73]. OCT4-pg4, which is abnormally activated in HCC, can restrain the inhibitory effects of miR-145 on OCT4 by competing for the binding site with miR-145, thus increasing the expression level of OCT4 to promote HCC cell growth and tumorigenesis in vitro and in vivo [74].
In summary, pseudogenes can protect the mRNAs of the parental genes or of genes possessing the same MREs from degradation by competitively interacting with suppressive miRNAs, indicating a parallel expression pattern between the pseudogene and the miRNA target gene. Moreover, these findings add a new layer of posttranscriptional regulation of target genes and shed novel light on the treatment of certain diseases.
Production of lncRNAs
In addition to RNA transcripts of antisense RNAs and siRNAs, pseudogenes can produce long noncoding RNAs (lncRNAs), as supported by data from next-generation sequencing analysis [31]. LncRNAs are characterized as a class of ncRNAs with sequence lengths over 200 nucleotides that exert a series of functions in biological processes, such as transcription, translation and epigenetic modification [75, 76]. By producing lncRNAs, pseudogenes can modulate gene expression in a lncRNA-like manner. However, studies in this field are still in their infancy; indeed, only a small number of the pseudogene-derived lncRNAs have been found [77], and very little is known about their functions. As an example, the lncRNA of murine Oct4P4 can bind to suppressor of variegation 3-9 homolog 1 (SUV39H1) HMTase to form a complex that recruits histone trimethylated at lysine 9 (H3K9me3) and Heterochromatin Protein 1A (HP1a) to the Oct4 promoter region, leading to a silencing effect on Oct4 expression [78]. This is a relatively clear mechanism. SUMO1P3 is a pseudogene of SUMO that can generate lncRNAs, though the detailed functions of these SUMO1P3-derived lncRNAs remain unclear [10]. The clinical significance of these pseudogene-derived lncRNAs is also largely unknown (Figure 3F).
Interaction with RNA-binding proteins (RBPs)
RBPs, which play two entirely opposite roles in mRNA expression as stabilizing or destabilizing factors, serve as critical regulators at the posttranscriptional level. By functioning as stabilizing factors, RBPs help protect mRNAs from degradation; as destabilizing factors, RBPs alleviate the stability of target mRNAs. As a pseudogene presents high sequence similarity to its counterpart, the RNA transcripts of a pseudogene can both interact with RBPs and compete with the counterpart for binding sites in RBPs. Therefore, according to the roles of RBPs, pseudogenes function as positive or negative regulators to help stabilize or destabilize target mRNAs when their RBPs are bound to the pseudogene [70, 79] (Figure 3G).
Coexpression of myosin light chain kinase pseudogene 1 (MYLKP1), a pseudogene of MYLK with smMLCK, an encoded isoform of MYLK, promotes reduced smMLCK mRNA stability, suggesting potential competition between the RNA transcripts of a pseudogene and its counterpart for RNA-stabilizing RBPs [80]. This may be the reason that MYLKP1 negatively regulates its parental gene MYLK expression, is positively correlated with multiple tumors and increases tumor cell proliferation. Overall, there is little evidence for positive effects on target gene expression of interaction of a pseudogene with RBPs.
Function of pseudogenes at the protein level
Encoding a protein or peptide
According to its identification, a pseudogene is labeled with “pseudo” due to its deficiency in protein coding. Nevertheless, some fully processed pseudogenes maintain or regain this capability, in contrast to the majority of identified pseudogenes.
The first known protein-coding pseudogene was phosphoglycerate mutase 3, pseudogene (PGAM3), which was formed by the PGAM1 retrotransposon. The protein-coding ability of PGAM3 had not been verified until Betran et al. [6] identified it through polymorphism and expression data. Another classic example of a protein-producing pseudogene is OCT4-pg1, which contains an entire open reading frame (ORF) that produces a protein that is localized to the nucleus and does not have similar activities as its parental gene OCT4 [81].
In addition to the entire functional protein encoded by an entire ORF, a pseudogene may generate a peptide without full function because of mutations, especially if the pseudogene encodes a premature stop codon; e.g., BRAFP1, located on chromosome Xq13, is interrupted by many stop codons that prevent it from being translated into a fully functional protein. The longest peptide, with a total of 244 amino acids, has a sequence that is highly homologous to the CR1 domain of and cooperates with the BRAF protein to promote MAPK signaling [82], leading to enhanced tumorigenesis in thyroid tumors. Notably, pseudogene-derived proteins or peptides can be treated as “antigens” by the human immune system to induce immune responses [83], which may lead to novel and promising biomarkers or therapeutic targets for certain diseases (Figure 4).
Disease Involvement
Due to the incredible development of next-generation sequencing technology, a large number of pseudogenes have gradually been discovered. Despite some “dying” pseudogenes that have been demonstrated to be nonfunctional, there is sufficient evidence to confirm that pseudogenes harbor various functions at the DNA, RNA, and protein levels, participating in the modulation of target gene expression, particularly their parental genes. Therefore, these molecules are involved in the development and progression of certain diseases, especially cancer. In addition, pseudogenes present their own expression patterns in different species, with a connection to disease diagnosis and prognosis (Table 1). Moreover, many studies have attempted to overexpress or inhibit specific pseudogenes to detect their effects on different diseases in vitro and in vivo (Table 2), providing clues for clinical reference to formulate reasonable treatments. Although pseudogene studies are only beginning to be initiated, they have revealed broad participation of pseudogenes and great clinical value in diseases.
A pseudogene should no longer be treated as a “nonfunctional” element. The critical criterion for judging whether a gene is “functional” or not is predominantly according to its encoding-protein ability. In fact, some pseudogenes harbor a complete open reading frame (ORF) to produce mRNAs. Therefore, these pseudogenes can produce proteins that exert parental gene-like or parental gene-unlike functions. In addition, a small number of pseudogenes can be transcribed as fragments of entire mRNAs, generating different peptides that can induce immune responses or cooperate with parental genes. Because pseudogenes have the potential to produce proteins, as opposed to the traditional opinions that pseudogenes are “nonfunctional”, a reasonable nomenclature is required to reidentify these special types of sequences.
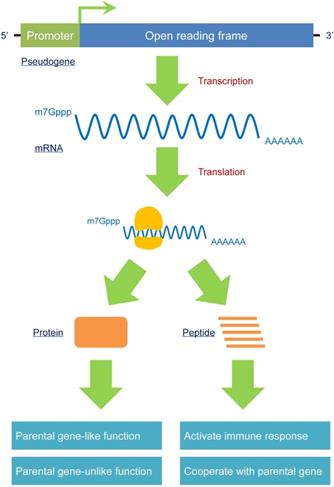
Pseudogenes widely participate in the pathogenesis of different diseases
| Diseases | Pseudogenes | Species | Tissues | Sponge Targets | Expression Pattern | Overall Survival | Potential Functions / Applications | References |
|---|---|---|---|---|---|---|---|---|
| HCC | ANXA2P2 | Homo Sapiens | Liver | - | Upregulated | Decline | Induces migration and invasion of HCC cells in vitro | Wang et al. (2019) [84] |
| RACGAP1P | Homo Sapiens | Cell Line | miR-15-5p | Upregulated | Decline | Sequestrates miR-15-5p from RACGAP1; Activates RhoA/ERK signaling | Wang et al. (2019) [85] | |
| RP11-564D11.3 | Homo Sapiens | Liver | miR-9-5p; miR-101-3p; miR-200 family | Upregulated | Decline | Sequestrates miR-9-5p, miR-101-3p, and miR-200 family to target VEGFA to initiate and promote HCC | Song et al. (2019) [86] | |
| SUMO1P3 | Homo Sapiens | Liver; Cell Line | - | Upregulated | Decline | Promotes cell proliferation, migration, invasion and radiation resistant of HCC in vitro | Zhou et al. (2019) [87] | |
| UBE2CP3 | Homo Sapiens | Liver; Cell Line | - | Upregulated | Decline | Induces angiogenesis functions of HUVECs by activating ERK/HIF-1α/p70S6K/VEGFA signaling | Lin et al. (2018) [88] | |
| OCT4-pg1 | Homo Sapiens | Liver; Cell Line | - | Upregulated | - | Promotes proliferation of HCC cells by inducing activation of AKT | Pan et al. (2018) [89] | |
| OCT4-pg4 | Homo Sapiens | Liver; Cell Line | miR-145 | Upregulated | Decline | Serves as a miR-145 decoy to increase its parental gene OCT4 expression to promote HCC | Wang et al. (2013) [74] | |
| PTENP1 | Homo Sapiens | Liver; Cell Line | miR-193a-3p | Downregulated | Elevation | Sequestrates miR-193a-3p to suppress cell growth, migration and invasion of HCC via PTEN pathway | Qian et al. (2017) [90] | |
| UBE2CP3 | Homo Sapiens | Liver; Cell Line | - | Upregulated | Decline | Promotes epithelial to mesenchymal transition (EMT) in vitro and in vivo | Cao et al. (2017) [91] | |
| INTS6P1 | Homo Sapiens | Liver; Cell Line | miR-17-5p | Downregulated | - | Serves as a miR-17-5p decoy to induce its parental gene INTS6 expression to inhibit HCC | Peng et al. (2015) [92] | |
| E2F3P1 | Homo Sapiens | Liver | - | - | - | The A allele of rs9909601 in E3F3P1 is positively correlated with a better prognosis than the G allele | Pan et al. (2014) [39] Liu et al. (2013) [93] | |
| BC | PTTG3P | Homo Sapiens | Breast | miR-129-5p; miR-376c-3p; miR-383-5p | Upregulated | Decline | Serves as a miRNA decoy to exert an oncogenic role via a miRNA-mRNA regulatory network | Lou et al. (2019) [94] |
| PTENP1 | Homo Sapiens | Breast | - | Downregulated | - | Inhibits growth and migration and enhances doxorubicin sensitivity in ER-negative cells; Increases ER-positive cell growth and decreases ERα | Yndestad et al. (2018) [95] | |
| CKS1BP7 | Homo Sapiens | Breast | - | Upregulated | - | Interacts with IGF1R to enhance cell proliferation | Liu et al. (2018) [96] | |
| CYP4Z2P | Homo Sapiens | Breast; Cell Line | miR-125a-3p | Upregulated | - | Sequesters miR-125a-3p to trigger a hTERT-dependent apoptotic inhibition; Induces phosphorylation of ERK1/2 and PI3K/Akt to enhance tumor-angiogenesis | Li et al. (2017) [97] Zheng et al. (2015) [98] Zheng et al. (2014) [99] | |
| HMGA1P7 | Homo Sapiens | Breast Cell Line | miR-15; miR-16; miR-214; miR-761 | Upregulated | - | Acts as a ceRNA to promote H19 and Igf2 expression to induce MCF-7 cell proliferation in vitro | De Martino et al. (2016) [100] | |
| GC | NANOGP8 | Homo Sapiens | Cell Line | - | Upregulated | - | Binds to the promoter region of DBC1 to induce cell proliferation and suppress apoptosis | Li et al. (2019) [101] |
| GBAP1 | Homo Sapiens | Stomach | miR-212-3p | - | - | Serves as a miR-212-3p decoy to induce GBA; SNP rs2990245 in GBAP1 influences the DNA methylation status of its own promoter region | Ma et al. (2019) [102] | |
| PTENP1 | Homo Sapiens | Stomach; Cell Line | - | Downregulated | - | G allele of rs7853346 in PTENP1 is negatively correlated with the risk of GC; Suppresses GC proliferation, migration and invasion | Ge et al. (2017) [103] Guo et al. (2016) [104] | |
| OCT4-pg1 | Homo Sapiens | Stomach; Cell Line | - | Upregulated | Decline | Elevates expression of various growth factors to enhance GC cell proliferation and angiogenesis | Hayashi et al. (2015) [105] | |
| SUMO1P3 | Homo Sapiens | Stomach | - | Upregulated | Decline | Positively correlates with GC growth, differentiation and lymphatic metastasis | Mei et al. (2013) [10] | |
| RCC | DUXAP8 | Homo Sapiens | Kidney; Cell Line | - | Upregulated | Decline | Alters miR-126/CED-9 signaling to promote RCC cell proliferation and migration | Huang et al. (2018) [106] |
| ccRCC | PTENP1 | Homo Sapiens | Kidney; Cell Line | miR-21 | Downregulated | Elevation | Serves as a miR-21 decoy to increase PTEN; Inhibits cell growth, migration, invasion and sensitivity to cisplatin and gemcitabine | Yu et al. (2014) [107] |
| LUAD | CSDAP1 | Homo Sapiens | Lung; Cell Line | - | Upregulated | Decline | Positively correlates with LUAD occurrence, development and prognosis | Xu et al. (2018) [108] |
| CHIAP2 | Homo Sapiens | Lung; Cell Line | miR-873-3p; miR-3614-5p | Downregulated | - | Serves as a miRNA decoy to regulate NFATC2 or GSK3β in WNT signaling pathway | Shang et al. (2019) [109] | |
| NSCLC | DUXAP8 | Homo Sapiens | Lung; Cell Line | - | Upregulated | Decline | Recruits LSD1 and EZH2 to the promoters of EGR1 and RHOB to enhance NSCLC malignant phenotypes | Sun et al. (2017) [110] |
| DUXAP10 | Homo Sapiens | Lung; Cell Line | - | Upregulated | Decline | Interacts with LSD1 to epigenetically silence LATS2 and RRAD to induce tumorigenesis in NSCLC | Wei et al. (2017) [111] | |
| CRC | MYLKP1 | Homo Sapiens | Colon; Cell Line | - | Upregulated | - | Suppresses its parental gene MYLK expression; Two SNPs, rs12490683 and rs12497343 in MYLKP1 significantly elevate the risk of CRC | Lynn et al. (2018) [112] Han et al. (2011) [80] |
| SUMO1P3 | Homo Sapiens | Colon; Cell Line | - | Upregulated | Decline | Induces expression of cyclin D1, Vimentin and VEGFA and decreases E-cadherin to promote tumorigenesis and angiogenesis of CRC | Zhang et al. (2017) [113] | |
| TPTE2P1 | Homo Sapiens | Colon; Cell Line | - | Upregulated | Decline | Induces cell cycle progression and inhibits apoptosis of CRC cells in vitro and in vivo | Dai et al. (2019) [114] | |
| EEC | HMGA1P6 HMGA1P7 | Homo Sapiens | Endometrium | - | Upregulated | Decline | Positively correlates with expression of its parental gene HMGA1 and EEC progression | Palumbo Junior et al. (2019) [115] |
| UCS | HMGA1P6 | Homo Sapiens | Uterus | let-7a; miR-26a; miR-16; miR-214 | Upregulated | - | Serves as a miRNA decoy implicated in downregulation of the HMGA1-targeting miRNAs, contributing to HMGA1 overexpression | Brunetti et al. (2019) [116] |
| OCS | HMGA1P6 HMGA1P7 | Homo Sapiens | Ovary | let-7a; miR-26a; miR-16; miR-214 | Upregulated | - | Serves as miRNA decoys implicated in downregulation of the HMGA1-targeting miRNAs, contributing to HMGA1 overexpression | Esposito et al. (2014) [72] Brunetti et al. (2019) [116] |
| GBM | DUXAP8 | Homo Sapiens | Brain; Cell Line | - | Upregulated | Decline | Promotes cell proliferation and colony formation of GBM in vitro | Zhao et al. (2019) [117] |
| LGMNP1 | Homo Sapiens | Cell Line | - | Upregulated | - | Reduces DNA damage processes and apoptosis to help acquire radiotherapy resistance | Xu et al. (2019) [118] | |
| PT | HMGA1P6 HMGA1P7 | Homo Sapiens | Pituitary; Cell Line | - | Upregulated | - | Serve as ceRNAs to induce parental gene HMGA1 expression and increase cell proliferation and migration | Esposito et al. (2015) [119] |
| CC | OCT4-pg1 | Homo Sapiens | Cervix; Cell Line | - | Upregulated | - | Induces cell proliferation and migration and inhibits apoptosis of CC cells in vitro and in vivo | Yu et al. (2019) [120] |
| PDAC | DUXAP8 | Homo Sapiens | Pancreas; Cell Line | - | Upregulated | Decline | Serves as a scaffold for EZH2 and LSD1 to epigenetically silence CDKN1A and KLF2 | Lian et al. (2018) [121] |
| SUMO1P3 | Homo Sapiens | Pancreas; Cell Line | - | Upregulated | Decline | Induces development of EMT to enhance cell proliferation, migration and invasion in vitro | Tian et al. (2018) [122] | |
| ESCC | TUSC2P | Homo Sapiens | Esophagus; Cell Line | miR-17-5p; miR-520a-3p; miR-608; miR-611 | Downregulated | Elevation | Plays a tumor suppressive role in a miRNA-binding manner to induce a cell growth and invasion inhibition | Liu et al. (2018) [123] |
| FTH1P3 | Homo Sapiens | Esophagus; Cell Line | - | Upregulated | - | Enhances cell proliferation, migration, and invasion by activating Sp1 and NF-κB | Yang et al. (2018) [124] | |
| DLBCL | Braf-rs1 | Murine | Spleen; Cell Line | miR-134; miR-543; miR-653 | Upregulated | Decline | Functions in a ceRNA manner to elevate BRAF expression and to regulate MAPK signaling and cell proliferation in vitro and in vivo | Karreth et al. (2015) [73] |
| BRAFP1 | Homo Sapiens | Spleen; Cell Line | miR-30a; miR-182; miR-876; miR-590 | Upregulated | Decline | Functions in a ceRNA manner to elevate BRAF expression and to regulate MAPK signaling and cell proliferation in vitro and in vivo | Karreth et al. (2015) [73] | |
| AML | OCT4-pg1 | Homo Sapiens | Blood | - | Downregulated | Elevation | Contributes to the diagnosis and prognosis of AML | Yi et al. (2019) [125] |
| CML | OCT4-pg1 | Homo Sapiens | Cell Line | - | - | - | Interacts directly with OCT-4, SOX2, and NANOG and indirectly with ABC transporters | Lettnin et al. (2019) [126] |
| TC | HMGA1P6 HMGA1P7 | Homo Sapiens | Thyroid; Cell Line | miR-15; miR-16; miR-214; miR-761 | Upregulated | - | Serve as miRNA decoys to induce parental gene HMGA1 and cancer-related genes expression to promote malignant phenotypes of cells in vitro | Esposito et al. (2014) [72] |
| OSCC | FTH1P3 | Homo Sapiens | Oral; Cell Line | - | Upregulated | Decline | Promotes cell proliferation, migration and invasion by inducing PI3K/Akt/GSK3β/Wnt/β-catenin signaling | Liu et al. (2018) [127] |
| BA | ANXA2P3 | Homo Sapiens | Liver; Cell Line | - | Upregulated | - | Induces cell cycle progression and growth but inhibits apoptosis by activating ANXA2/ANXA2P3 signaling | Nuerzhati et al. (2019) [128] |
| SPE | HK2P1 | Homo Sapiens | Decidua; Cell Line | miR-6887-3p | Downregulated | - | Promotes glycolytic metabolism, and HESC decidualization by sequestering miR-6887-3p | Lv et al. (2018) [129] |
| PGK1P2 | Homo Sapiens | Decidua | miR-330-5p | Downregulated | - | Sequesters miR-330-5p to affect decidualization by regulating angiogenesis and glycolysis metabolism | Tong et al. (2018) [130] | |
| Schizophrenia | NDUFV2P1 | Homo Sapiens | Brain; Cell Line | - | Upregulated | - | Negatively correlates with pre- and mature NDUFV2 and with CoI-driven cellular respiration | Bergman et al. (2018) [131] |
| ASD | lncLRFN5-10 | Homo Sapiens | Cell Line | - | Downregulated | - | Positively affects expression of non-deleted LRFN5 | Cappuccio et al. (2019) [132] |
| OA | PMS2L2 | Homo Sapiens | Cell Line | miR-203 | Downregulated | - | Serves as a miR-203 decoy to block Wnt/β-catenin and JAK/STAT signaling pathway | Li et al. (2019) [133] |
| AD | PTENP1 | Homo Sapiens | Aorta; Cell Line | miR-21 | Upregulated | - | Induces PTEN to promote apoptosis and to inhibit HASMC growth by competing for miR-21 | Lai et al. (2019) [134] |
| MM | PDIA3P | Homo Sapiens | Bone Marrow; Cell Line | - | Upregulated | Decline | Interacts with c-Myc to bind to G6PD promoter and to induce PPP flux to affect cell growth and drug resistant | Yang et al. (2018) [135] |
Abbreviations: AD: aortic dissection; AML: acute myelocytic leukemia; ASD: autism spectrum disorder; BA: biliary atresia; BC: breast cancer; CC: cervical cancer; ccRCC: clear cell renal cell carcinoma; ceRNA: competing endogenous RNA; CML: chronic myelocytic leukemia; CRC: colorectal cancer; DLBCL: diffuse large B cell lymphoma; EEC: endometrioid endometrial carcinomas; EMT: epithelial to mesenchymal transition; ESCC: esophageal squamous cell carcinoma; GBM: glioblastoma; GC: gastric cancer; HASMC: human aortic smooth muscle cell; HCC: hepatocellular carcinoma; HESC: human endometrial stromal cell; HUVEC: human umbilical vein endothelial cell; LUAD: lung adenocarcinoma; miRNA: microRNA; MM: multiple myeloma; mRNA: messenger RNA; NSCLC: non-small cell lung cancer; OA: osteoarthritis; OCS: ovarian carcinosarcomas; OSCC: oral squamous cell carcinoma; PDAC: pancreatic ductal adenocarcinoma; PT: pituitary tumor; RCC: renal cell carcinoma; SNP: single nucleotide polymorphism; SPE: severe preeclampsia; TC: thyroid carcinoma; UCS: uterine carcinosarcomas.
Overexpression or knockdown effects of pseudogenes in vitro and in vivo
| Pseudogenes | Treatments | Diseases | Effects | References |
|---|---|---|---|---|
| ANXA2P2 | Knockdown | HCC | Inhibits migration and invasion of HCC cells in vitro | Wang et al. (2019) [84] |
| RACGAP1P | Overexpression | HCC | Sequestrates miR-15-5p to trigger RhoA/ERK signaling; Promotes cell growth and migration of HCC in vitro and in vivo | Wang et al. (2019) [85] |
| SUMO1P3 | Knockdown | HCC | Suppresses proliferation, migration, invasion and enhances radio-sensitivity of HCC cells | Zhou et al. (2019) [87] |
| UBE2CP3 | Overexpression | HCC | Enhances HUVEC proliferation, migration and tube formation by inducing ERK/HIF-1a/p70S6K/VEGFA signaling | Lin et al. (2018) [88] |
| OCT4-pg1 | Overexpression | HCC | Promotes HCC cell proliferation by activating AKT in vitro | Pan et al. (2018) [89] |
| OCT4-pg4 | Knockdown | HCC | Suppresses cell proliferation and colony formation in vitro and in vivo | Wang et al. (2013) [74] |
| PTENP1 | Overexpression | HCC | Inhibits HCC growth, migration and invasion via the PTEN pathway in vitro and in vivo | Qian et al. (2017) [90] |
| UBE2CP3 | Knockdown | HCC | Suppresses the epithelial to mesenchymal transition in vitro and in vivo | Cao et al. (2017) [91] |
| INTS6P1 | Knockdown | HCC | Induces HCC growth and migration in vitro and in vivo | Peng et al. (2015) [92] |
| PTENP1 | Overexpression | BC | Promotes ER-positive cell growth and decreases PTEN1 and ERα; Inhibits growth and migration and enhances PTEN1 in ER-negative cells | Yndestad et al. (2018) [95] |
| CYP4Z2P | Knockdown | BC | Releases miR-125a-3p to inhibit a hTERT-mediated apoptotic repression | Li et al. (2017) [97] |
| NANOGP8 | Knockdown | GC | Suppresses DBC1 to inhibit cell proliferation and promote apoptosis | Li et al. (2019) [101] |
| OCT4-pg1 | Overexpression | GC | Promotes expression levels of different growth factors to induce GC cell proliferation and angiogenesis in vitro and in vivo | Hayashi et al. (2015) [105] |
| DUXAP8 | Knockdown | RCC | Increases miR-126 to inhibit CED-9 expression to suppress RCC cell growth and migration in vitro | Huang et al. (2018) [106] |
| PTENP1 | Overexpression | ccRCC | Sequesters miR-21 to induce PTEN expression to suppress cell growth, migration, and invasion and increase sensitivity to cisplatin and gemcitabine in vitro and in vivo | Yu et al. (2014) [107] |
| CHIAP2 | Overexpression | LUAD | Sequesters miR-873-3p and miR-3614-5p to inhibit cell proliferation and invasion | Shang et al. (2019) [109] |
| DUXAP8 | Knockdown | NSCLC | Inhibits recruitment of LSD1 and EZH2 to the promoter regions of EGR1 and RHOB to suppress NSCLC malignant phenotypes in vitro and in vivo | Sun et al. (2017) [110] |
| DUXAP10 | Knockdown | NSCLC | Promotes transcriptions of LATS2 and RRAD to restrain NSCLC cell growth, migration and invasion in vitro and in vivo | Wei et al. (2017) [111] |
| MYLKP1 | Overexpression | CRC | Suppresses expression of MYLK and promotes cell proliferation and migration | Lynn et al. (2018) [112] Han et al. (2011) [80] |
| SUMO1P3 | Knockdown | CRC | Suppresses expression levels of cyclin D1, Vimentin, and VEGFA and enhances E-cadherin to reduce CRC malignant behavior in vitro and in vivo | Zhang et al. (2017) [113] |
| TPTE2P1 | Knockdown | CRC | Promotes cell cycle arrest at S phase and induces apoptosis by activating the BCL2/Caspase 3 signaling cascade in vitro and in vivo | Dai et al. (2019) [114] |
| DUXAP8 | Knockdown | GBM | Suppresses cell proliferation and colony formation in GBM in vitro | Zhao et al. (2019) [117] |
| LGMNP1 | Overexpression | GBM | Reduces DNA damage processes and apoptosis to resist radiotherapy | Xu et al. (2019) [118] |
| HMGA1P6 HMGA1P7 | Overexpression | PT | Functions as a decoy for HMGA1-targeting miRNAs to promote HMGA1 expression and induce AtT20 cell proliferation and migration | Esposito et al. (2015) [119] |
| OCT4-pg1 | Knockdown | CC | Suppresses cell proliferation and migration and promotes apoptosis of CC cells in vitro and in vivo | Yu et al. (2019) [120] |
| DUXAP8 | Knockdown | PDAC | Increases expression levels of CDKN1A, and KLF2 to inhibit cell proliferation and promote apoptosis in vitro and in vivo | Lian et al. (2018) [121] |
| SUMO1P3 | Knockdown | PDAC | Reverses the process of EMT to inhibit cell proliferation, migration and invasion in vitro | Tian et al. (2018) [122] |
| TUSC2P | Overexpression | ESCC | Suppresses cell growth and invasion in a miRNA-binding manner in vitro | Liu et al. (2018) [123] |
| FTH1P3 | Knockdown | ESCC | Inhibits cell growth, migration and invasion by silencing Sp1 and NF-κB in vitro | Yang et al. (2018) [124] |
| Braf-rs1 | Overexpression | DLBCL | Sequesters miRNAs to activate MAPK signaling and cell proliferation in vitro and in vivo | Karreth et al. (2015) [73] |
| BRAFP1 | Overexpression | DLBCL | Sequesters miRNAs to activate MAPK signaling and cell proliferation in vitro and in vivo | Karreth et al. (2015) [73] |
| OCT4-pg1 | Knockdown | CML | Reduces expression and activity of OCT4 as well as ABCB1 and activates ALOX5 and ABCC1 to render cell sensitive to chemotherapy | Lettnin et al. (2019) [126] |
| HMGA1P6 HMGA1P7 | Overexpression | TC | Sequesters HMGA1-targeting miRNAs to increase cell proliferation, migration and invasion and suppress apoptosis in vitro | Esposito et al. (2014) [72] |
| FTH1P3 | Knockdown | OSCC | Suppresses cell proliferation, migration and invasion by reducing activation of PI3K/Akt/GSK3β/Wnt/β-catenin signaling | Liu et al. (2018) [127] |
| ANXA2P3 | Knockdown | BA | Suppresses proliferation, increases apoptosis and induces cell cycle arrest in G1 phase in vitro by inhibiting ANXA2/ANXA2P3 signaling | Nuerzhati et al. (2019) [128] |
| HK2P1 | Knockdown | SPE | Inhibits glucose uptake, lactate production and decidualization in HESCs by releasing miR-6887-3p | Lv et al. (2018) [129] |
| PTENP1 | Overexpression | AD | Increases expression of PTEN to induce apoptosis and inhibit proliferation of HASMCs by sequestering miR-21 in vitro and in vivo | Lai et al. (2019) [134] |
Abbreviations: AD: aortic dissection; BA: biliary atresia; BC: breast cancer; CC: cervical cancer; ccRCC: clear cell renal cell carcinoma; CML: chronic myelocytic leukemia; CRC: colorectal cancer; DLBCL: diffuse large B cell lymphoma; EMT: epithelial to mesenchymal transition; ESCC: esophageal squamous cell carcinoma; GBM: glioblastoma; GC: gastric cancer; HASMC: human aortic smooth muscle cell; HCC: hepatocellular carcinoma; HESC: human endometrial stromal cell; HUVEC: human umbilical vein endothelial cell; LUAD: lung adenocarcinoma; NSCLC: non-small cell lung cancer; OSCC: oral squamous cell carcinoma; PDAC: pancreatic ductal adenocarcinoma; PT: pituitary tumor; RCC: renal cell carcinoma; SPE: severe preeclampsia; TC: thyroid carcinoma.
Actually, pseudogenes have strong potential to serve as key regulators in certain diseases, as based on recent studies presented in the tables above due to the following reasons. 1) Wide involvement in diseases: Current studies demonstrate broad pseudogene participation in the pathogenesis and pathology of diseases involving nearly every system and organ of the human body. 2) Close correlations with cancer: The majority of pseudogene studies, nearly 80%, concentrate on the relationship between pseudogenes and cancer, and multiple cancers, such as GBM, BC, HCC, GC, CRC and RCC, are induced by disorders caused by pseudogene expression or function. 3) Prognosis-related expression patterns: A number of pseudogenes have been clinically proven to be expressed in a specific manner for predicting the prognosis of patients with several diseases, indicating their involvement in disease. 4) Effects on cellular biology: Pseudogenes can have a global effect on cellular behavior, including the cell cycle, apoptosis, proliferation, migration, invasion, metabolism, drug resistance and radiotherapy resistance. In addition, the cell microenvironment and angiogenesis can be influenced by pseudogenes. 5) A series of functions in disease modulation: Of note, the ceRNA function of pseudogenes in disease regulation is dominant. Simultaneously, pseudogenes serving as a scaffold to interact with RBPs, triggering specific epigenetic modifications, e.g., DNA methylation and initiating multiple signaling pathways such as PI3K/Akt, Wnt, EMT, JAK/STAT and MAPK have been reported. 6) A single pseudogene in multiple disease regulation: Intriguingly, several pseudogenes can regulate and correlate closely with the diagnosis and prognosis of more than a single disease, e.g., PTENP1 in HCC, BC, GC, ccRCC and AD, SUMO1P3 in HCC, GC, CRC and PDAC, and HMGA1P6/7 in EEC, PT, TC, BC, UCS and OCS. Notably, an increasing number of studies have gradually demonstrated an irreplaceable and connectable role of pseudogenes in the development and progression of diseases.
Clinical Perspective: A Promising Biotarget
Diagnosis: A pseudogene is a potential biomarker for disease diagnosis
To alleviate the morbidity and mortality of diseases, especially of several lesions that are typically asymptomatic at the early stage, as well as some lesions that seriously develop rapidly, sensitive biomarkers are of great significance and urgency for diagnosis to create an optimized therapeutic time window for patients. Because of the tremendous efforts from researchers, in particular the emergence of high-throughput sequencing analysis in pseudogene studies, thousands of pseudogenes have been identified and found to play roles in the etiology and pathology of certain diseases. Accordingly, these pseudogenes are likely to be regarded as diagnostic markers. Of note, pseudogenes intrinsically exhibit some characteristics that are helpful in disease diagnosis. 1) Broad-spectrum distribution: Pseudogenes can be detected in a variety of organisms, as well as organs, tissues and blood [30], which is beneficial for acquiring and enhancing the diversity and objectivity of diagnostic indexes for specific diseases. 2) Disease subtype-unique expression: Pseudogenes can differentiate one disease subtype from another because some pseudogenes are only expressed in a single disease subtype [9]; thus, they act as a specific disease signature to assist in diagnosis. 3) Tissue-specific expression: Pseudogenes can be applied to distinguish normal tissues from lesion tissues by differential expression, indicating their great power in disease diagnosis and differential diagnosis [10, 30]. Intriguingly, the expression profile of multiple pseudogenes in a group can be recognized as a signature with a diagnostic value [136]. 4) Dissimilar counterpart expression: In some cases, the expression level of RNA transcript of a pseudogene is not always parallel to its counterpart but is significant for differentiation, especially in several species that only harbor the RNA transcript of the pseudogene [137]. 5) Evolutionary conservation: Regardless of the evolutionary distance, pseudogenes are highly conserved, providing an advantageous condition for its measurement and evidence to investigate the probable mechanisms of some zoonoses. In summary, with proper and reasonable development and utilization, these five properties are likely to allow pseudogenes to serve as sensitive and promising biomarkers in disease diagnosis in the future.
Prognosis: A pseudogene is a possible indicator of life expectancy
Consistent with diagnosis, prognosis is an equally critical aspect that should be considered in the clinic because it determines the best choice of therapy as well as the life expectancy of patients. As a potential biomarker for disease diagnosis, pseudogenes are being proven to be valuable for prognosis [138]. The expression level of a single pseudogene can be regarded as helpful in evaluating overall survival (OS). For example, ccRCC patients with low levels of PTENP1 show a shorter OS rate than do those with high PTENP1 levels [107]; overexpression of OCT4-pg1 in GC due to aberrant amplification can result in a poor rate of life expectancy in GC patients [105]. In addition, SNPs in the pseudogene sequence correlate with prognosis; e.g., HCC patients who carry the GG allele of E2F3P1 show a worse OS than do those carrying the GA/AA allele [39]. Moreover, by combining pseudogene expression with the tumor grade or stage, pseudogenes can indirectly predict OS in several diseases, especially cancer. Intriguingly, a certain number of pseudogenes can be combined as a signature that helps stratify disease risk, as do the clues provided by research in kidney renal clear cell carcinoma (KIRC) [136]. Furthermore, recent studies have indicated that pseudogenes are more amenable for determining the prognosis of several diseases than are traditional signatures, such as mRNAs, miRNAs and epigenetic modifications [18], thus elevating the prognostic value of pseudogenes to the next level. Taken together, pseudogenes exhibit enormous potential as effective prognosis indicators of diseases. Future investigations should focus on finding more prognosis-related pseudogenes and using a combination of pseudogenes and traditional signatures or a combination of multiple pseudogenes as signatures to increase efficiency and accuracy in clinical applications.
Therapeutics: A pseudogene is a promising approach for therapeutic strategy
Although substantial therapeutic methods, especially drugs, are rapidly emerging in recent years, several issues remain that impair their efficiency, such as off-target effects and drug resistance. New and effective biotargets are needed to overcome these obstacles. Taking the features of pseudogenes into account, it is feasible to exploit pseudogenes to develop therapeutic strategies in a novel field for the following reasons. First, a pseudogene is an ideal target for biological treatment. Because they widely expressed in a series of organisms and different specimens, pseudogenes are reported to be involved in multiple processes, including physiology and pathology, are important for homeostasis. Therefore, aberrantly expressed pseudogenes can be used as biological targets for treatment. Second, pseudogenes are a resource for therapy. By sequestering target miRNAs through MREs, pseudogenes can abolish the pathogenic effects of miRNAs or elements with MREs, e.g., RBPs. Additionally, pseudogenes can restrain the pathogenic effects of their counterparts by producing antisense RNAs or siRNAs. Notably, pseudogenes can generate to several lncRNAs that probably exert “lncRNA” functions in disease prevention. Hence, developing and optimizing sequences to create more pseudogene analogs that better facilitate the miRNA “decoy”, along with the antisense RNA, siRNA and lncRNA “producer” functions, may be an ideal strategy. Third, Transcribed pseudogenes can be used as vaccines. One study has shown that peptides from transcribed pseudogenes can be recognized as “antigens” that activate the innate or adaptive immune response in the human body [83]. Based on this evidence, disease-specific transcribed pseudogenes can be redesigned to produce peptides with high antigenicity and immunogenicity as well as low toxicity to induce a strong and rapid immune response. In conclusion, pseudogenes provide a novel method and strategy for enriching treatments for certain diseases, especially those that are not sensitive to or resist traditional therapies in the clinic.
Detection: An Integrated Method
With detection based on DNA sequencing analysis of the whole human genome by GENCODE, over 60,000 total genes have been discovered. However, because of the high similarity in DNA sequences, it is challenging to distinguish pseudogenes from parental genes at the DNA level. Recently, multiple pipelines have been developed and applied to measure pseudogene DNA, such as PseudoPipe [139], PseudoFinder and RetroFinder [140], which markedly enhance the detection efficiency and accuracy of pseudogene DNA. Two comprehensive databases, Encyclopedia of DNA Elements (ENCODE) [141] and Functional Annotation of Mammals (FANTOM) [142], which were built on integration of a series of public pipelines, are currently considered gold standards for pseudogene DNA detection. In fact, as detection methods are constantly being developed, the number of newly found pseudogenes in the genomes of different species will continue to increase.
For pseudogene RNA detection, RNA sequencing analysis is regarded as the first choice that provides precise and thorough detection of all pseudogenes, from subtype to quantity, in a transcriptome, thus creating a plethora of novel information for further exploration of the known and unknown aspects of pseudogene. Moreover, with the rapid development of bioinformatics pipelines, a large number of pseudogene RNA transcripts have been found in different conditions, especially in cancer [136, 143, 144]. Microarrays and quantitative real-time polymerase chain reaction (qRT-PCR) are two general methods that have higher sensitivities and specificities but lower costs than RNA sequencing. Nevertheless, extreme care should be taken to ensure that the probes used for microarrays and the primers used for qRT-PCR are specific enough to avoid amplification of parental genes or nonspecific templates. Notably, northern blotting has been utilized for measuring transcribed pseudogenes [29]. Furthermore, in situ hybridization assays, such as ISH and FISH, have an advantage for investigating the spatiotemporal distribution of pseudogene RNA transcripts. To better understand the intrinsic relationship between a pseudogene and its parental gene or to separate these two factors, gene-editing technology, e.g., the clustered regularly interspaced short palindromic repeats-cas9 (CRISPR-Cas9) system, can be employed to knockout the parental gene without an off-target effect. Additionally, the protein expression levels assessed via western blotting can directly reflect the functional RNA transcripts of pseudogenes at the translational level. Because each method has both advantages and disadvantages in pseudogene DNA or RNA detection, the combined use of multiple methods to acquire a more thorough and accurate result should be the trend, as opposed to a single strategy, in future pseudogene studies.
Limitations, Challenges and Perspectives
It has been confirmed that the majority of DNA sequences in the human genome do not encode proteins and that the genome is partially composed of pseudogenes. Without a coding function, pseudogenes have been regarded as “junk DNA” or “genomic fossils” for a long period of time, and substantial efforts have been made into devising strategies to eliminate pseudogenes when its parental coding gene was being analyzed [5, 145]. However, the real nature of the pseudogene is far more than “trash” or “nonfunctional”. Indeed, recent studies have gradually shed light on the function and involvement of pseudogenes in physiological and pathological conditions. Pseudogenes serve as a function-combined machine that plays different roles at the DNA, RNA or protein level, with participation in the gene regulation network at the transcriptional and posttranscriptional levels. Although pseudogenes are evolutionally conserved, they act as a gene reservoir that effectively allows the genome to carry out novel functions. In addition, a small number of pseudogenes are able to encode proteins, and this finding questions the reasonability of the “pseudo” prefix that conceals the real functions of pseudogenes. “Pseudogene” is a term relative to the parental gene, suggesting sequential variance rather than function. Furthermore, with accumulating evidence confirming pseudogene functions, it is appropriate to create novel nomenclature to better reflect the intrinsic property of these sequences.
Nonetheless, some limitations exist in recent pseudogene studies that future works should focus on. 1) A unified and standard rule for pseudogene naming is needed. In fact, the current pseudogene naming system is relatively chaotic and does not have a unified principle, which is likely to generate errors in gene annotation, especially during sequencing analysis. 2) A certain number of pseudogene investigations have only revealed the differential expression of a single or cluster of pseudogenes in several diseases, whereas their functions and mechanisms have not been fully discussed. 3) The majority of current pseudogene studies concentrate on the correlation between pseudogenes and cancer, and a shortage of evidence on other diseases is notable in this field. 4) Although a substantial number of pseudogenes have been reported to be involved in some diseases, only a small fraction of them meet the requirements of a diagnostic marker or therapy target. Moreover, more in vitro experiments, in vivo experiments and clinical trials are needed for potential markers to demonstrate their value in the clinic. 5) More attempts should be made to elucidate other pseudogene functions involved in the development and progression of diseases, as opposed to ceRNA behaviors. 6) Additionally, more focus on the regulatory relationships between pseudogenes and other genes in the genome without a restriction to parental genes is needed. 7) Although some studies have reported a few clues regarding the role of pseudogenes in epigenetic modifications, in particular DNA methylation [61, 62], multiple questions remain for other regulation patterns, such as histone modification and chromatin remodeling. Therefore, pseudogene investigations are still in their infancy, and knowledge should be increased.
In addition to these limitations, pseudogene studies are inevitably confronted with the following challenges. First, it is difficult to identify a cluster of pseudogenes with no protein-coding ability because this type of pseudogene cannot be traced via its RNA or protein products. Second, due to the high sequence similarity between a pseudogene and its parental gene, highly specific primers and antibodies that can effectively differentiate them are urgently needed to help with detection. Third, accurately evaluating the outcomes of high-throughput sequencing analysis and matching the data to pseudogene locations hinders their use, especially without a uniform naming principle. Fourth, more precise methods should be introduced for pseudogene studies, such as luciferase reporter assays, northern blotting and CRISPR-Cas9 technology, as qRT-PCR has poor specificity because of the high sequence homology. Furthermore, bioinformatics analyses of pseudogenes, particularly those that can predict the clinical value of pseudogenes, are lacking. Developing databases that are more clinically applicable is of significance for further pseudogene investigations. In this case, many obstacles to pseudogene studies still need to be addressed in the future.
Individualized medicine, which is mainly based on the theory that a specific treatment for a certain patient should be formulated in accordance with his or her own detailed information contained in the genome and epigenome, is critical to improve the therapeutic efficiency for some diseases of unknown etiology and that are not sensitive to or are resistant to traditional therapy, especially cancer. Moreover, with the rapid emergence of high-throughput sequencing technologies, including multiple omics studies, the precision of disease subtype classification and evidence for the utilization of drugs or drug combinations, which relies on variation in gene profiles, has greatly increased. Notably, in this scenario, sensitive biomarkers for disease diagnosis and treatment appear to be crucial because substantial biotargets can be obtained at one time. In fact, pseudogenes are highly valuable in disease diagnosis and prognosis due to their own expression patterns; in some cases, pseudogenes even display higher power than do mRNAs and miRNAs [136], suggesting that pseudogenes are quite appropriate as biomarkers. Additionally, pseudogenes have great potential to be translated into feasible therapeutic targets because of their various properties, particularly ceRNA functions. In summary, although our current knowledge on pseudogenes is preliminary, it is expected that pseudogenes may help to establish novel strategies for clinical applications with regard to disease diagnosis, prognosis and therapeutics, thus accelerating the development of individualized medicine.
Conclusions
(1) Pseudogenes should no longer be regarded as “junk” or “relics” because multiple functions of pseudogenes at the DNA, RNA and protein levels have recently been discovered, especially their ability to encode functional proteins, which seriously contradicts their previous “nonfunctional” identification. Herein, we shed light on the details of their functions and roles in gene regulation at the molecular level.
(2) Pseudogenes are a product of gene mutation and serve as a gene “repository” and can generally be classified into three categories according to distinct biogenesis processes, storing and expanding genetic information.
(3) Pseudogenes possess a series of clinic-associated features, including wide and uneven distribution, spatiotemporal and unique expression patterns and evolutionary conservation, which allow them to potentially be involved in disease diagnosis, prognosis and therapeutics as promising biomarkers or biotargets, suggesting their high value in clinical applications, even in individualized medicine.
(4) Current pseudogene detection methods have been largely improved at the DNA and RNA levels, in particular with the emergence of high-throughput sequencing analysis. However, pseudogene studies are still in their infancy and affected by various limitations and challenges that need further investigation.
Abbreviations
3'-UTR: 3'-untranslated region; ACYL3: O-acyltransferase like, pseudogene; BRAF: B-Raf proto-oncogene, serine/threonine kinase; BRAFP1: BRAF pseudogene 1; BRCA1: BRCA1 DNA repair associated; ccRCC: clear cell renal cell carcinoma; ceRNA: competing endogenous RNA; CNS: central nervous system; CRISPR-Cas9: clustered regularly interspaced short palindromic repeats cas9; DNMT3A: DNA methyltransferase 3 alpha; E2F3: E2F transcription factor 3; ENCODE: Encyclopedia of DNA Elements; EZH2: enhancer of zeste 2 polycomb repressive complex 2 subunit; FANTOM: Functional Annotation of Mammals; FER1L4: fer-1-like family member 4 (pseudogene); FISH: fluorescence in situ hybridization; FUT2: fucosyltransferase 2; G9A: euchromatic histone lysine methyltransferase 2; GC: gastric cancer; H3K27me3: histone trimethylated at lysine 27; H3K9me3: histone trimethylated at lysine 9; HCC: hepatocellular carcinoma; HP1a: Heterochromatin Protein 1A; HTR7: 5-hydroxytryptamine receptor 7; INTS6: integrator complex subunit 6; INTS6P1: integrator complex subunit 6 pseudogene 1; ISH: in situ hybridization; KIRC: kidney renal clear cell carcinoma; KLK4: kallikrein related peptidase 4; KLKP1: kallikrein pseudogene 1; lncRNA: long noncoding RNA; MAPK: mitogen-activated protein kinase; MGA: MAX dimerization protein MGA; miRNA: microRNA; MRE: miRNA response element; MYLK: myosin light chain kinase; MYLKP1: myosin light chain kinase pseudogene 1; NANOG: Nanog homeobox; NANOGP8: Nanog homeobox retrogene P8; NEK8: NIMA related kinase 8; NOS: nitric oxide synthase; OCT4: POU class 5 homeobox 1; ORF: open reading frame; PARP: poly (ADP-ribose) polymerase; PGAM1: phosphoglycerate mutase 1; PGAM3: phosphoglycerate mutase processed gene; PMS2: PMS1 homolog 2, mismatch repair system component; PMS2CL: PMS2 C-terminal-like pseudogene; PPM1K: protein phosphatase, Mg2+/Mn2+ dependent 1K; PsiBRCA1: BRCA1 pseudogene 1; psiTPTE22: TPTE pseudogene 1; PTEN: phosphatase and tensin homolog; PTENP1: phosphatase and tensin homolog pseudogene 1; PTPN12: protein tyrosine phosphatase non-receptor type 12; qRT-PCR: quantitative reverse transcription polymerase chain reaction; RBP: RNA binding protein; RNAi: RNA interference; SEC1P: secretory blood group 1, pseudogene; siRNA: small interfering RNA; SNP: single-nucleotide polymorphism; SUMO1: small ubiquitin like modifier 1; SUMO1P3: SUMO1 pseudogene 3; SUV39H1: suppressor of variegation 3-9 homolog 1; TPTE: transmembrane phosphatase with tensin homology.
Acknowledgements
Our work was supported by grants 81773003, 81472633 and 81800185 from the National Natural Science Foundation of China.
Author Contributions
X.C. conceived the project. X.C., T.W. and A.-G.Y. designed and directed the project. X.C., L.W., W.W., W.-J.X. and T.W. contributed to the writing of the manuscript. X.C. and L.W. designed the figures. X.C., W.W. and W.-J.X. designed the tables. T.W. and A.-G.Y. supervised the literature review. All the authors have read and approved the final version for publication.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J. et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860-921
2. Jacq C, Miller JR, Brownlee GG. A pseudogene structure in 5S DNA of Xenopus laevis. Cell. 1977;12:109-20
3. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-8
4. Muro EM, Mah N, Andrade-Navarro MA. Functional evidence of post-transcriptional regulation by pseudogenes. Biochimie. 2011;93:1916-21
5. Brent MR. Steady progress and recent breakthroughs in the accuracy of automated genome annotation. Nat Rev Genet. 2008;9:62-73
6. Betran E, Wang W, Jin L, Long M. Evolution of the phosphoglycerate mutase processed gene in human and chimpanzee revealing the origin of a new primate gene. Mol Biol Evol. 2002;19:654-63
7. Zhu J, Sanborn JZ, Diekhans M, Lowe CB, Pringle TH, Haussler D. Comparative genomics search for losses of long-established genes on the human lineage. PLoS Comput Biol. 2007;3:e247
8. Pink RC, Wicks K, Caley DP, Punch EK, Jacobs L, Carter DR. Pseudogenes: pseudo-functional or key regulators in health and disease? RNA. 2011;17:792-8
9. Gasi Tandefelt D, Boormans J, Hermans K, Trapman J. ETS fusion genes in prostate cancer. Endocr Relat Cancer. 2014;21:R143-52
10. Mei D, Song H, Wang K, Lou Y, Sun W, Liu Z. et al. Up-regulation of SUMO1 pseudogene 3 (SUMO1P3) in gastric cancer and its clinical association. Med Oncol. 2013;30:709
11. Proudfoot N. Pseudogenes. Nature. 1980;286:840-1
12. Liu Y, Harrison PM, Kunin V, Gerstein M. Comprehensive analysis of pseudogenes in prokaryotes: widespread gene decay and failure of putative horizontally transferred genes. Genome Biol. 2004;5:R64
13. Li WH, Gojobori T, Nei M. Pseudogenes as a paradigm of neutral evolution. Nature. 1981;292:237-9
14. Torrents D, Suyama M, Zdobnov E, Bork P. A genome-wide survey of human pseudogenes. Genome Res. 2003;13:2559-67
15. Emadi-Baygi M, Sedighi R, Nourbakhsh N, Nikpour P. Pseudogenes in gastric cancer pathogenesis: a review article. Brief Funct Genomics. 2017;16:348-60
16. Zhang ZD, Frankish A, Hunt T, Harrow J, Gerstein M. Identification and analysis of unitary pseudogenes: historic and contemporary gene losses in humans and other primates. Genome Biol. 2010;11:R26
17. Li W, Yang W, Wang XJ. Pseudogenes: pseudo or real functional elements? J Genet Genomics. 2013;40:171-7
18. Lu XJ, Gao AM, Ji LJ, Xu J. Pseudogene in cancer: real functions and promising signature. J Med Genet. 2015;52:17-24
19. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57-74
20. Zhang Z, Gerstein M. Large-scale analysis of pseudogenes in the human genome. Curr Opin Genet Dev. 2004;14:328-35
21. McDonell L, Drouin G. The abundance of processed pseudogenes derived from glycolytic genes is correlated with their expression level. Genome. 2012;55:147-51
22. Rogalla P, Blank C, Helbig R, Wosniok W, Bullerdiek J. Significant correlation between the breakpoints of rare clonal aberrations in benign solid tumors and the assignment of HMGIY retropseudogenes. Cancer Genet Cytogenet. 2001;130:51-6
23. Harrison PM, Zheng D, Zhang Z, Carriero N, Gerstein M. Transcribed processed pseudogenes in the human genome: an intermediate form of expressed retrosequence lacking protein-coding ability. Nucleic Acids Res. 2005;33:2374-83
24. Zheng D, Frankish A, Baertsch R, Kapranov P, Reymond A, Choo SW. et al. Pseudogenes in the ENCODE regions: consensus annotation, analysis of transcription, and evolution. Genome Res. 2007;17:839-51
25. Poliseno L. Pseudogenes: newly discovered players in human cancer. Sci Signal. 2012;5:re5
26. Suo G, Han J, Wang X, Zhang J, Zhao Y, Dai J. Oct4 pseudogenes are transcribed in cancers. Biochem Bioph Res Co. 2005;337:1047-51
27. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033-8
28. Poliseno L, Haimovic A, Christos PJ, Vega YSdMEC, Shapiro R, Pavlick A. et al. Deletion of PTENP1 pseudogene in human melanoma. J Invest Dermatol. 2011;131:2497-500
29. Koda Y, Soejima M, Wang B, Kimura H. Structure and expression of the gene encoding secretor-type galactoside 2-alpha-L-fucosyltransferase (FUT2). Eur J Biochem. 1997;246:750-5
30. Lui KY, Peng HR, Lin JR, Qiu CH, Chen HA, Fu RD. et al. Pseudogene integrator complex subunit 6 pseudogene 1 (INTS6P1) as a novel plasma-based biomarker for hepatocellular carcinoma screening. Tumour Biol. 2016;37:1253-60
31. Kalyana-Sundaram S, Kumar-Sinha C, Shankar S, Robinson DR, Wu YM, Cao X. et al. Expressed pseudogenes in the transcriptional landscape of human cancers. Cell. 2012;149:1622-34
32. Kawamura N, Nimura K, Nagano H, Yamaguchi S, Nonomura N, Kaneda Y. CRISPR/Cas9-mediated gene knockout of NANOG and NANOGP8 decreases the malignant potential of prostate cancer cells. Oncotarget. 2015;6:22361-74
33. Lin M, Pedrosa E, Shah A, Hrabovsky A, Maqbool S, Zheng D. et al. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PloS One. 2011;6:e23356
34. Chiefari E, Iiritano S, Paonessa F, Le Pera I, Arcidiacono B, Filocamo M. et al. Pseudogene-mediated posttranscriptional silencing of HMGA1 can result in insulin resistance and type 2 diabetes. Nat Commun. 2010;1:40
35. Tsitsiou E, Williams AE, Moschos SA, Patel K, Rossios C, Jiang X. et al. Transcriptome analysis shows activation of circulating CD8+ T cells in patients with severe asthma. J Allergy Clin Immunol. 2012;129:95-103
36. Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919-31
37. Zheng X, Tang H, Zhao X, Sun Y, Jiang Y, Liu Y. Long non-coding RNA FTH1P3 facilitates uveal melanoma cell growth and invasion through miR-224-5p. PloS One. 2017;12:e0184746
38. Lyn D, Cherney BW, Lalande M, Berenson JR, Lichtenstein A, Lupold S. et al. A duplicated region is responsible for the poly(ADP-ribose) polymerase polymorphism, on chromosome 13, associated with a predisposition to cancer. Am J Hum Genet. 1993;52:124-34
39. Pan Y, Sun C, Huang M, Liu Y, Qi F, Liu L. et al. A genetic variant in pseudogene E2F3P1 contributes to prognosis of hepatocellular carcinoma. J Biomed Res. 2014;28:194-200
40. Shen L, Du M, Wang C, Gu D, Wang M, Zhang Q. et al. Clinical significance of POU5F1P1 rs10505477 polymorphism in Chinese gastric cancer patients receving cisplatin-based chemotherapy after surgical resection. Int J Mol Sci. 2014;15:12764-77
41. Byun DS, Cho K, Ryu BK, Lee MG, Park JI, Chae KS. et al. Frequent monoallelic deletion of PTEN and its reciprocal associatioin with PIK3CA amplification in gastric carcinoma. Int J Cancer. 2003;104:318-27
42. Schmitz J, Piskurek O, Zischler H. Forty million years of independent evolution: a mitochondrial gene and its corresponding nuclear pseudogene in primates. J Mol Evol. 2005;61:1-11
43. Marques AC, Tan J, Lee S, Kong L, Heger A, Ponting CP. Evidence for conserved post-transcriptional roles of unitary pseudogenes and for frequent bifunctionality of mRNAs. Genome Biol. 2012;13:R102
44. Khachane AN, Harrison PM. Assessing the genomic evidence for conserved transcribed pseudogenes under selection. BMC Genomics. 2009;10:435
45. Porter KA, Duffy EB, Nyland P, Atianand MK, Sharifi H, Harton JA. The CLRX.1/NOD24 (NLRP2P) pseudogene codes a functional negative regulator of NF-kappaB, pyrin-only protein 4. Genes Immun. 2014;15:392-403
46. Millson A, Lewis T, Pesaran T, Salvador D, Gillespie K, Gau CL. et al. Processed pseudogene confounding deletion/duplication assays for SMAD4. J Mol Diagn. 2015;17:576-82
47. Noll A, Raabe CA, Churakov G, Brosius J, Schmitz J. Ancient traces of tailless retropseudogenes in therian genomes. Genome Biol Evol. 2015;7:889-900
48. Bustamante CD, Nielsen R, Hartl DL. A maximum likelihood method for analyzing pseudogene evolution: implications for silent site evolution in humans and rodents. Mol Biol Evol. 2002;19:110-7
49. Khachane AN, Harrison PM. Strong association between pseudogenization mechanisms and gene sequence length. Biol Direct. 2009;4:38
50. Pei B, Sisu C, Frankish A, Howald C, Habegger L, Mu XJ. et al. The GENCODE pseudogene resource. Genome Biol. 2012;13:R51
51. Taniguchi Y, Katsumata Y, Koido S, Suemizu H, Yoshimura S, Moriuchi T. et al. Cloning, sequencing, and chromosomal localization of two tandemly arranged human pseudogenes for the proliferating cell nuclear antigen (PCNA). Mamm Genome. 1996;7:906-8
52. Mighell AJ, Smith NR, Robinson PA, Markham AF. Vertebrate pseudogenes. FEBS lett. 2000;468:109-14
53. Vanin EF. Processed pseudogenes: characteristics and evolution. Annu Rev Genet. 1985;19:253-72
54. Vinckenbosch N, Dupanloup I, Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci U S A. 2006;103:3220-5
55. Cooke SL, Shlien A, Marshall J, Pipinikas CP, Martincorena I, Tubio JM. et al. Processed pseudogenes acquired somatically during cancer development. Nat Commun. 2014;5:3644
56. Poliseno L, Marranci A, Pandolfi PP. Pseudogenes in human cancer. Front Med (Lausanne). 2015;2:68
57. Ganster C, Wernstedt A, Kehrer-Sawatzki H, Messiaen L, Schmidt K, Rahner N. et al. Functional PMS2 hybrid alleles containing a pseudogene-specific missense variant trace back to a single ancient intrachromosomal recombination event. Hum Mutat. 2010;31:552-60
58. Wang J, Pitarque M, Ingelman-Sundberg M. 3'-UTR polymorphism in the human CYP2A6 gene affects mRNA stability and enzyme expression. Biochem Biophys Res Commun. 2006;340:491-7
59. Puget N, Gad S, Perrin-Vidoz L, Sinilnikova OM, Stoppa-Lyonnet D, Lenoir GM. et al. Distinct BRCA1 rearrangements involving the BRCA1 pseudogene suggest the existence of a recombination hot spot. Am J Hum Genet. 2002;70:858-65
60. Korneev SA, Park JH, O'Shea M. Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J Neurosci. 1999;19:7711-20
61. Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Grander D. et al. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol. 2013;20:440-6
62. Hawkins PG, Morris KV. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription. 2010;1:165-75
63. Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673-8
64. Chan WL, Yuo CY, Yang WK, Hung SY, Chang YS, Chiu CC. et al. Transcribed pseudogene psiPPM1K generates endogenous siRNA to suppress oncogenic cell growth in hepatocellular carcinoma. Nucleic Acids Res. 2013;41:3734-47
65. Guo X, Lin M, Rockowitz S, Lachman HM, Zheng D. Characterization of human pseudogene-derived non-coding RNAs for functional potential. PloS One. 2014;9:e93972
66. Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S. et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534-8
67. Wen YZ, Zheng LL, Liao JY, Wang MH, Wei Y, Guo XM. et al. Pseudogene-derived small interference RNAs regulate gene expression in African Trypanosoma brucei. Proc Natl Acad Sci U S A. 2011;108:8345-50
68. Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA. et al. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57
69. Guo X, Zhang Z, Gerstein MB, Zheng D. Small RNAs originated from pseudogenes: cis- or trans-acting? PLoS Comput Biol. 2009;5:e1000449
70. Hu X, Yang L, Mo YY. Role of pseudogenes in tumorigenesis. Cancers (Basel). 2018:10
71. Chen DL, Lu YX, Zhang JX, Wei XL, Wang F, Zeng ZL. et al. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics. 2017;7:4836-49
72. Esposito F, De Martino M, Petti MG, Forzati F, Tornincasa M, Federico A. et al. HMGA1 pseudogenes as candidate proto-oncogenic competitive endogenous RNAs. Oncotarget. 2014;5:8341-54
73. Karreth FA, Reschke M, Ruocco A, Ng C, Chapuy B, Leopold V. et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161:319-32
74. Wang L, Guo ZY, Zhang R, Xin B, Chen R, Zhao J. et al. Pseudogene OCT4-pg4 functions as a natural micro RNA sponge to regulate OCT4 expression by competing for miR-145 in hepatocellular carcinoma. Carcinogenesis. 2013;34:1773-81
75. Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol. 2013;45:1895-910
76. Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963-9
77. Feng J, Yang G, Liu Y, Gao Y, Zhao M, Bu Y. et al. LncRNA PCNAP1 modulates hepatitis B virus replication and enhances tumor growth of liver cancer. Theranostics. 2019;9:5227-45
78. Scarola M, Comisso E, Pascolo R, Chiaradia R, Marion RM, Schneider C. et al. Epigenetic silencing of Oct4 by a complex containing SUV39H1 and Oct4 pseudogene lncRNA. Nat Commun. 2015;6:7631
79. Wen X, Liu X, Mao YP, Yang XJ, Wang YQ, Zhang PP. et al. Long non-coding RNA DANCR stabilizes HIF-1alpha and promotes metastasis by interacting with NF90/NF45 complex in nasopharyngeal carcinoma. Theranostics. 2018;8:5676-89
80. Han YJ, Ma SF, Yourek G, Park YD, Garcia JG. A transcribed pseudogene of MYLK promotes cell proliferation. FASEB J. 2011;25:2305-12
81. Zhao S, Yuan Q, Hao H, Guo Y, Liu S, Zhang Y. et al. Expression of OCT4 pseudogenes in human tumours: lessons from glioma and breast carcinoma. J Pathol. 2011;223:672-82
82. Zou M, Baitei EY, Alzahrani AS, Al-Mohanna F, Farid NR, Meyer B. et al. Oncogenic activation of MAP kinase by BRAF pseudogene in thyroid tumors. Neoplasia. 2009;11:57-65
83. Hendrickson RC, Cicinnati VR, Albers A, Dworacki G, Gambotto A, Pagliano O. et al. Identification of a 17beta-hydroxysteroid dehydrogenase type 12 pseudogene as the source of a highly restricted BALB/c Meth A tumor rejection peptide. Cancer Immunol Immunother. 2010;59:113-24
84. Wang QS, Shi LL, Sun F, Zhang YF, Chen RW, Yang SL. et al. High expression of ANXA2 pseudogene ANXA2P2 promotes an aggressive phenotype in hepatocellular carcinoma. Dis Markers. 2019;2019:9267046
85. Wang MY, Chen DP, Qi B, Li MY, Zhu YY, Yin WJ. et al. Pseudogene RACGAP1P activates RACGAP1/Rho/ERK signalling axis as a competing endogenous RNA to promote hepatocellular carcinoma early recurrence. Cell Death Dis. 2019;10:426
86. Song H, Yang J, Zhang Y, Zhou J, Li Y, Hao X. Integrated analysis of pseudogene RP11-564D11.3 expression and its potential roles in hepatocellular carcinoma. Epigenomics. 2019;11:267-80
87. Zhou Y, He P, Xie X, Sun C. Knockdown of SUMO1P3 represses tumor growth and invasion and enhances radiosensitivity in hepatocellular carcinoma. Mol Cell Biochem. 2019;450:125-34
88. Lin J, Cao S, Wang Y, Hu Y, Liu H, Li J. et al. Long non-coding RNA UBE2CP3 enhances HCC cell secretion of VEGFA and promotes angiogenesis by activating ERK1/2/HIF-1alpha/VEGFA signalling in hepatocellular carcinoma. J Exp Clin Cancer Res. 2018;37:113
89. Pan Y, Zhan L, Chen L, Zhang H, Sun C, Xing C. POU5F1B promotes hepatocellular carcinoma proliferation by activating AKT. Biomed Pharmacother. 2018;100:374-80
90. Qian YY, Li K, Liu QY, Liu ZS. Long non-coding RNA PTENP1 interacts with miR-193a-3p to suppress cell migration and invasion through the PTEN pathway in hepatocellular carcinoma. Oncotarget. 2017;8:107859-69
91. Cao SW, Huang JL, Chen J, Hu YW, Hu XM, Ren TY. et al. Long non-coding RNA UBE2CP3 promotes tumor metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma. Oncotarget. 2017;8:65370-85
92. Peng H, Ishida M, Li L, Saito A, Kamiya A, Hamilton JP. et al. Pseudogene INTS6P1 regulates its cognate gene INTS6 through competitive binding of miR-17-5p in hepatocellular carcinoma. Oncotarget. 2015;6:5666-77
93. Liu L, Liu Y, Liu J, Zhai X, Wen J, Xie K. et al. Genetic variants in pseudogene E2F3P1 confer risk for HBV-related hepatocellular carcinoma in a Chinese population. J Biomed Res. 2013;27:215-9
94. Lou W, Ding B, Fan W. High expression of pseudogene PTTG3P indicates a poor prognosis in human breast cancer. Mol Ther Oncolytics. 2019;14:15-26
95. Yndestad S, Austreid E, Skaftnesmo KO, Lonning PE, Eikesdal HP. Divergent activity of the pseudogene PTENP1 in ER-positive and negative breast cancer. Mol Cancer Res. 2018;16:78-89
96. Liu Y, Wang W, Li Y, Sun F, Lin J, Li L. CKS1BP7, a pseudogene of CKS1B, is co-amplified with IGF1R in breast cancers. Pathol Oncol Res. 2018;24:223-9
97. Li C, Zheng L, Xin Y, Tan Z, Zhang Y, Meng X. et al. The competing endogenous RNA network of CYP4Z1 and pseudogene CYP4Z2P exerts an anti-apoptotic function in breast cancer. FEBS Lett. 2017;591:991-1000
98. Zheng L, Li X, Gu Y, Lv X, Xi T. The 3'UTR of the pseudogene CYP4Z2P promotes tumor angiogenesis in breast cancer by acting as a ceRNA for CYP4Z1. Breast Cancer Res Treat. 2015;150:105-18
99. Zheng L, Li X, Gu Y, Ma Y, Xi T. Pseudogene CYP4Z2P 3'UTR promotes angiogenesis in breast cancer. Biochem Biophys Res Commun. 2014;453:545-51
100. De Martino M, Forzati F, Marfella M, Pellecchia S, Arra C, Terracciano L. et al. HMGA1P7-pseudogene regulates H19 and Igf2 expression by a competitive endogenous RNA mechanism. Sci Rep. 2016;6:37622
101. Li L, Feng R, Fei S, Cao J, Zhu Q, Ji G. et al. NANOGP8 expression regulates gastric cancer cell progression by transactivating DBC1 in gastric cancer MKN-45 cells. Oncol Lett. 2019;17:555-63
102. Ma G, Liu H, Du M, Zhang G, Lin Y, Ge Y. et al. A genetic variation in the CpG island of pseudogene GBAP1 promoter is associated with gastric cancer susceptibility. Cancer. 2019;125:2465-73
103. Ge Y, He Y, Jiang M, Luo D, Huan X, Wang W. et al. Polymorphisms in lncRNA PTENP1 and the risk of gastric cancer in a Chinese population. Dis Markers. 2017;2017:6807452
104. Guo X, Deng L, Deng K, Wang H, Shan T, Zhou H. et al. Pseudogene PTENP1 suppresses gastric cancer progression by modulating PTEN. Anticancer Agents Med Chem. 2016;16:456-64
105. Hayashi H, Arao T, Togashi Y, Kato H, Fujita Y, De Velasco MA. et al. The OCT4 pseudogene POU5F1B is amplified and promotes an aggressive phenotype in gastric cancer. Oncogene. 2015;34:199-208
106. Huang T, Wang X, Yang X, Ji J, Wang Q, Yue X. et al. Long non-coding RNA DUXAP8 enhances renal cell carcinoma progression via downregulating miR-126. Med Sci Monit. 2018;24:7340-7
107. Yu G, Yao W, Gumireddy K, Li A, Wang J, Xiao W. et al. Pseudogene PTENP1 functions as a competing endogenous RNA to suppress clear-cell renal cell carcinoma progression. Mol Cancer Ther. 2014;13:3086-97
108. Xu T, Li D, He Y, Zhang F, Qiao M, Chen Y. The expression level of CSDAP1 in lung cancer and its clinical significance. Oncol Lett. 2018;16:4361-6
109. Shang J, Wang Z, Chen W, Yang Z, Zheng L, Wang S. et al. Pseudogene CHIAP2 inhibits proliferation and invasion of lung adenocarcinoma cells by means of the WNT pathway. J Cell Physiol. 2019;234:13735-46
110. Sun M, Nie FQ, Zang C, Wang Y, Hou J, Wei C. et al. The pseudogene DUXAP8 promotes non-small-cell lung cancer cell proliferation and invasion by epigenetically silencing EGR1 and RHOB. Mol Ther. 2017;25:739-51
111. Wei CC, Nie FQ, Jiang LL, Chen QN, Chen ZY, Chen X. et al. The pseudogene DUXAP10 promotes an aggressive phenotype through binding with LSD1 and repressing LATS2 and RRAD in non small cell lung cancer. Oncotarget. 2017;8:5233-46
112. Lynn H, Sun X, Ayshiev D, Siegler JH, Rizzo AN, Karnes JH. et al. Single nucleotide polymorphisms in the MYLKP1 pseudogene are associated with increased colon cancer risk in African Americans. PloS One. 2018;13:e0200916
113. Zhang LM, Wang P, Liu XM, Zhang YJ. LncRNA SUMO1P3 drives colon cancer growth, metastasis and angiogenesis. Am J Transl Res. 2017;9:5461-72
114. Dai X, Xie Y, Dong M, Zhao J, Yu H, Zhou B. et al. The long noncoding RNA TPTE2P1 promotes the viability of colorectal cancer cells. J Cell Biochem. 2019;120:5268-76
115. Palumbo Junior A, de Sousa VPL, Esposito F, De Martino M, Forzati F, Moreira FCB. et al. Overexpression of HMGA1 figures as a potential prognostic factor in endometrioid endometrial carcinoma (EEC). Genes (Basel). 2019:10
116. Brunetti M, Agostini A, Staurseth J, Davidson B, Heim S, Micci F. Molecular characterization of carcinosarcomas arising in the uterus and ovaries. Oncotarget. 2019;10:3614-24
117. Zhao X, Hao S, Wang M, Xing D, Wang C. Knockdown of pseudogene DUXAP8 expression in glioma suppresses tumor cell proliferation. Oncol Lett. 2019;17:3511-6
118. Xu H, Chen B, Xing J, Wei Z, Liu C, Qiu Y. et al. Upregulation of LGMNP1 confers radiotherapy resistance in glioblastoma. Oncol Rep. 2019;41:3435-43
119. Esposito F, De Martino M, D'Angelo D, Mussnich P, Raverot G, Jaffrain-Rea ML. et al. HMGA1-pseudogene expression is induced in human pituitary tumors. Cell Cycle. 2015;14:1471-5
120. Yu J, Zhang J, Zhou L, Li H, Deng ZQ, Meng B. The octamer-binding transcription factor 4 (OCT4) pseudogene, POU domain class 5 transcription factor 1B (POU5F1B), is upregulated in cervical cancer and down-regulation inhibits cell proliferation and migration and induces apoptosis in cervical cancer cell lines. Med Sci Monit. 2019;25:1204-13
121. Lian Y, Yang J, Xiao C, Hu X, Xu H. DUXAP8, a pseudogene derived lncRNA, promotes growth of pancreatic carcinoma cells by epigenetically silencing CDKN1A and KLF2. Cancer Commun (Lond). 2018;38:64
122. Tian C, Jin Y, Shi S. Long non-coding RNA SUMO1P3 may promote cell proliferation, migration, and invasion of pancreatic cancer via EMT signaling pathway. Oncol Lett. 2018;16:6109-15
123. Liu F, Gong R, He B, Chen F, Hu Z. TUSC2P suppresses the tumor function of esophageal squamous cell carcinoma by regulating TUSC2 expression and correlates with disease prognosis. BMC Cancer. 2018;18:894
124. Yang L, Sun K, Chu J, Qu Y, Zhao X, Yin H. et al. Long non-coding RNA FTH1P3 regulated metastasis and invasion of esophageal squamous cell carcinoma through SP1/NF-kB pathway. Biomed Pharmacother. 2018;106:1570-7
125. Yi J, Zhou LY, Yi YY, Zhu X, Su XY, Zhao Q. et al. Low expression of pseudogene POU5F1B affects diagnosis and prognosis in acute myeloid leukemia (AML). Med Sci Monit. 2019;25:4952-9
126. Lettnin AP, Wagner EF, Carrett-Dias M, Dos Santos Machado K, Werhli A, Canedo AD. et al. Silencing the OCT4-PG1 pseudogene reduces OCT-4 protein levels and changes characteristics of the multidrug resistance phenotype in chronic myeloid leukemia. Mol Biol Rep. 2019;46:1873-84
127. Liu M, Gao X, Liu CL. Increased expression of lncRNA FTH1P3 promotes oral squamous cell carcinoma cells migration and invasion by enhancing PI3K/Akt/GSK3b/ Wnt/beta-catenin signaling. Eur Rev Med Pharmacol Sci. 2018;22:8306-14
128. Nuerzhati Y, Dong R, Song Z, Zheng S. Role of the long noncoding RNA-Annexin A2 pseudogene 3/Annexin A2 signaling pathway in biliary atresiaassociated hepatic injury. Int J Mol Med. 2019;43:739-48
129. Lv H, Tong J, Yang J, Lv S, Li WP, Zhang C. et al. Dysregulated pseudogene HK2P1 may contribute to preeclampsia as a competing endogenous RNA for hexokinase 2 by impairing decidualization. Hypertension. 2018;71:648-58
130. Tong J, Yang J, Lv H, Lv S, Zhang C, Chen ZJ. Dysfunction of pseudogene PGK1P2 is involved in preeclampsia by acting as a competing endogenous RNA of PGK1. Pregnancy Hypertens. 2018;13:37-45
131. Bergman O, Karry R, Milhem J, Ben-Shachar D. NDUFV2 pseudogene (NDUFV2P1) contributes to mitochondrial complex I deficits in schizophrenia. Mol Psychiatry. 2018
132. Cappuccio G, Attanasio S, Alagia M, Mutarelli M, Borzone R, Karali M. et al. Microdeletion of pseudogene chr14.232.a affects LRFN5 expression in cells of a patient with autism spectrum disorder. Eur J Hum Genet. 2019;27:1475-80
133. Li X, Yu M, Chen L, Sun T, Wang H, Zhao L. et al. LncRNA PMS2L2 protects ATDC5 chondrocytes against lipopolysaccharide-induced inflammatory injury by sponging miR-203. Life Sci. 2019;217:283-92
134. Lai Y, Li J, Zhong L, He X, Si X, Sun Y. et al. The pseudogene PTENP1 regulates smooth muscle cells as a competing endogenous RNA. Clin Sci (Lond). 2019;133:1439-55
135. Yang X, Ye H, He M, Zhou X, Sun N, Guo W. et al. LncRNA PDIA3P interacts with c-Myc to regulate cell proliferation via induction of pentose phosphate pathway in multiple myeloma. Biochem Biophys Res Commun. 2018;498:207-13
136. Han L, Yuan Y, Zheng S, Yang Y, Li J, Edgerton ME. et al. The Pan-Cancer analysis of pseudogene expression reveals biologically and clinically relevant tumour subtypes. Nat Commun. 2014;5:3963
137. Zhang J, Wang X, Li M, Han J, Chen B, Wang B. et al. NANOGP8 is a retrogene expressed in cancers. FEBS J. 2006;273:1723-30
138. Wang H, Liang L, Dong Q, Huan L, He J, Li B. et al. Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-kappaB pathway in hepatocellular carcinoma. Theranostics. 2018;8:2814-29
139. Zhang Z, Carriero N, Zheng D, Karro J, Harrison PM, Gerstein M. PseudoPipe: an automated pseudogene identification pipeline. Bioinformatics. 2006;22:1437-9
140. Zheng D, Gerstein MB. A computational approach for identifying pseudogenes in the ENCODE regions. Genome Biol. 2006;7(Suppl 1):S13 1-0
141. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775-89
142. Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N. et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559-63
143. Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199-208
144. Welch JD, Baran-Gale J, Perou CM, Sethupathy P, Prins JF. Pseudogenes transcribed in breast invasive carcinoma show subtype-specific expression and ceRNA potential. BMC Genomics. 2015;16:113
145. Williams TK, Yeo CJ, Brody J. Does this band make sense? Limits to expression based cancer studies. Cancer Lett. 2008;271:81-4
Author contact
![]() Corresponding authors: Tao Wang, Department of Medical Genetics and Developmental Biology, Fourth Military Medical University, #169 Changle West Road, Xi'an, Shaanxi, 710032, P.R. China. Tel.: 86-29-84772709; Fax: 86-29-83253816; E-mail: wangtedu.cn OR An-Gang Yang, State Key Laboratory of Cancer Biology, Department of Immunology, Fourth Military Medical University, #169 Changle West Road, Xi'an, Shaanxi, 710032, P.R. China. Tel.: 86-29-84772703; Fax: 86-29-83253816; E-mail: agyangedu.cn
Corresponding authors: Tao Wang, Department of Medical Genetics and Developmental Biology, Fourth Military Medical University, #169 Changle West Road, Xi'an, Shaanxi, 710032, P.R. China. Tel.: 86-29-84772709; Fax: 86-29-83253816; E-mail: wangtedu.cn OR An-Gang Yang, State Key Laboratory of Cancer Biology, Department of Immunology, Fourth Military Medical University, #169 Changle West Road, Xi'an, Shaanxi, 710032, P.R. China. Tel.: 86-29-84772703; Fax: 86-29-83253816; E-mail: agyangedu.cn
 Global reach, higher impact
Global reach, higher impact