13.3
Impact Factor
Theranostics 2020; 10(9):4150-4167. doi:10.7150/thno.42735 This issue Cite
Review
F-box proteins and cancer: an update from functional and regulatory mechanism to therapeutic clinical prospects
1. Department of Thoracic Surgery, Cancer Hospital of China Medical University, Liaoning Cancer Hospital & Institute, Shenyang 110042, China.
2. CAS Key Laboratory of Separation Science for Analytical Chemistry, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China.
* Equal contribution
Received 2019-12-4; Accepted 2020-2-4; Published 2020-3-4
Abstract
E3 ubiquitin ligases play a critical role in cellular mechanisms and cancer progression. F-box protein is the core component of the SKP1-cullin 1-F-box (SCF)-type E3 ubiquitin ligase and directly binds to substrates by various specific domains. According to the specific domains, F-box proteins are further classified into three sub-families: 1) F-box with leucine rich amino acid repeats (FBXL); 2) F-box with WD 40 amino acid repeats (FBXW); 3) F-box only with uncharacterized domains (FBXO). Here, we summarize the substrates of F-box proteins, discuss the important molecular mechanism and emerging role of F-box proteins especially from the perspective of cancer development and progression. These findings will shed new light on malignant tumor progression mechanisms, and suggest the potential role of F-box proteins as cancer biomarkers and therapeutic targets for future cancer treatment.
Keywords: F-box, E3 ligase, ubiquitin, substrate, cancer progression
Introduction
Ubiquitination is one of the key post translational modification which is regulated by cascade of three component enzymes including ubiquitin activating E1 enzyme, ubiquitin conjugating E2 enzyme and ubiquitin-protein E3 ligase. In the ubiquitination system, ubiquitin (Ubi) is a polypeptide of 76 amino acids in length, it is activated by an E1, delivered to an E2 by the E1, and finally an E3 interacts with the Ubi-loaded E2 and recognizes a specific “motif” at the substrate and links the self-lysine residue to lysine residues at the target protein. Ubiquitylation is initially described as a process that induces substrate degradation and erases the unfavorable products [1]. Subsequently, different consequences have also been identified, such as signal transduction, enzyme stabilization or activation. Emerging evidences have revealed that ubiquitination participates in nearly all kinds of biological processes including cell cycle, transcription, and various signaling pathways. Dysregulations of ubiquitination will induce multiple diseases, including neurodegenerative diseases, inflammatory disorders and various types of cancers especially.
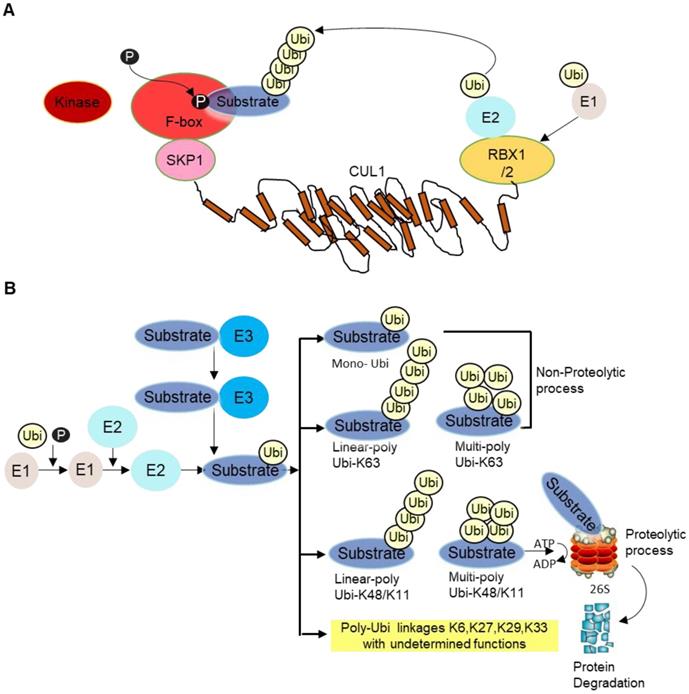
E3 ligase is the core component of the ubiquitination cascade, because they control the substrate specificity and binds to the substrates directly. There are hundreds of E3 ubiquitin ligases in humans [2]. Among them, the SKP1-cullin 1(CUL1)-F‑box (SCF) E3 ligase complex, is so far the best-characterized E3 ligase family. The SCF complex consists of four subunits, an adaptor protein SKP1, a RING finger protein RBX1/2, a scaffold protein CUL1, and one variable F‑box protein that recognizes specific substrates [3]. CUL1 binds to SKP1 and F-box protein at its N-terminus and to the RING protein RBX1/2 at its C-terminus. F-box protein generally recognizes phosphorylated substrate and presents it for ubiquitination (Figure 1A). Then, different types of ubiquitination will occur depending on number and types of Ubi(s) linked to substrates. Lysine 48 (K48) as well as K11 linked Ubi chains are proteolytic in nature whereas K63-linked and mono-Ubi are non-proteolytic in nature (Figure 1B). Although not all F-box proteins are well-characterized, multiple F-box proteins like SKP2, FBXW7, FBXO4, FBXO32 have been linked to cancer development, progression as well as cancer cachexia [4].
Here, we will discuss the molecular regulatory mechanisms, especially those involved in cancers, for F-box proteins, summarize the F-box protein relevant small compound inhibitors, and envision the future perspectives of F-box protein targeted cancer treatment.
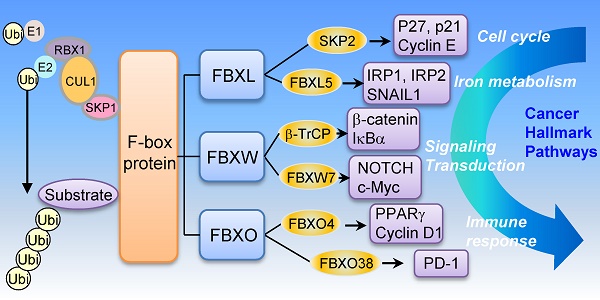
Classification of F-box proteins
F-box proteins are broadly classified into three sub-families: i) FBXL: F-box with leucine rich amino acid repeats, ii) FBXW: F-box with WD 40 amino acid repeats, iii) FBXO: F-box only with uncharacterized domains. There are about 22 FBXLs, 10 FBXWs and 37 FBXOs in the human genome at present.
A SCF complex. This complex comprises of scaffold CUL1, SKP1, RBX1/2 and F-box receptor. The substrate is phosphorylated by specific kinase enzyme and recognized by the substrate recognition domain. Ubi is transferred from E2 to E3 ligase/F-box proteins in coordination with RBX1/2 for proteosomal degradation. B Different forms of ubiquitination. After the substrate is presented to F-box protein, different types of ubiquitination occur depending on the number and types of Ubi/Ubis presented to substrates, namely mono-ubiquitination (i.e., single ubiquitin is added to substrate), linear poly Ubi-K63/K48/K11 (many Ubis are added one after another along a line format at lysine K63/K48/K11 locus of Ubis) and multi-poly-Ubi-K63 (Ubis are added in multilayers at lysine K63/K48/K11 locus). Some other Ubi-types are yet undetermined, such as K6, K27, K29, K33, etc. Ubi-K48/K11 types are proteolytic whereas Ubi-K63 and mono-Ubi are non-proteolytic in nature. The substrate is mainly processed at the 26S proteasome complex for ubiquitination guided proteasomal degradation.
Substrate recognition mechanism
F-box proteins generally recognize substrates modified by proper post-translational modification, especially phosphorylation. For example, substrates of FBXW7 contain the conserved CDC4 phospho-degron sequence 4-X‑pThr (or pSer)‑Pro‑Pro‑X‑pSer (or pThr, Glu or Asp) (X represents any amino acid) [5, 6]. Only when these amino acids get phosphorylated, FBXW7 can recognize and ubiquitinate the substrates for degradation. In some other cases, dephosphorylated degrons can also be recognized and ubiquitinated. When phospho-Tyr-655 is dephosphorylated by protein Tyr phosphatase L1 (PTPL1), p85β binds to FBXL2 and gets ubiquitinated [7]. In addition, the substrates can also be modified by glycosylation or mannose oligosaccharide [5, 6, 8, 9]. For instance, FBXO6 ubiquitinates the glycosylated degron in T-cell receptor alpha chain [9], and FBXO2 ubiquitinates N‑linked high-mannose oligosaccharides of precursor β1 integrin [8] of the respective substrates. Very rarely, the substrates are modified by lysine acetylation or tyrosine phosphorylation. Of note, the ubiquitination process can be nullified (or deubiquitinated) by deubiquitinating enzymes with evidences in various types of mammalian cell systems [10].
F-box proteins are involved in multiple cancer hallmark pathways
Biological functions of F-box proteins are well characterized in cells, mouse models and human cancer tissues. Many F-box proteins are found to act as either direct tumor suppressors/oncogenes or indirect cancer regulators. Their key functions largely depend on their ubiquitination abilities on substrates involved in cancer hallmark pathways, including cell cycle, DNA damage, epithelial-mesenchymal transition (EMT) as well as multiple signaling pathways like AKT/PI3K, BMP, p53, NRF2, AMPK/mTOR, AKT, NF-κB and Hippo pathway [7, 11-21] all of which can contribute to tumor growth, proliferation, progression, metastasis and invasion. Functions of F-box proteins and the corresponding substrates are being described briefly in Table 1. In the following parts, we will introduce each individual F-box protein with respect to their molecular function and potential clinical utility in cancer.
FBXL family
SKP2
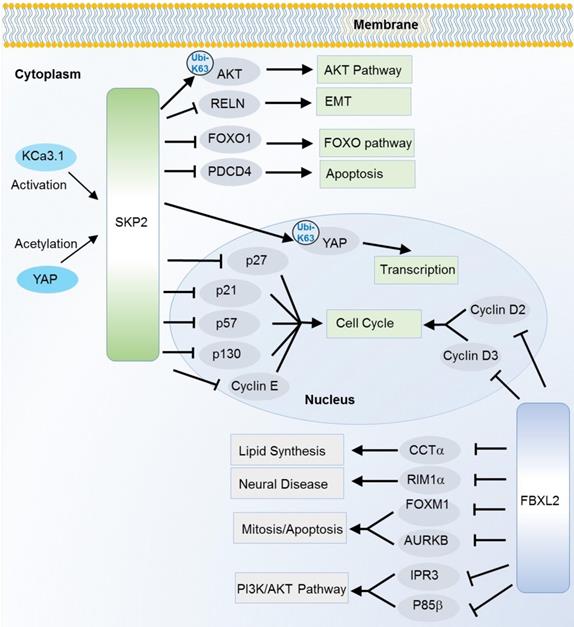
SKP2, also known as FBXL1, is the most characterized oncogene among F-box members. It is especially well known as a cell cycle regulator that can induce degradation of various cell cycle regulators. Of them, p27, a cyclin-dependent kinase (CDK) inhibitor is one of the best known substrates since the first discovery in 1999 [22]. Later on, a collection of CDK inhibitors including p21 [23], p27 [22] and p57 [24], and other cell cycle regulators like cyclin E [25], c-Myc [26] and p130 [27] are successively identified as the SKP2 substrates. Interestingly, SKP2 not only induces degradation of c-Myc, but also activates c-Myc target genes as a transcription cofactor [26]. Recently, a potential up-stream regulator of SKP2 in cell cycle is also revealed. The intermediate conductance calmodulin/calcium-activated potassium channel (KCa3.1) activates SKP2 and promotes cell proliferation, invasion and metastasis by degrading p21 and p27 through SKP2 [16]. In addition to the cell cycle pathway, SKP2 also participates in other cancer hallmark pathways such as FOXO, AMPK/mTOR, AKT, apoptosis and Hippo signaling pathway [15-18, 20, 21, 28-32] (Figure 2). SKP2 can ubiquitinate and induce the degradation of the transcription factor FOXO1 which possesses a tumor suppressor function [33]. SKP2 can regulate the AMPK pathway via degradation of coactivator-associated arginine methyltransferase 1 (CARM1), a member of protein arginine methyltransferase [18]. In response to DNA damage, SKP2 enhances phosphorylation and ubiquitination of programmed cell death protein 4 (PDCD4) to inhibit apoptosis with subsequent increase in cell growth and proliferation of breast cancer cells [34]. In addition to the degradation effect (mainly mediated by K48 linkage Ubis), SKP2 can also regulated the non-degradation polyubiquitination (mainly mediated by K63 linkage Ubis). The Hippo signaling key factor YAP was stabilized by SKP2 through the K63-linkage ubiquitination [35]. Recently, SKP2 has also been found to promote the K63-linkage mediated ubiquitination and activation of AKT, a key factor that conveys growth factor signals from cell outside to inside [36].
Probably because of its proteolysis effects on its substrates like p21, p27, PDCD4 and FOXO1, a large part of which are tumor suppressors, SKP2 mainly play oncogenic functions. Correspondingly, SKP2 up-regulation has been observed in many cancers like malignant oral cancer [37], colorectal cancer [38], hepatocellular carcinoma [18], and breast cancer [34]. Besides, the high expression of SKP2 has been implicated as a poor prognosis indicator in several types of cancers [39-41].
FBXL2
FBXL2 is also known for its active role in cell cycle. However, the ubiquitination substrates are unknown until 2012 when cyclin D2, D3 and Aurora kinase B (AURKB) were successively recognized as the substrates [42-44]. Meanwhile, unlike many F-box proteins, FBXL2 does not always recognize the phosphodegron of its substrates. For cyclin D2 and D3, FBXL2 target on their calmodulin-binding motifs. As mentioned above, FBXL2 also regulates PI3K/AKT pathway through targeting and degrading the dephosphorylated p85β (a catalytic subunit of PI3K pathway) [7]. Besides, FBXL2 can ubiquitinate phosphorylated forkhead box M1 (FOXM1), a transcriptional factor which regulates the expression of several cell cycle genes including cyclin B1 and D1 [45] (Figure 2). Due to its ubiquitin mediated degradation of cell cycle activators like cyclin D2, cyclin D3 and FOXM1, the FBXL2 mainly play a tumor suppressive role in several types of cancers like gastric cancer and leukemia [42, 43, 45]. However, its tumor suppressive role is controversial. In a recent study, inhibition of FBXL2 may also promote apoptosis and limit tumor growth in PTEN-null cancers where PTEN has been identified as a counteractor of FBXL2 in binding with IP3R3 (a major player in Ca+ dependent apoptosis) for ubiquitin mediated degradation [11].
FBXL3
FBXL3 mainly participates in mammalian oscillating circadian clock system through ubiquitinating two inhibitors of CLOCK-BMAL1 complex, cryptochrome-1 (CRY1) and cryptochrome-2 (CRY2), since the simultaneous discovery by several research groups in 2007 [46-49]. FBXL3 mediated degradation of CRY1 and CRY2 can re-activate the CLOCK-BMAL1 complex and increase the protein levels of period circadian protein homolog 1 (PER1) and 2 (PER2), two circadian clock regulators with tumor suppressor activity [49]. Nearly ten years later, another unexpected regulatory function between FBXL3 and CRY2 is found that FBXL3 in cooperation with CRY2 targets an oncogenic substrate c-Myc to inhibit uncontrolled tumor cell growth [50]. Recently, phosphorylated Tousled-like kinase (TLK2) is also found to be ubiquitinated by FBXL3 in cooperation with CRY2 in a cell cycle regulating mechanism [51]. Thus, FBXL3 is a remarkable molecular connection between circadian and cell cycle, and with tumor suppressive potentials.
Substrates of SKP2 and FBXL2 in various cancer relevant cellular functions and pathways. K63-linkage poly-ubiquitinations are annotated on the arrows, the others are K48-linkage ubiquitinations.
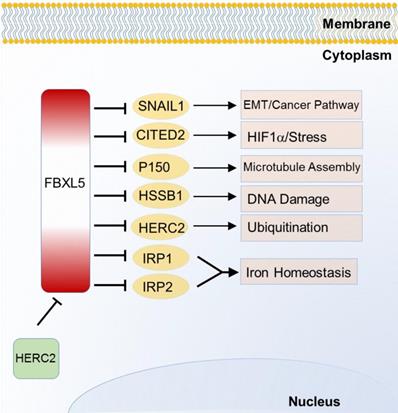
Substrates of FBXL5 in iron homeostasis and various cancer relevant cellular functions and pathways.
FBXL4
There are two revealed substrates, lysine demethylase 4A (KDM4A, also called JMJD2A) and GABAA receptor resistant to dieldrin [10], for FBXL4 at present. FBXL4 was firstly found to regulate replication time by degrading the substrate KDM4A in 2011 [52]. Later on, it was also revealed to regulate the timing of sleep through ubiquitin mediated degradation of RDL [53]. Despite the limited knowledge about its substrates, it is a potential tumor suppressor. Loss of FBXL4 gene is associated with advanced tumor stage and poor survival in prostate cancer [54]. Detection of deleted variants of FBXL4 in circulating tumor cells suggests it as a potential prognostic biomarker [54]. FBXL4 is also associated with mitochondrial DNA depletion syndrome and intellectual disabilities [55].
FBXL5
FBXL5 is the first SCF E3 ligase identified to regulate homeostasis or iron metabolism. Iron regulatory protein 1 (IRP1) and 2 (IRP2), two post transcriptional regulatory genes, can control and maintain cellular iron uptake, use, release and storage. FBXL5 was found to target and degrade IRP1 and IRP2 through ubiquitination in 2009 [56]. Unlike other F-box protein members, FBXL5 possesses an iron and oxygen binding hemerythrin domain that acts as a specific motif-dependent regulator for FBXL5-self differential stability [56]. Self-renewal of hematopoietic stem cell without FBXL5 can no longer survive due to cellular iron overload [57]. Recently, FBXL5 is found to be ubiquitinated by HECT and RLD domain containing E3 ubiquitin protein ligase 2 (HERC2) for proteasomal destruction. When FBXL5-HERC2 interaction is blocked, stability and abundance of FBXL5 is increased with lower intracellular Fe2+ load [58]. Emerging evidences of FBXL5-IRP2 axis suggest its potential therapeutic implication in cancer and hematopoietic stem cells [56, 57]. Additionally, FBXL5 also triggers chromosomal instability by degrading p150 which is required for binding to dynein and microtubules [59]. Recent studies also find that FBXL5 targets on Snail homolog 1 (SNAIL1) [60], Cbp/p300-interacting transactivator 2 (CITED2) [61] and human single-strand DNA binding proteins 1 (HSSB1) [62] which are respectively involved in EMT, HIF signaling pathway and DNA damage response (Figure 3).
FBXL7
The most well-known substrate of FBXL7 is Aurora A kinase (AURKA) [63], a pivotal regulator of mitosis. Interestingly, the ubiquitination between FBXL7 and AURKA only occurs during mitosis although FBXL7 co-localizes with AURKA throughout cell cycle [63]. FBXL7 can also regulate mitochondrial function by ubiquitinating survivin for degradation [64]. Interestingly, AURKA restricts the ubiquitination of survivin by tightly regulating FBXL7, thereby promoting gastric cancer resistance to drug [65]. Transcript level of FBXL7 is very high and associated with poor prognosis and unfavorable response to paclitaxel-based chemotherapy in ovarian cancer patients [66]. In addition, FBXL7 is a target of FBXL18 for polyubiquitination and proteasomal degradation to regulate the cell cycle progression [67].
FBXL12
FBXL12 also regulates cell cycle. It can induce calcium/calmodulin-dependent protein kinase (CaMKI) polyubiquitination guided proteasomal degradation to attenuate p27 phosphorylation and disrupt cyclin D1/CDK4 complex assembly and G1 arrest in lung epithelia [68]. FBXL12 can also augment p21 by mixed-type ubiquitination, including both K48 and K63 linked Ubi chains [69]. On the other hand, FBXL12 is mostly distributed in thymus and regulates the T-cell differentiation. FBXL12 regulates transition or T cell differentiation from CD4+CD8+ cells into CD4-CD8+/CD4+CD8- cells through degradation of aldehyde dehydrogenase 3 (ALDH3). The level of FBXL12 diminishes as T cells (CD4+CD8+ cells) progress into CD4-CD8+/CD4+CD8- cells, suggesting the key role of FBXL12-ALDH3 axis in the maturation of undifferentiated thymocytes [70]. Besides, FBXL12 ubiquitinates and degrades one sub-unit of the Ku heterodimer, Ku80, a key regulator for the nonhomologous end joining double strand break repair pathway [71].
FBXL13
FBXL13 is abundant at centrosome and is associated with chromosomal stability. It interacts with centrosome associated proteins Centrin-2, Centrin-3, CEP152 and CEP192 [72]. Of these proteins, accumulation of CEP192 isoform is harmful to cells by increasing centrosome over-duplication that can promote cancer cell invasion and metastasis. FBXL13 targets CEP192 for proteasomal degradation to lower centrosomal γ-tubulin and disrupt microtubule array formation.
FBXL14
FBXL14 mainly regulates the EMT pathway by degradation of the EMT inducers SNAIL1 [73] and Twist-related protein 1 (TWIST1) [74]. In pancreatic cancer, liver kinase B1 (LKB1) promotes the ubiquitination of SNAIL1 by FBXL14, suggesting LKB1/FBXL14/SNAIL1 axes a potential therapeutic target [75]. One key oncogene c-Myc is also ubiquitinated by FBXL14 for proteasomal degradation, and this ubiquitination can be reversed back by a deubiquitinase USP13 in glioma stem cells. The antagonistic relation between USP13 and FBXL14 deserves deep studies for further clinical and therapeutic applications [76]. Additionally, FBXL14 targets and degrades CUB domain-containing protein 1 (CDCP1) to reduce its stability and prevent CDCP1 target genes involved in breast cancer metastasis [77]. FBXL14 even reaches neuronal differentiation by targeting C-terminal WRPW motif in a Notch signaling factor, hairy and enhancer of split 1 (HES1) [78].
FBXL17
FBXL17 degrades suppressor of fused homolog (SUFU) to release glioma-associated oncogene (GLI) from the SUFU domain for proper Hedgehog signaling pathway. Lack of FBXL17 often causes defective Hedgehog signaling, a characteristic of impaired cancer cell proliferation and medulloblastoma tumor growth [79]. FBXL17 is also a regulator of NRF2 oxidative stress pathway by degradation of transcription regulator protein BACH1 [14]. In addition, FBXL17 is a quality control factor for dimeric BTB complexes [80].
FBXL18
As mentioned above, FBXL18 can mediate the ubiquitination and degradation of FBXL7, thus indirectly impact cell cycle progression [67]. In another study, FBXL18 inhibits apoptosis and exerts an oncogenic function through K63-linked ubiquitination of AKT in glioma [15]. Recently, FBXL18 is found to ubiquitinate xeroderma pigmentosum group B complementing protein (XPB) where the CDK7 triggers Ser90 phosphorylation of XPB and presents XPB to FBXL18 [81].
FBXL19
FBXL19 is one F-box protein showing self-induced ubiquitination. An acetyltransferase CBP catalyzes acetylation of FBXL19. Stability of FBXL19 is increased with the level of CBP and vice versa [82]. Additionally, FBXL19 targets lysine-166 of Rac family small GTPase 3 (RAC3) for proteasomal degradation to regulate TGFβ1-induced E-cadherin down-regulation in esophageal cancer cells [83]. In lung epithelial cells, FBXL19 induces ubiquitination and degradation of ras homolog family member A (RhoA) by binding cytoplasmic small GTPase at lysine-135 of RhoA. Consequently, phosphorylation of p27 and cell proliferation is reduced. Of note, phosphorylation of RhoA is mediated by protein kinase ERK2. Thus, FBXL19 regulates the cell proliferation and cytoskeleton rearrangement [84].
The other FBXLs
Although FBXL is the most comprehensively described F-box family protein, there are still several FBXLs with only few substrates. FBXL10 and FBXL11, although contain the FBXL domains, are better known as two histone demethylases KDM2B and KDM2A. Their ubiquitination substrates are still unclear. FBXL6, FBXL8, FBXL9 and FBXL16 are orphan E3s without any known substrates at present. Our previous computational study predicts some substrates for the FBXLs including the orphan ones, e.g., voltage-dependent anion-selective channel protein 2 (VDAC2) and cyclin-A2 are predicted as substrates of FBXL6 [85]. FBXL15 can regulate BMP signaling pathway by degradating its ubiquitinated substrate SMAD ubiquitination regulatory factor 1 (SMURF1) [12]. FBXL20 can ubiquitinate Vacuolar protein-sorting 34 (Vps34), a regulator involved in autophagy and receptor degradation [86] , and E-cadherin for degradation [87]. Like FBXL3, FBXL21 also targets on both CRY1 and CRY2 for degradation, but its interaction mainly occurs in the cytoplasm, and it antagonizes the degradation induced by FBXL3 in the nucleus [88].
FBXW family
β-TrCP (FBXW1 or FBXW11)
β-TrCP has two main isoforms β-TrCP1 (also called FBXW1) and β-TrCP2 (also called FBXW11) [89]. Although specific substrates of different isoforms are observed, most of the substrates are common between the two main isoforms. The functional specificity of these isoforms is yet to be elucidated. Here, we shall use β-TrCP to refer to both of them, or otherwise specified.
Way back to 1990s, β-TrCP was found as a regulator of β-catenin and was one main regulator of cell viability. β-TrCP recognizes the Ser33 and Ser37 of β-catenin phosphorylated by glycogen synthase kinase 3β (GSK3β) [90, 91]. It also interacts with the phosphorylated domains in IκBα and mediates IκBα ubiquitination, thus activating the NF-κB pathway [92, 93]. When DNA damage or stalled DNA replication occurs, the activated checkpoint kinase-1 (CHK1) and 2 (CHK2) trigger hyperphosphorylation of cell division cycle 25A (CDC25A), then β-TrCP targets CDC25A for ubiquitin-mediated proteolysis, and delays the cell cycle progression. Thus, β-TrCP regulates normal cell cycle progression and acts like cell cycle check-points [94]. Oncogenic transformation and neural differentiation are also controlled by β-TrCP through targeting and ubiquitinating RE1-silencing transcription factor (REST). Over-expression of β-TrCP is commonly found in cancers with low level of REST [95]. High level or truncated REST in cancer cells causes genomic instability which leads to oncogenic cellular transformation. In few cases, β-TrCP also acts like a tumor suppressor gene. The SCF-β-TrCP dependent ubiquitination guided degradation of REST during G2 phase increases the optimum time for activation of spindle check points [96]. β-TrCP also stimulates GSK3β mediated apoptosis where GSK3β phosphorylates the protein, induced myeloid leukemia cell differentiation protein Mcl-1 (MCL1), and the MCL1 is then recognized by β-TrCP for proteasomal degradation [97]. Another substrate of β-TrCP is NRF2 which is also GSK3β dependent [98]. β-TrCP with the UbcH5 ubiquitin-conjugating enzyme which helps to form heterotypic polyubiquitin chains on c-Myc can induce ubiquitination mediated stabilization of c-Myc [99]. β-TrCP degrades phosphorylated LPIN1, a factor of fatty acid biosynthesis. Thus, the role of β-TrCP becomes clear in lipid metabolic homeostasis [100] (Figure 4).
Substrates of FBXW family proteins β-TrCP and FBXW7 in various cancer relevant cellular functions and pathways.
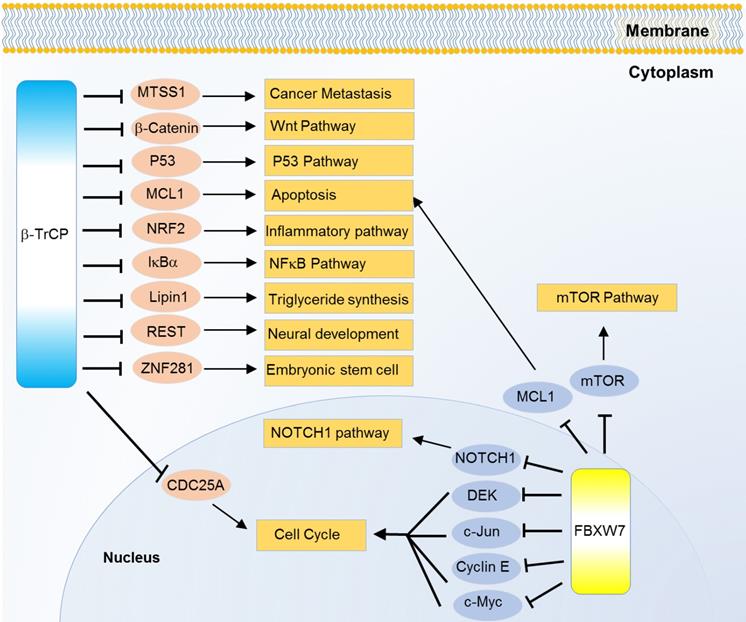
Some isoforms specific mechanisms are also revealed. β-TrCP, especially β-TrCP1, and IκB kinase 2 (IKKβ) can, in part, regulate the loss of function of p53. In this case, IKKβ phosphorylates p53 at ser362 and ser366 positions, then β-TrCP1 recruits the phosphorylated p53 for ubiquitination guided degradation [101]. Besides, β-TrCP1 promotes another F-box protein FBXW2 ubiquitination, and so does FBXW2 to SKP2, then the β-TrCP1-FBXW2-SKP2 axis presents an oncogene-tumor suppressor-oncogene cascade that controls cancer cell growth [102]. β-TrCP2 but not β-TrCP1 can mediate the ubiquitination and degradation of ZNF281, thus inhibiting the progression of colorectal cancer [103]. In addition, mainly β-TrCP1, can ubiquitinate and degrade MTSS1 in prostate and breast cancers [104] (Figure 4). Notably, β-TrCP1 and β-TrCP2 are observed to target each other for degradation in a recent study and β-TrCP2 preferentially degrades DEPTOR and REDD1, two inhibitors of mTORC1, thereby inhibiting autophagy and promoting cell growth [105].
FBXW7
FBXW7 (also known as hCdc4, SEL10) is another well characterized member of F-box family with WD40 repeat. There are three FBXW7 isoforms, FBXW7α, FBXW7β, and FBXW7γ in mammalian cells. These isoforms have different cellular localizations: FBXW7α is localized in the nucleoplasm, FBXW7β is in cytoplasm, and FBXW7γ is nucleolar. FBXW7α is most ubiquitously one and performs most of the recognized functions. Here, we mainly use FBXW7 to represent FBXWα. Since the first identified oncogenic substrate cyclin E [106], FBXW7 is recognized to ubiquitinate multiple oncogenic substrates like NOTCH1 [107], JUN [108], c-Myc [109], mTOR [110], MCL1 [111] and DEK [112] for degradation (Figure 4). Meanwhile, mutation mediated down regulation of FBXW7 is common in various types of cancers, especially T cell acute lymphatic leukemia and cholangiocarcinoma [113]. Notably, FBXW7 mutation reduces the binding affinity to NOTCH and knocked-out FBXW7 increases the level of NOTCH1-NICD, c-Myc as well as HIF-1α activity in chronic lymphoid leukemia [114]. Growth and progression of cholangiocarcinoma cells can also be regulated by FBXW7. FBXW7, in some aspects, is a p53 dependent tumor suppressor gene. In a search to rule out the relation between FBXW7 and p53, several putative DNA response elements were respectively identified at the FBXW7α, FBXW7β and FBXW7γ isoforms. siRNA knocked-down FBXW7 MEF (under p53+/- condition) cells show growth advantage than controls, and p53-/- MEF cells show similar growth like controls [115].
The other FBXWs
Some of the other FBXWs also participate in cancer relevant processes. FBXW2 can target on SKP2 for degradation, thus stabilizing the substrates of SKP2 [102]. FBXW2 also ubiquitinates β-catenin for degradation [116]. FBXW5 mediates the ubiquitination and subsequent degradation of a tumor suppressor TSC2 [117]. FBXW5 also regulates cell cycle by ubiquitination and subsequent proteasomal degradation of spindle assembly abnormal protein 6 (SASS6) [118] and epidermal growth factor receptor kinase substrate 8 (EPS8) [119] during S and G2 phase respectively. FBXW8 can ubiquitinate and degrade MAP4K1, thereby affecting cell proliferation and differentiation [120]. Substrates of the other FBXWs remain to be explored.
FBXO family
FBXO1
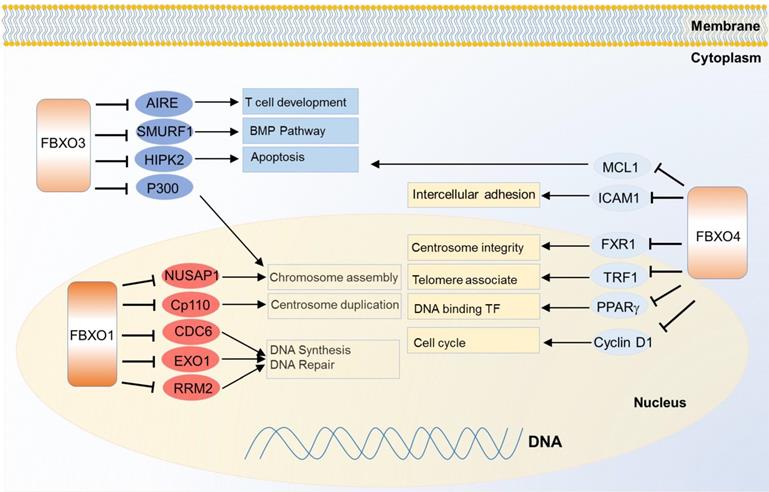
FBXO1 is also called cyclin F because it contains a cyclin box domain, however, it also functions through SCF E3 ligase complex. It mainly localizes in the nucleus, and participates in centrosome duplication and DNA repair. The first identified substrate of FBXO1 is centriolar coiled-coil protein of 110 kDa (CP110) which is necessary for centrosome duplication, and the FBXO1 mediates degradation of CP110 [121]. It also controls the maintenance of genome stability by degradation of ribonucleoside-diphosphate reductase subunit M2 (RRM2), which converts ribonucleotide to deoxyribonucleotide required for DNA replication and DNA repair [122]. Nucleolar and spindle-associated protein 1 (NUSAP1), a cell-cycle-regulated microtubule-binding protein involved in chromosome assembly, is one substrate of FBXO1 as well [123]. In another case, FBXO1 degrades eukaryotic DNA replication protein CDC6 and blocks DNA replication at the end of mitosis, thus inhibiting the progress of error DNA synthesis to attain genomic stability [124] (Figure 5). Meanwhile, down regulation of FBXO1 is associated with advanced tumor stage, poor survival and accelerated tumor growth in hepatocellular carcinoma [125].
FBXO3
FBXO3 may generate effects on cancer cells through multiple pathways. It was identified to regulate apoptosis by degradation of two transcription factor co-activators homeodomain-interacting protein kinase 2 (HIPK2) and p300. Meanwhile, protein PML protects them from the degradation without influencing on their ubiquitinations, thus PML, HIPK2 and FBXO3 cooperatively activating p53 dependent transactivation [126]. FBXO3 also participates in immune and inflammatory regulation. It stimulates cytokine secretion from human inflammatory cells by destabilizing the phosphorylating FBXL2, a TRAF inhibitor. TRAF is generally involved in responses ranging from tissue injury to cytokine release [127]. FBXO3 regulates T cell development by degrading autoimmune regulator (AIRE) which helps eliminate auto-reactive T cells during development, and it increases the AIRE's binding affinity to the positive transcription elongation factor b (P-TEFb) to properly monitor the transcription and directs proper expression of AIRE-responsive tissue-specific antigens in the thymus [128]. Besides, FBXO3 can regulate BMP signaling through ubiquitination guided degradation of SMURF1 [129].
FBXO4
FBXO4 also plays important functions in cancer. One of the best known substrates is cyclin D1 since the discovery in 2006. FBXO4 promotes ubiquitin-mediated degradation of Thr286-phosphorylated cyclin D1 [130]. Correspondingly, FBXO4 dysfunction can contribute to cyclin D1 overexpression and promote malignance in a large fraction of human cancers like melanoma [131] and esophageal cancer [132]. Another critical substrate of FBXO4 is telomeric repeat binding factor 1 (TRF1), a negative regulator of telomere length. FBXO4 regulates the ubiquitin-dependent degradation of TRF1 through an atypical small GTPase domain, thereby promoting telomere elongation [133]. In two recent studies, there emerges a feedback loop mechanism to balance the level of both FBXO4 and one of its substrate, fragile X mental retardation syndrome-related protein 1 (FXR1), in both head and neck squamous cell carcinoma and prostate cancer [134, 135]. FBXO4 also shows tumor suppressive functions in breast cancer and lung cancer through ubiquitin dependent degradation of intercellular adhesion molecule 1 (ICAM1) [136] and MCL1 [137] respectively. Besides, FBXO4 mediates ubiquitination of peroxisome proliferator-activated receptor gamma (PPARγ) with cooperation of heat shock 20 kDa-like protein p20 (HSP20) [138] (Figure 5).
FBXO6
FBXO6 regulates CHK1 ubiquitination and degradation and this may influence drug sensitivity to cisplatin [139, 140]. FBXO6 can also control the endoplasmic reticulum stress induced apoptosis by targeting endoplasmic oxidoreductin-1-like protein (Ero1L) for degradation [141]. Recently, an evidence has emerged that FBXO6 regulates the genomic stability via chromosome arrangement by monitoring two substrates, mitotic arrest deficient 2-like protein 1 (MAD2) and BUB1-related protein 1 (BUBR1) [142].
Substrates of FBXO family proteins FBXO1, FBXO3 and FBXO4 in various cancer relevant cellular functions and pathways.
FBXO7
FBXO7 has shown oncogenic potentials [143]. However, different from most F-box proteins, most of FBXO7's interacting proteins are not its ubiquitination substrates. For instance, FBXO7 interacts with CDK6, and the interaction is necessary to regulate entry of cell cycle. In a nude mouse experiment, over-expression of FBXO7 transformed the murine fibroblast into tumorigenic cells in a CDK6 dependent manner [144]. In another study, knock- down of FBXO7 increases the cell proliferation with reduced cell size and mitotic time [145]. Besides, over-expressing of FBXO7 increases the development of T cell lymphoma in p53 null cells. Thus, FBXO7 negatively regulates the proliferation and differentiation in a p53 dependent manner [143]. Although most of the above oncogenic functions are independent of ubiquitination, some cancer relevant substrates are also revealed. For example, FBXO7 can control apoptosis by ubiquitination guided degradation of human inhibitor of apoptosis protein 1 (cIAP1) whose function is increasingly involved in cell cycle and signaling pathway [146].
FBXO11
FBXO11 is most likely a tumor suppressor. It can target on a collection of oncogenic substrates. For instance, B-cell lymphoma 6 protein (BCL6), the product of a proto-oncogene, is ubiquitinated and degraded by FBXO11 and the FBXO11 gene is frequently deleted or mutated in diffuse large B-cell lymphomas [147]. EMT and metastasis factor of SNAIL1 and critical oncogenic protein HIF-1α can also ubiquitinated by FBXO11 for degradation [148, 149]. In addition, FBXO11 is also known as neddylating E3 ligase, which covalently conjugates NEDD8 to its substrates. One neddylation substrate is p53, FBXO11 can inactivates p53 by inhibiting its nuclear translocation [150]. Moreover, a lower expression of FBXO11 implicated poor prognosis in cancer patients [151].
FBXO31
FBXO31 may also act as a tumor suppressor. It is encoded in the 16q24.3 region in which the loss of heterozygosity is observed in various cancers like breast, ovarian, hepatocellular and prostate cancers. The current identified substrates of FBXO31 include cyclin D1 [152], SNAIL [153], MDM2 (an E3 ligase of p53) [154] and mitogen-activated protein kinase kinase 6 (MKK6) [155], all of which are oncogenic or promotive to tumorigenesis. FBXO31 mediates ubiquitination and degradation of these substrates and thereby inhibiting cancer cell development and progression.
FBXO32
FBXO32 may regulate cancer relevant processes through several targets. It degrades Krueppel-like factor 4 (KLF4) to suppress the breast cancer progression via p38 mitogen-activated protein kinase pathway [156]. Like β-TRCP, FBXO32 also degrades IκBα to activate NF-κB, and this degradation happens even under genotoxic and inflammatory stress [157]. In another study, FBXO32 ubiquitinates and degrades C-terminal-binding protein 1 (CTBP1), thereby controlling EMT activation [158]. Besides, activation of FBXO32 (also known as Atrogin-1) has been linked to cancer induced cachexia [159].
FBXO45
Lastly, we will update briefly about the role of FBXO45 in cancer. Low level of FBXO45 in gastric cancer tissues was found to be associated with the low survival rate of gastric cancer patients regardless of lymph node metastasis [160]. FBXO45 also degrades prostate apoptosis response protein 4 (PAR4) to block selective cell death and promotes cancer cell proliferation and survival [161]. FBXO45 can target p73 in vitro and in vivo to regulate the apoptosis mediated by p53 [162]. Besides, FBXO45 is also involved in neural development by degradation of the substrate N-Cadherin [163].
The other FBXOs
Substrates of the other FBXOs are less described, and only limited substrates or interactors are revealed. FBXO5, better knew as early mitotic inhibitor 1 (EMI1), can function as both substrate and inhibitor of the anaphase-promoting complex (APC/CCDH1), which is also an E3 ligase, to start the cell cycle. FBXO10 mediates ubiquitination and degradation of an antiapoptotic protein BCL2. FBXO21 mediates the ubiquitylation and proteasomal degradation of EP300-interacting inhibitor of differentiation 1 (EID1) [164]. FBXO22 mediates the ubiquitin-dependent degradation of key sarcomeric proteins, such as alpha-actinin (ACTN2) and filamin-C (FLNC) [165]. Notably, FBXO38 mediates the ubiquitination and degradation of the substrate programmed cell death protein 1 (PD-1), a promising cancer immunotherapy target, thus regulating T-cells-mediated immunity [166]. Many FBXO members like FBXO15, FBXO16, FBXO20 (also known as LMO7), FBXO24, etc., are still orphan E3 ligases, none of their substrates are characterized.
Micro-RNAs regulating F-box proteins
Micro-RNAs (mi-RNAs) refer to a class of small regulatory RNAs that control the multiple signaling factors including E3 ubiquitin enzyme of F-box proteins. Here, we have discussed some miRNAs which regulate F-box proteins. The miR-203 promotes cell cycle exit and long term cell proliferation by significant inhibition of SKP2 expression in genetically engineered mouse model [171]. And also, miR-378 can bind and down regulate the expression of SKP2 to control diabetic neuropathy [31]. Interestingly, miRNA-181d targets 3'-UTR of FBXL3 and stabilizing c-Myc expression and increased glucose consumption and lactate production in colorectal cancer [172]. Besides, miR-4735-3p reduces the expression of FBXL3 and suppresses cell proliferation and migration in small cell lung cancer [173]. The miR-1306-3p promotes cancer cell progression and metastasis by directly targeting FBXL5 through suppressing snail degradation in hepatocellular carcinoma [174].
FBXW7 is a tumor suppressor strongly suppressed the cancer cells proliferation, however the role of FBXW7 can be significantly inhibited by several mi-RNAs including miR-25, miR-92, miR-182, miR194, miR-223 and miR-503 in gastric, esophageal, colorectal, breast and cervical cancer cells [175-179]. The expression of FBXO11 can be reduced by miR-21 with subsequent BCL6 elevation in melanoma, prostate cancer and glioma [180, 181]. miR-218 suppresses FBXW8 expression and inhibits the cell proliferation in human choriocarcinoma cells [182], and miR-29c negatively regulates FBXO31 in gastric cancer cells [183]. Even here we have discussed the published interaction between microRNAs and F-box proteins, however there are still many non-discovered microRNA regulating F-box proteins, and have to be elucidated.
Targets/substrates of F-box proteins and their biological functions
| F-box protein | Localization | Role | Substrates | Biological functions |
|---|---|---|---|---|
| FBXL gene family | ||||
| SKP2 | Nucleus,Cytoplasm | Oncogene | RELN | EMT [16] |
| CARM1 | AMPK pathway [18] | |||
| AKT (K63) | AKT Pathway [167] | |||
| PDCD4 | Apoptosis [34] | |||
| P21 | Cell cycle [23] | |||
| P27 | Cell cycle [22] | |||
| P130 | Cell cycle [27] | |||
| c-Myc | Cell cycle [26] | |||
| Cyclin E | Cell cycle [25] | |||
| FOXO1 | FOXO signaling pathway [33] | |||
| YAP1 (K63) | Transcription of target genes [35] | |||
| FBXL2 | Cytoplasm, Membrane | Potential tumor suppressor | Cyclin D | Cell cycle checkpoints[42, 43] |
| AURKB | Cell cycle checkpoints[44] | |||
| p85β | PI3K pathway [7] | |||
| IP3R3 | PI3K pathway [11] | |||
| FOXM1 | Cell proliferation [45] | |||
| FBXL3 | Nucleus, Cytoplasmic | Potential tumor suppressor | CRY1,CRY2 | Circadian clock system [49] |
| c-Myc | Cell cycle progression, apoptosis and cellular transformation [50] | |||
| TLK2 | Cell cycle [51] | |||
| FBXL4 | Cytoplasm, Mitochondrion, Nucleus | Potential tumor suppressor | KDM4A | Replication time [52] |
| RDL | Timing of sleep [53] | |||
| FBXL5 | Cytoplasm, Perinuclear region | Potential tumor suppressor | IRP1,IRP2 | Iron metabolism [56] [168] |
| p150 | Genome stability[59] | |||
| SNAIL1 | EMT [60] | |||
| CITED2 | HIF pathway [61] | |||
| hSSB1 | DNA repair [62] | |||
| FBXL7 | Cytoskeleton | Potential oncogene | AURKA | Cell cycle [63] |
| Survivin | Apoptosis [65] | |||
| FBXL12 | Cytoplasm, Nucleus | Unclear | ALDH3 | T cell development [70] |
| Ku80 | Non homologues end joining double strand break repair mechanism [169] | |||
| CaMKI | Cell cycle [68] | |||
| p21 | Cell cycle progression at G1 [69] | |||
| FBXL13 | Cytoskeleton, Cytoplasm | Potential oncogene | CEP192 | Centrosome duplication [72] |
| FBXL14 | Cytoplasm | Potential tumor suppressor | CDCP1 | Tyrosine phosphorylation-dependent regulation of cellular events [77] |
| SNAIL1 | EMT [75] | |||
| c-Myc | Cell cycle progression, apoptosis and cellular transformation [76] | |||
| HES1 | Neuron development [78] | |||
| TWIST1 | EMT [74] | |||
| FBXL15 | Cytoplasm | Unclear | SMURF1 | BMP signaling pathway [12] |
| FBXL17 | Nucleus, Cytoplasm | Potential oncogene | SUFU | Hedgehog signal pathway[79] |
| BACH1 | NRF2 oxidative stress pathway [14] | |||
| FBXL18 | Cytoplasm, Nucleus | Potential oncogene | XPB | Transcription [81] |
| AKT(K63) | AKT pathway [15] | |||
| FBXL19 | Cytoplasm | Potential tumor suppressor | RAC3 | TGFβ1-induced E-cadherin down-regulation [83] |
| RhoA | Cell proliferation and cytoskeleton rearrangement [84] | |||
| FBXL20 | Cytoplasm | Potential oncogene | E-cadherin | Wnt signaling pathway [87] |
| Vps34 | Autophagy [86] | |||
| FBXL21 | Cytoplasm, Nucleus | Unclear | CRY1,CRY2 | Circadian clock system [88] |
| FBXW gene family | ||||
| β-TrCP | Nucleus, Cytoplasm | Generally oncogene & tumor suppressor in a few cases | β-catenin | Cell viability [90] |
| IκBα | NF-κB pathway [93] | |||
| CDC25A | Cell cycle [94] | |||
| REST | Spindle check points [96] | |||
| MCL1 | Anti-apoptotic (a member of the Bcl-2 family) [97] | |||
| p53 | p53 pathway [101] | |||
| c-Myc | Apoptosis [99] | |||
| Lipin1 | Fatty acid biosynthesis [100] | |||
| MTSS1 | Tumour suppression [104] | |||
| NRF2 | NRF2 pathway [98] | |||
| FBXW2 | β-TrCP-FBXW2-SKP2 axis [102] | |||
| ZNF281 | Colorectal cancer progression [103] | |||
| DEPTOR, REDD1 | Autophagy [105] | |||
| FBXW7 | FBXWα: Nucleoplasm, FBXWβ: Cytoplasm, FBXWγ:Nucleolus | Tumor suppressor | Cyclin E | Cell cycle [106] |
| mTOR | mTOR signaling pathway [110] | |||
| NOTCH1 | NOTCH1 signaling [114] | |||
| c-Jun and DEK | Closed circularity of DNA, cell cycle progression [112] | |||
| MCL1 | Apoptosis [111] | |||
| c-Myc | Cell proliferation [109] | |||
| FBXW2 | Cytoplasm | Potential tumor suppressor | SKP2 | β-TrCP-FBXW2-SKP2 axis [102] |
| β-catenin | [116] | |||
| FBXW5 | Cytoplasm, Nucleus | Unclear | SASS6 | Centrosome duplication [118] |
| EPS8 | Cell proliferation and motility [119] | |||
| TSC2 | Tuberous sclerosis [117] | |||
| FBXW8 | Golgi apparatus, Cytoplasm | Unclear | MAP4K1 | MAPK pathway [120] |
| FBXO gene family | ||||
| FBXO1 | Nucleus, Cytoplasm, Cytoskeleton | Potential tumor suppressor | RRM2 | DNA replication and DNA repair synthesis [122] |
| CP110 | Centrosome duplication; Genomic integrity [121] | |||
| NUSAP1 | Chromosome assembly [123] | |||
| CDC6 | Early steps of DNA replication [124] | |||
| FBXO3 | Nucleus, Cytoplasm | Unclear | AIRE | T cell development [128] |
| HIPK2,p300 | Transcription [126] | |||
| SMRUF1 | BMP signaling pathway [129] | |||
| FBXO4 | Cytoplasm | Tumor suppressor | TRF1 | Cell cycle [131] |
| MCL1 | Apoptosis [137] | |||
| FXR1 | RNA binding protein and Fragile X syndrome [134] | |||
| ICAM1 | Intercellular adhesion [136] | |||
| PPARγ | Adipocyte differentiation [138] | |||
| Cyclin D1 | Cell cycle [130] | |||
| FBXO6 | Cytoplasm | Unclear | CHK1 | Cisplatin sensitivity [140] |
| MAD2,BUBR1 | Spindle checkpoint [142] | |||
| Ero1L | Apoptosis [141] | |||
| FBXO7 | Cytoplasm, Mitochondrion, Nucleus | Potential oncogene | cIAP1 | Inhibition of apoptosis [146] |
| FBXO10 | Cytoplasm | Potential tumor suppressor | BCL1 | Apoptosis [170] |
| FBXO11 | Chromosome, Nucleus | Potential tumor suppressor | HIF-1α | HIF-1αsignaling pathway [143] |
| BCL6 | B-cells differentiation [147] | |||
| SNAIL | EMT [148] | |||
| FBXO21 | Cytoplasm | Unclear | EID1 | Cell cycle [164] |
| FBXO22 | Cytoplasm, Nucleus, Z disc | Unclear | ACTN2, FLNC | Contractile function [165] |
| FBXO31 | Cytoplasm, Cytoskeleton | Potential tumor suppressor | SNAIL1 | EMT [153] |
| Cyclin D1 | Cell cycle [152] | |||
| MDM2 | p53-mediated growth arrest [154] | |||
| MKK6 | MAPK pathway [155] | |||
| FBXO32 | Cytoplasm, Nucleus | Unclear | KLF4 | Apoptosis [156] |
| IκBα | NF-κB pathway [157] | |||
| CTBP1 | EMT [158] | |||
| FBXO38 | Cytoplasm, Nucleus | Potential tumor suppressor | PD-1 | Immunity of T cells [166] |
| FBXO45 | Postsynaptic cell membrane , Cell junction, Synapse | Unclear | N-Cadherin | Neuronal differentiation [163] |
| PAR4 | Apoptosis [161] | |||
| p73 | Apoptosis [162] | |||
Note: other unlisted F-box proteins have no known substrates at present. The potential roles of F-box proteins, mostly context dependent, are just based on current limited studies.
Small molecules and compounds are effective as inhibitors of F-box gene functions
Owing to their interactions with multiple cancer hallmark pathways, F-box proteins have been regarded as potential cancer therapeutic targets, and many small compounds have been applied to intervene F-box proteins (Table 2). SKP2 is one of the most promising targets, with various compounds showing effectiveness in inhibiting SKP2. Compound A and compound 25 interfere the binding between SKP2 and SKP1 in the SCF complex, and subsequently increases accumulation of p21, p27 and other SKP2 substrates, thereby promoting cell apoptosis [184, 185]. SMIP004 can reduce the SKP2 expression in prostate cancer [186]. In addition to SKP2, recognition of p27 during ubiquitination also depends on an accessory protein, Cdc kinase subunit 1 (CKS1). Compounds C1, C2, C16 and C20 have the ability to bind a structural pocket developed between SKP2 and CKS1 and block the interaction in metastatic melanoma, prostate, breast, ovarian and lung cancer [187]. Besides, natural products such as curcumin, quercetin, lycopene, silibinin, EGCG and vitamin D can inhibit the expression of SKP2 in breast and prostate cancer [188-190].
β-TrCP is another promising target. GS143 disrupts interaction between phospho-IκBα and β-TrCP, thereby suppressing IκBα ubiquitylation [191]. Generally, erioflorin inhibits the interaction between β-TrCP and a tumor suppressor PDCD4 in cancer [192]. STG28 also modulates the expression of β-TrCP and β-catenin citing its potential chemotherapeutic efficacies [193].
In addition to SKP2 and β-TrCP, some small compounds targeting on FBXW7, FBXO3, FBXL2, and FBXL3 are under study as well. SINE KPT-185 can block nuclear export of FBXW7 and enhances nuclear retention of FBXW7 to degrade NOTCH1 [194]. Oridonin increases level of FBXW7 and activates GSK3 to enhance c-Myc turnover in leukaemia and lymphoma [195]. BC1215 inhibits the binding of FBXO3 and FBXL2 to the target [109]. Another compound BC1258 has property to block binding between FBXO3 and FBXL2 and stabilize FBXL2 to promote Aurora B degradation [44]. KL001 competes for binding in the FAD pocket of CRYs and prevents FBXL3 binding in disorders including sleep disorder, cancer, cardiovascular and metabolic diseases [196] (Table 2).
Recently, chemists have developed a proteolysis-targeting chimeras (PROTACs) technology that can induce targeted protein degradation by the ubiquitin-proteasome system. PROTACs focused on drug resistance and 'undruggable' targets research. Till now less than ten E3 ubiquitin ligases have been exploited for targeted protein degradation [197]. With advances in this technology, more potential F-box proteins will be discovered.
Details of different molecules and compounds which target F-box proteins
| Target | Compound | Identified functions |
|---|---|---|
| SKP2 | Compound A | Disrupts SKP2-SKP1 interaction and prevents ubiquitination of p27 [184] |
| SKP2 | SMIP004 | Targets SKP2 and down-regulation SKP2[186] |
| SKP2 | C1, C2, C16, C20 | Binds to a pocket formed by SKP2 and CKS1 to block substrate binding [187] |
| SKP2 | Compound 25/ SZL-P1-41 | Binds to SKP2 and prevents SKP2-SKP1 interaction [185] |
| SKP2 | Curcumin, Quercetin, Lycopene, Silibinin, EGCG, EGCG, Vitamin-D | Blocks SKP2 expression [188-190] . |
| SKP2 | NSC689857 NSC681152 | Blocks the SKP2-CKS1 interaction and p27 ubiquitination in vitro [198] |
| β-TrCP | GS143 | Interfere interaction between phospho-IkBα and β-TrCP, suppress IkBα ubiquitylation [191] |
| β-TrCP | Erioflorin | Blocks the interaction of β-TrCP to PDCD4 [192] |
| β-TrCP | STG28 | Modulates the expression of β-TrCP and β-catenin [193] |
| FBXW7 | SINE KPT-185 | Inhibits transport of FBXW7, increases nuclear FBXW7 level and degrades NOTCH1 [194] |
| FBXW7 | Oridonin | Increases FBXW7 level, activates GSK3 and facilitates c-Myc degradation [195] |
| FBXW7 | Genistein | Down-regulates miR-223 level and elevates its target FBXW7 level [199] |
| FBXW7 | SCF-12 | Interferes substrate binding pocket and impede recognition of phosphodegron on substrates [6] |
| FBXL2 | BC-1215 | Inhibits FBXO3 and FBXL2 binding [44] |
| FBXL2 | BC-1258 | Inhibits binding FBXO3 and FBXL2; stabilizes FBXL2 and promotes AURKB Degradation [44] |
| FBXL3 | KL001 | Competes for binding in the FAD pocket of CRYs and prevents FBXL3 binding [200] |
| FBXO3 | BC1215 | Inhibits the substrate binding to FBXO3 [44] |
Concluding Remarks: Future prospect and therapeutic implications of F-box gene family
Going through the updates on F-box protein mediated ubiquitination mechanisms, it is very clear that F-box proteins play key roles in vital cellular functions namely, cell cycle, genome instability, signaling pathway and apoptosis in normal as well as tumor cells. Despite an initial limited knowledge on the role of F-box proteins, multiple F-box proteins have shown either oncogenic or tumor suppressive potentials in certain types of cancer (Table 1). The clinical implications and therapeutic or prognostic importance have emerged widely and been improved in the current decade.
Although many F-box proteins have been proposed as potential cancer therapeutic targets, none of them have entered into clinical research. Large challenges remain for F-box protein targeted cancer therapy. It is obvious that most of cancer relevant pathways are controlled by multiple F-box proteins rather than only one of them, for instance, the cell cycle pathway alone is influenced by at least 10 F-box proteins (Table 1), implying the potential cooperative mechanism between different F-box proteins in regulating the cellular functions. Even the same substrates can be ubiquitinated by multiple F-box proteins, e.g., MCL1 can be degraded by β-TrCP, FBXW7 and FBXO4 (Table 1) under different contexts. Drugs simply inhibiting of one specific F-box protein are probably not enough to control cancer cell development or progression, since various compensative mechanisms exist. Additionally, it should be noted that one F-box protein (e.g., SKP2, FBXW7, etc.) generally target on multiple different substrates involved in different pathways. Before taking the F-box proteins as therapeutic targets, we must make sure the intervention will not cause unexpected consequences by influencing the multifunctional substrates. What's more, most of the ubiquitination interactions are context dependent and varied with different cancer types, the up-stream pathways or co-regulators. The corresponding therapeutic effects are also determined by their context-dependent functions. Consequently, a more comprehensive understanding about the interactome with respect to F-box proteins as well as other E3 ligases is necessary. Future drug development should pay more attention to the complicated interactome and regulatory mechanism of F-box proteins, the drugs are better to target on specific interactions between F-box proteins and substrates or co-regulators that play key functions in certain types of cancer.
Advanced state-of-art technology and evolving scientific advances across various models have poured a concrete confidence for discovery of novel mechanisms played by F-box proteins. Physical interaction mechanism study has now placed a new scope to identify the detailed structure and topology of the interacting partners or domains. Identification of upstream target genes and compounds has led us to a new therapeutic strategy for future. Before stepping at the clinical door, biological suitability and efficacy of the drugs or compounds needs more rigorous investigations across wide range of cell systems and animal models. Combination of systems and in-depth molecular biology will promote the illustration about the unknown mechanisms left with undetermined F-box proteins. More importantly, investigation of large pool of human clinical samples is warranted. The most challenging aspect considering the future clinical trials will be the great genetic diversity to all corners of populations in this world. Taken together, we should always keep in mind all the aforesaid findings and issues when we stick and step on this way.
Acknowledgements
The authors gratefully acknowledge the funding support from National Natural Science Foundation of China grants (No. 81672440, No. 31701156, No. 81972625), Innovation program of science and research from the DICP, CAS (DICP TMSR201601, DICP ZZBS201803) and supported by grants from Department of Liaoning Science and Technology, titled with The Construction of Liaoning Cancer Research Center (Lung Cancer) (1564992449013); and titled with Precise diagnosis and treatment and optimization of clinical pathway for malignant tumor based on molecular markers-the research of precise treatment and optimization of clinical pathway for lung cancer (2019020176-JH1/103-02); Central financial fund for promoting medical service and safeguarding capability (Capability construction of medical and health organizations) -subsidy to the Construction of Provincial Key Specialty; Research grant to introduced talents of Liaoning Cancer Hospital.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425-79
2. Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14:369-81
3. Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438-49
4. Sukari A, Muqbil I, Mohammad RM, Philip PA, Azmi AS. F-BOX proteins in cancer cachexia and muscle wasting: Emerging regulators and therapeutic opportunities. Semin Cancer Biol. 2016;36:95-104
5. Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD. et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514-21
6. Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243-56
7. Kuchay S, Duan S, Schenkein E, Peschiaroli A, Saraf A, Florens L. et al. FBXL2- and PTPL1-mediated degradation of p110-free p85beta regulatory subunit controls the PI(3)K signalling cascade. Nat Cell Biol. 2013;15:472-80
8. Yoshida Y, Chiba T, Tokunaga F, Kawasaki H, Iwai K, Suzuki T. et al. E3 ubiquitin ligase that recognizes sugar chains. Nature. 2002;418:438-42
9. Yoshida Y, Tokunaga F, Chiba T, Iwai K, Tanaka K, Tai T. Fbs2 is a new member of the E3 ubiquitin ligase family that recognizes sugar chains. J Biol Chem. 2003;278:43877-84
10. Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12:1-222
11. Kuchay S, Giorgi C, Simoneschi D, Pagan J, Missiroli S, Saraf A. et al. PTEN counteracts FBXL2 to promote IP3R3- and Ca(2+)-mediated apoptosis limiting tumour growth. Nature. 2017;546:554-8
12. Cui Y, He S, Xing C, Lu K, Wang J, Xing G. et al. SCFFBXL15 regulates BMP signalling by directing the degradation of HECT-type ubiquitin ligase Smurf1. EMBO J. 2011;30:2675-89
13. Sato K, Kusama Y, Tategu M, Yoshida K. FBXL16 is a novel E2F1-regulated gene commonly upregulated in p16INK4A- and p14ARF-silenced HeLa cells. Int J Oncol. 2010;36:479-90
14. Tan MK, Lim HJ, Bennett EJ, Shi Y, Harper JW. Parallel SCF adaptor capture proteomics reveals a role for SCFFBXL17 in NRF2 activation via BACH1 repressor turnover. Mol Cell. 2013;52:9-24
15. Zhang S, Chen Q, Liu Q, Li Y, Sun X, Hong L. et al. Hippo Signaling Suppresses Cell Ploidy and Tumorigenesis through Skp2. Cancer Cell. 2017;31:669-84 e7
16. Du Y, Song W, Chen J, Chen H, Xuan Z, Zhao L. et al. The potassium channel KCa3.1 promotes cell proliferation by activating SKP2 and metastasis through the EMT pathway in hepatocellular carcinoma. Int J Cancer. 2019;145:503-16
17. Ding L, Wang C, Cui Y, Han X, Zhou Y, Bai J. et al. S-phase kinase-associated protein 2 is involved in epithelial-mesenchymal transition in methotrexate-resistant osteosarcoma cells. Int J Oncol. 2018;52:1841-52
18. Wei X, Li X, Yan W, Zhang X, Sun Y, Zhang F. SKP2 Promotes Hepatocellular Carcinoma Progression Through Nuclear AMPK-SKP2-CARM1 Signaling Transcriptionally Regulating Nutrient-Deprived Autophagy Induction. Cell Physiol Biochem. 2018;47:2484-97
19. Shapira M, Kakiashvili E, Rosenberg T, Hershko DD. The mTOR inhibitor rapamycin down-regulates the expression of the ubiquitin ligase subunit Skp2 in breast cancer cells. Breast Cancer Res. 2006;8:R46
20. Tan Y, Zhou G, Wang X, Chen W, Gao H. USP18 promotes breast cancer growth by upregulating EGFR and activating the AKT/Skp2 pathway. Int J Oncol. 2018;53:371-83
21. Liu K, Zhang L, Zhao Q, Zhao Z, Zhi F, Qin Y. et al. SKP2 attenuates NF-kappaB signaling by mediating IKKbeta degradation through autophagy. J Mol Cell Biol. 2018;10:205-15
22. Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193-9
23. Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21(Cip1) in S phase. J Biol Chem. 2003;278:25752-7
24. Kamura T, Hara T, Kotoshiba S, Yada M, Ishida N, Imaki H. et al. Degradation of p57(Kip2) mediated by SCFSkp2 - dependent ubiquitylation. P Natl Acad Sci USA. 2003;100:10231-6
25. Yeh KH, Kondo T, Zheng JY, Tsvetkov LM, Blair J, Zhang H. The F-box protein SKP2 binds to the phosphorylated threonine 380 in cyclin E and regulates ubiquitin-dependent degradation of cyclin E. Biochem Bioph Res Co. 2001;281:884-90
26. von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C. et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11:1189-200
27. Bhattacharya S, Garriga J, Calbo J, Yong T, Haines DS, Grana X. SKP2 associates with p130 and accelerates p130 ubiquitylation and degradation in human cells. Oncogene. 2003;22:2443-51
28. Shapira M, Kakiashvili E, Rosenberg T, Hershko DD. Correction to: The mTOR inhibitor rapamycin down-regulates the expression of the ubiquitin ligase subunit Skp2 in breast cancer cells. Breast Cancer Res. 2018;20:68
29. Koyama Y, Sumie S, Nakano Y, Nagao T, Tokumaru S, Michinaga S. Endothelin-1 stimulates expression of cyclin D1 and S-phase kinase-associated protein 2 by activating the transcription factor STAT3 in cultured rat astrocytes. J Biol Chem. 2019;294:3920-33
30. Deng T, Yan G, Song X, Xie L, Zhou Y, Li J. et al. Deubiquitylation and stabilization of p21 by USP11 is critical for cell-cycle progression and DNA damage responses. Proc Natl Acad Sci U S A. 2018;115:4678-83
31. Tsai YC, Kuo PL, Kuo MC, Hung WW, Wu LY, Chang WA. et al. The Interaction of miR-378i-Skp2 Regulates Cell Senescence in Diabetic Nephropathy. J Clin Med. 2018:7
32. Pratheeshkumar P, Siraj AK, Divya SP, Parvathareddy SK, Begum R, Melosantos R. et al. Downregulation of SKP2 in Papillary Thyroid Cancer Acts Synergistically With TRAIL on Inducing Apoptosis via ROS. J Clin Endocrinol Metab. 2018;103:1530-44
33. Huang H, Regan KM, Wang F, Wang D, Smith DI, van Deursen JM. et al. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc Natl Acad Sci U S A. 2005;102:1649-54
34. Li C, Du L, Ren Y, Liu X, Jiao Q, Cui D. et al. SKP2 promotes breast cancer tumorigenesis and radiation tolerance through PDCD4 ubiquitination. J Exp Clin Cancer Res. 2019;38:76
35. Yao F, Zhou Z, Kim J, Hang Q, Xiao Z, Ton BN. et al. SKP2- and OTUD1-regulated non-proteolytic ubiquitination of YAP promotes YAP nuclear localization and activity. Nat Commun. 2018;9:2269
36. Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z. et al. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012;149:1098-111
37. Gstaiger M, Jordan R, Lim M, Catzavelos C, Mestan J, Slingerland J. et al. Skp2 is oncogenic and overexpressed in human cancers. P Natl Acad Sci USA. 2001;98:5043-8
38. Li JQ, Wu F, Masaki T, Kubo A, Fujita J, Dixon DA. et al. Correlation of Skp2 with carcinogenesis, invasion, metastasis, and prognosis in colorectal tumors. Int J Oncol. 2004;25:87-95
39. Yang G, Ayala G, De Marzo A, Tian WH, Frolov A, Wheeler TM. et al. Elevated Skp2 protein expression in human prostate cancer: Association with loss of the cyclin-dependent kinase inhibitor p27 and PTEN and with reduced recurrence-free survival. Clin Cancer Res. 2002;8:3419-26
40. Shigemasa K, Gu LJ, O'Brien TJ, Ohama K. Skp2 overexpression is a prognostic factor in patients with ovarian adenocarcinoma. Clin Cancer Res. 2003;9:1756-63
41. Zhang WW, Cao LL, Sun ZJ, Xu J, Tang L, Chen WW. et al. Skp2 is over-expressed in breast cancer and promotes breast cancer cell proliferation. Cell Cycle. 2016;15:1344-51
42. Chen BB, Glasser JR, Coon TA, Zou C, Miller HL, Fenton M. et al. F-box protein FBXL2 targets cyclin D2 for ubiquitination and degradation to inhibit leukemic cell proliferation. Blood. 2012;119:3132-41
43. Chen BB, Glasser JR, Coon TA, Mallampalli RK. F-box protein FBXL2 exerts human lung tumor suppressor-like activity by ubiquitin-mediated degradation of cyclin D3 resulting in cell cycle arrest. Oncogene. 2012;31:2566-79
44. Chen BB, Glasser JR, Coon TA, Mallampalli RK. Skp-cullin-F box E3 ligase component FBXL2 ubiquitinates Aurora B to inhibit tumorigenesis. Cell Death Dis. 2013;4:e759
45. Li LQ, Pan D, Chen H, Zhang L, Xie WJ. F-box protein FBXL2 inhibits gastric cancer proliferation by ubiquitin-mediated degradation of forkhead box M1. FEBS Lett. 2016;590:445-52
46. Siepka SM, Yoo SH, Park J, Song WM, Kumar V, Hu YN. et al. Circadian mutant overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011-23
47. Yamanaka I, Koinuma S, Shigeyoshi Y, Uchiyama Y, Yagita K. Presence of robust circadian clock oscillation under constitutive over-expression of mCry1 in rat-1 fibroblasts. FEBS Letters. 2007;581:4098-102
48. Godinho SIH, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L. et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897-900
49. Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI. et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900-4
50. Huber AL, Papp SJ, Chan AB, Henriksson E, Jordan SD, Kriebs A. et al. CRY2 and FBXL3 Cooperatively Degrade c-MYC. Mol Cell. 2016;64:774-89
51. Correia SP, Chan AB, Vaughan M, Zolboot N, Perea V, Huber AL. et al. The circadian E3 ligase complex SCF(FBXL3+CRY) targets TLK2. Sci Rep. 2019;9:198
52. Van Rechem C, Black JC, Abbas T, Allen A, Rinehart CA, Yuan GC. et al. The SKP1-Cul1-F-box and leucine-rich repeat protein 4 (SCF-FbxL4) ubiquitin ligase regulates lysine demethylase 4A (KDM4A)/Jumonji domain-containing 2A (JMJD2A) protein. J Biol Chem. 2011;286:30462-70
53. Li Q, Li Y, Wang X, Qi JX, Jin X, Tong HW. et al. Fbxl4 Serves as a Clock Output Molecule that Regulates Sleep through Promotion of Rhythmic Degradation of the GABA(A) Receptor. Current Biology. 2017;27:3616 -+
54. Stankiewicz E, Mao X, Mangham DC, Xu L, Yeste-Velasco M, Fisher G. et al. Identification of FBXL4 as a Metastasis Associated Gene in Prostate Cancer. Sci Rep. 2017;7:5124
55. Ballout RA, Al Alam C, Bonnen PE, Huemer M, El-Hattab AW, Shbarou R. FBXL4-Related Mitochondrial DNA Depletion Syndrome 13 (MTDPS13): A Case Report With a Comprehensive Mutation Review. Front Genet. 2019;10:39
56. Salahudeen AA, Thompson JW, Ruiz JC, Ma HW, Kinch LN, Li Q. et al. An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science. 2009;326:722-6
57. Muto Y, Nishiyama M, Nita A, Moroishi T, Nakayama KI. Essential role of FBXL5-mediated cellular iron homeostasis in maintenance of hematopoietic stem cells. Nat Commun. 2017;8:16114
58. Moroishi T, Yamauchi T, Nishiyama M, Nakayama KI. HERC2 Targets the Iron Regulator FBXL5 for Degradation and Modulates Iron Metabolism. J Biol Chem. 2014;289:16430-41
59. Zhang N, Liu J, Ding X, Aikhionbare F, Jin C, Yao X. FBXL5 interacts with p150Glued and regulates its ubiquitination. Biochem Biophys Res Commun. 2007;359:34-9
60. Vinas-Castells R, Frias A, Robles-Lanuza E, Zhang K, Longmore GD, Garcia de Herreros A. et al. Nuclear ubiquitination by FBXL5 modulates Snail1 DNA binding and stability. Nucleic Acids Res. 2014;42:1079-94
61. Machado-Oliveira G, Guerreiro E, Matias AC, Facucho-Oliveira J, Pacheco-Leyva I, Braganca J. FBXL5 modulates HIF-1alpha transcriptional activity by degradation of CITED2. Arch Biochem Biophys. 2015;576:61-72
62. Chen ZW, Liu B, Tang NW, Xu YH, Ye XY, Li ZM. et al. FBXL5-mediated degradation of single-stranded DNA-binding protein hSSB1 controls DNA damage response. Nucleic Acids Res. 2014;42:11560-9
63. Coon TA, Glasser JR, Mallampalli RK, Chen BB. Novel E3 ligase component FBXL7 ubiquitinates and degrades Aurora A, causing mitotic arrest. Cell Cycle. 2012;11:721-9
64. Liu Y, Lear T, Iannone O, Shiva S, Corey C, Rajbhandari S. et al. The Proapoptotic F-box Protein Fbxl7 Regulates Mitochondrial Function by Mediating the Ubiquitylation and Proteasomal Degradation of Survivin. J Biol Chem. 2015;290:11843-52
65. Kamran M, Long ZJ, Xu D, Lv SS, Liu B, Wang CL. et al. Aurora kinase A regulates Survivin stability through targeting FBXL7 in gastric cancer drug resistance and prognosis. Oncogenesis. 2017;6:e298
66. Chiu HW, Chang JS, Lin HY, Lee HH, Kuei CH, Lin CH. et al. FBXL7 Upregulation Predicts a Poor Prognosis and Associates with a Possible Mechanism for Paclitaxel Resistance in Ovarian Cancer. J Clin Med. 2018:7
67. Liu Y, Lear T, Zhao Y, Zhao J, Zou C, Chen BB. et al. F-box protein Fbxl18 mediates polyubiquitylation and proteasomal degradation of the pro-apoptotic SCF subunit Fbxl7. Cell Death Dis. 2015;6:e1630
68. Mallampalli RK, Kaercher L, Snavely C, Pulijala R, Chen BB, Coon T. et al. Fbxl12 triggers G1 arrest by mediating degradation of calmodulin kinase I. Cell Signal. 2013;25:2047-59
69. Tsuruta F, Takebe A, Haratake K, Kanemori Y, Kim J, Endo T. et al. SCFFbl12 Increases p21Waf1/Cip1 Expression Level through Atypical Ubiquitin Chain Synthesis. Mol Cell Biol. 2016;36:2182-94
70. Nita A, Nishiyama M, Muto Y, Nakayama KI. FBXL12 regulates T-cell differentiation in a cell-autonomous manner. Genes Cells. 2016;21:517-24
71. Postow L, Funabiki H. An SCF complex containing Fbxl12 mediates DNA damage-induced Ku80 ubiquitylation. Cell Cycle. 2013;12:587-95
72. Fung E, Richter C, Yang HB, Schaffer I, Fischer R, Kessler BM. et al. FBXL13 directs the proteolysis of CEP192 to regulate centrosome homeostasis and cell migration. EMBO Rep. 2018:19
73. Vinas-Castells R, Beltran M, Valls G, Gomez I, Garcia JM, Montserrat-Sentis B. et al. The Hypoxia-controlled FBXL14 Ubiquitin Ligase Targets SNAIL1 for Proteasome Degradation. J Biol Chem. 2010;285:3794-805
74. Hayano M, Yang WS, Corn CK, Pagano NC, Stockwell BR. Loss of cysteinyl-tRNA synthetase (CARS) induces the transsulfuration pathway and inhibits ferroptosis induced by cystine deprivation. Cell Death Differ. 2016;23:270-8
75. Song L, Guo J, Chang R, Peng X, Li J, Xu X. et al. LKB1 obliterates Snail stability and inhibits pancreatic cancer metastasis in response to metformin treatment. Cancer Sci. 2018;109:1382-92
76. Fang X, Zhou W, Wu Q, Huang Z, Shi Y, Yang K. et al. Deubiquitinase USP13 maintains glioblastoma stem cells by antagonizing FBXL14-mediated Myc ubiquitination. J Exp Med. 2017;214:245-67
77. Cui YH, Kim H, Lee M, Yi JM, Kim RK, Uddin N. et al. FBXL14 abolishes breast cancer progression by targeting CDCP1 for proteasomal degradation. Oncogene. 2018;37:5794-809
78. Chen F, Zhang C, Wu H, Ma Y, Luo X, Gong X. et al. The E3 ubiquitin ligase SCF(FBXL14) complex stimulates neuronal differentiation by targeting the Notch signaling factor HES1 for proteolysis. J Biol Chem. 2017;292:20100-12
79. Raducu M, Fung E, Serres S, Infante P, Barberis A, Fischer R. et al. SCF (Fbxl17) ubiquitylation of Sufu regulates Hedgehog signaling and medulloblastoma development. EMBO J. 2016;35:1400-16
80. Mena EL, Kjolby RAS, Saxton RA, Werner A, Lew BG, Boyle JM. et al. Dimerization quality control ensures neuronal development and survival. Science. 2018:362
81. Ueda M, Matsuura K, Kawai H, Wakasugi M, Matsunaga T. Spironolactone-induced XPB degradation depends on CDK7 kinase and SCF(FBXL18) E3 ligase. Genes Cells. 2019;24:284-96
82. Wei J, Dong S, Yao K, Martinez M, Fleisher PR, Zhao Y. et al. Histone acetyltransferase CBP promotes function of SCF FBXL19 ubiquitin E3 ligase by acetylation and stabilization of its F-box protein subunit. FASEB J. 2018;32:4284-92
83. Dong S, Zhao J, Wei J, Bowser RK, Khoo A, Liu Z. et al. F-box protein complex FBXL19 regulates TGFbeta1-induced E-cadherin down-regulation by mediating Rac3 ubiquitination and degradation. Mol Cancer. 2014;13:76
84. Wei J, Mialki RK, Dong S, Khoo A, Mallampalli RK, Zhao Y. et al. A new mechanism of RhoA ubiquitination and degradation: roles of SCF(FBXL19) E3 ligase and Erk2. Biochim Biophys Acta. 2013;1833:2757-64
85. Chen D, Liu X, Xia T, Tekcham DS, Wang W, Chen H. et al. A Multidimensional Characterization of E3 Ubiquitin Ligase and Substrate Interaction Network. iScience. 2019;16:177-91
86. Xiao J, Zhang T, Xu D, Wang H, Cai Y, Jin T. et al. FBXL20-mediated Vps34 ubiquitination as a p53 controlled checkpoint in regulating autophagy and receptor degradation. Genes Dev. 2015;29:184-96
87. Zhu J, Li K, Dong L, Chen Y. Role of FBXL20 in human colorectal adenocarcinoma. Oncol Rep. 2012;28:2290-8
88. Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M, Kozuka-Hata H. et al. FBXL21 Regulates Oscillation of the Circadian Clock through Ubiquitination and Stabilization of Cryptochromes. Cell. 2013;152:1106-18
89. Seo E, Kim H, Kim R, Yun S, Kim M, Han JK. et al. Multiple isoforms of beta-TrCP display differential activities in the regulation of Wnt signaling. Cell Signal. 2009;21:43-51
90. Hart M, Concordet JP, Lassot I, Albert I, del los Santos R, Durand H. et al. The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Curr Biol. 1999;9:207-10
91. Latres E, Chiaur DS, Pagano M. The human F box protein beta-Trcp associates with the Cul1/Skp1 complex and regulates the stability of beta-catenin. Oncogene. 1999;18:849-54
92. Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM. et al. Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature. 1998;396:590-4
93. Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 1999;13:270-83
94. Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV. et al. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87-91
95. Westbrook TF, Hu G, Ang XL, Mulligan P, Pavlova NN, Liang A. et al. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. 2008;452:370-4
96. Guardavaccaro D, Frescas D, Dorrello NV, Peschiaroli A, Multani AS, Cardozo T. et al. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature. 2008;452:365-9
97. Ding Q, He X, Hsu JM, Xia W, Chen CT, Li LY. et al. Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol. 2007;27:4006-17
98. Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, Cuadrado A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol. 2011;31:1121-33
99. Popov N, Schulein C, Jaenicke LA, Eilers M. Ubiquitylation of the amino terminus of Myc by SCF(beta-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nat Cell Biol. 2010;12:973-81
100. Shimizu K, Fukushima H, Ogura K, Lien EC, Nihira NT, Zhang J. et al. The SCFbeta-TRCP E3 ubiquitin ligase complex targets Lipin1 for ubiquitination and degradation to promote hepatic lipogenesis. Sci Signal. 2017:10
101. Xia Y, Padre RC, De Mendoza TH, Bottero V, Tergaonkar VB, Verma IM. Phosphorylation of p53 by IkappaB kinase 2 promotes its degradation by beta-TrCP. Proc Natl Acad Sci U S A. 2009;106:2629-34
102. Xu J, Zhou W, Yang F, Chen G, Li H, Zhao Y. et al. The β-TrCP-FBXW2-SKP2 axis regulates lung cancer cell growth with FBXW2 acting as a tumour suppressor. Nat Commun. 2017;8:14002 -
103. Zhu Y, Zhou Q, Zhu G, Xing Y, Li S, Ren N. et al. GSK-3beta phosphorylation-dependent degradation of ZNF281 by beta-TrCP2 suppresses colorectal cancer progression. Oncotarget. 2017;8:88599-612
104. Zhong J, Shaik S, Wan L, Tron AE, Wang Z, Sun L. et al. SCF beta-TRCP targets MTSS1 for ubiquitination-mediated destruction to regulate cancer cell proliferation and migration. Oncotarget. 2013;4:2339-53
105. Cui D, Dai X, Shu J, Ma Y, Wei D, Xiong X. et al. The cross talk of two family members of beta-TrCP in the regulation of cell autophagy and growth. Cell Death Differ. 2019
106. Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316-22
107. Wu G, Lyapina S, Das I, Li J, Gurney M, Pauley A. et al. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol Cell Biol. 2001;21:7403-15
108. Nateri AS, Riera-Sans L, Da Costa C, Behrens A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science. 2004;303:1374-8
109. Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H. et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116-25
110. Mao JH, Kim IJ, Wu D, Climent J, Kang HC, DelRosario R. et al. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321:1499-502
111. Mori A, Masuda K, Ohtsuka H, Shijo M, Ariake K, Fukase K. et al. FBXW7 modulates malignant potential and cisplatin-induced apoptosis in cholangiocarcinoma through NOTCH1 and MCL1. Cancer Sci. 2018;109:3883-95
112. Babaei-Jadidi R, Li N, Saadeddin A, Spencer-Dene B, Jandke A, Muhammad B. et al. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J Exp Med. 2011;208:295-312
113. Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A. et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006-12
114. Close V, Close W, Kugler SJ, Reichenzeller M, Yosifov DY, Bloehdorn J. et al. FBXW7 mutations reduce binding of NOTCH1, leading to cleaved NOTCH1 accumulation and target gene activation in CLL. Blood. 2019;133:830-9
115. Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI. et al. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775-9
116. Yang F, Sun Y. FBXW2 suppresses proliferation and invasion of lung cancer cells by targeting SKP2 and beta-catenin. Mol Cell Oncol. 2019;6:1607458
117. Hu J, Zacharek S, He YJ, Lee H, Shumway S, Duronio RJ. et al. WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Gene Dev. 2008;22:866-71
118. Puklowski A, Homsi Y, Keller D, May M, Chauhan S, Kossatz U. et al. The SCF-FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nat Cell Biol. 2011;13:1004-U291
119. Werner A, Disanza A, Reifenberger N, Habeck G, Becker J, Calabrese M. et al. SCFFbxw5 mediates transient degradation of actin remodeller Eps8 to allow proper mitotic progression. Nat Cell Biol. 2013;15:179-88
120. Wang H, Chen Y, Lin P, Li L, Zhou GS, Liu GC. et al. The CUL7/F-box and WD Repeat Domain Containing 8 (CUL7/Fbxw8) Ubiquitin Ligase Promotes Degradation of Hematopoietic Progenitor Kinase 1. J Biol Chem. 2014;289:4009-17
121. D'Angiolella V, Donato V, Vijayakumar S, Saraf A, Florens L, Washburn MP. et al. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138-42
122. D'Angiolella V, Donato V, Forrester FM, Jeong YT, Pellacani C, Kudo Y. et al. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149:1023-34
123. Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y. et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459-74
124. Walter D, Hoffmann S, Komseli ES, Rappsilber J, Gorgoulis V, Sorensen CS. SCF(Cyclin F)-dependent degradation of CDC6 suppresses DNA re-replication. Nat Commun. 2016;7:10530
125. Fu J, Qiu H, Cai M, Pan Y, Cao Y, Liu L. et al. Low cyclin F expression in hepatocellular carcinoma associates with poor differentiation and unfavorable prognosis. Cancer Sci. 2013;104:508-15
126. Shima Y, Shima T, Chiba T, Irimura T, Pandolfi PP, Kitabayashi I. PML activates transcription by protecting HIPK2 and p300 from SCFFbx3-mediated degradation. Mol Cell Biol. 2008;28:7126-38
127. Chen BB, Coon TA, Glasser JR, McVerry BJ, Zhao J, Zhao Y. et al. A combinatorial F box protein directed pathway controls TRAF adaptor stability to regulate inflammation. Nat Immunol. 2013;14:470-9
128. Shao W, Zumer K, Fujinaga K, Peterlin BM. FBXO3 Protein Promotes Ubiquitylation and Transcriptional Activity of AIRE (Autoimmune Regulator). J Biol Chem. 2016;291:17953-63
129. Li D, Xie P, Zhao F, Shu J, Li L, Zhan Y. et al. F-box protein Fbxo3 targets Smurf1 ubiquitin ligase for ubiquitination and degradation. Biochem Biophys Res Commun. 2015;458:941-5
130. Lin DI, Barbash O, Kumar KG, Weber JD, Harper JW, Klein-Szanto AJ. et al. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol Cell. 2006;24:355-66
131. Lee EK, Lian Z, D'Andrea K, Letrero R, Sheng W, Liu S. et al. The FBXO4 tumor suppressor functions as a barrier to BRAFV600E-dependent metastatic melanoma. Mol Cell Biol. 2013;33:4422-33
132. Lian Z, Lee EK, Bass AJ, Wong KK, Klein-Szanto AJ, Rustgi AK. et al. FBXO4 loss facilitates carcinogen induced papilloma development in mice. Cancer Biol Ther. 2015;16:750-5
133. Lee TH, Perrem K, Harper JW, Lu KP, Zhou XZ. The F-box protein FBX4 targets PIN2/TRF1 for ubiquitin-mediated degradation and regulates telomere maintenance. J Biol Chem. 2006;281:759-68
134. Qie S, Majumder M, Mackiewicz K, Howley BV, Peterson YK, Howe PH. et al. Fbxo4-mediated degradation of Fxr1 suppresses tumorigenesis in head and neck squamous cell carcinoma. Nat Commun. 2017;8:1534
135. Cao H, Gao R, Yu C, Chen L, Feng Y. The RNA-binding protein FXR1 modulates prostate cancer progression by regulating FBXO4. Funct Integr Genomics. 2019;19:487-96
136. Kang JH, Choi MY, Cui YH, Kaushik N, Uddin N, Yoo KC. et al. Regulation of FBXO4-mediated ICAM-1 protein stability in metastatic breast cancer. Oncotarget. 2017;8:83100-13
137. Feng C, Yang F, Wang J. FBXO4 inhibits lung cancer cell survival by targeting Mcl-1 for degradation. Cancer Gene Ther. 2017;24:342-7
138. Peng J, Li Y, Wang X, Deng S, Holland J, Yates E. et al. An Hsp20-FBXO4 Axis Regulates Adipocyte Function through Modulating PPARgamma Ubiquitination. Cell Rep. 2018;23:3607-20
139. Zhang YW, Brognard J, Coughlin C, You Z, Dolled-Filhart M, Aslanian A. et al. The F box protein Fbx6 regulates Chk1 stability and cellular sensitivity to replication stress. Mol Cell. 2009;35:442-53
140. Cai L, Li J, Zhao J, Guo Y, Xie M, Zhang X. et al. Fbxo6 confers drug-sensitization to cisplatin via inhibiting the activation of Chk1 in non-small cell lung cancer. FEBS letters. 2019;593:1827-36
141. Chen X, Duan LH, Luo PC, Hu G, Yu X, Liu J. et al. FBXO6-Mediated Ubiquitination and Degradation of Ero1L Inhibits Endoplasmic Reticulum Stress-Induced Apoptosis. Cell Physiol Biochem. 2016;39:2501-8
142. Xu HZ, Wang ZQ, Shan HZ, Zhou L, Yang L, Lei H. et al. Overexpression of Fbxo6 inactivates spindle checkpoint by interacting with Mad2 and BubR1. Cell Cycle. 2018;17:2779-89
143. Lomonosov M, Meziane el K, Ye H, Nelson DE, Randle SJ, Laman H. Expression of Fbxo7 in haematopoietic progenitor cells cooperates with p53 loss to promote lymphomagenesis. PLoS One. 2011;6:e21165
144. Laman H, Funes JM, Ye H, Henderson S, Galinanes-Garcia L, Hara E. et al. Transforming activity of Fbxo7 is mediated specifically through regulation of cyclin D/cdk6. EMBO J. 2005;24:3104-16
145. Meziane el K, Randle SJ, Nelson DE, Lomonosov M, Laman H. Knockdown of Fbxo7 reveals its regulatory role in proliferation and differentiation of haematopoietic precursor cells. J Cell Sci. 2011;124:2175-86
146. Chang YF, Cheng CM, Chang LK, Jong YJ, Yuo CY. The F-box protein Fbxo7 interacts with human inhibitor of apoptosis protein cIAP1 and promotes cIAP1 ubiquitination. Biochem Biophys Res Commun. 2006;342:1022-6
147. Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF. et al. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90-U100
148. Zheng HQ, Shen MH, Zha YL, Li WY, Wei Y, Blanco MA. et al. PKD1 Phosphorylation-Dependent Degradation of SNAIL by SCF-FBXO11 Regulates Epithelial-Mesenchymal Transition and Metastasis. Cancer Cell. 2014;26:358-73
149. Ju UI, Park JW, Park HS, Kim SJ, Chun YS. FBXO11 represses cellular response to hypoxia by destabilizing hypoxia-inducible factor-1 alpha mRNA. Biochem Bioph Res Co. 2015;464:1008-15
150. Abida WM, Nikolaev A, Zhao WH, Zhang WZ, Gu W. FBXO11 promotes the neddylation of p53 and inhibits its transcriptional activity. J Biol Chem. 2007;282:1797-804
151. Jin Y, Shenoy AK, Doernberg S, Chen H, Luo HC, Shen HX. et al. FBXO11 promotes ubiquitination of the Snail family of transcription factors in cancer progression and epidermal development. Cancer Lett. 2015;362:70-82
152. Santra MK, Wajapeyee N, Green MR. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459:722-5
153. Zou S, Ma C, Yang F, Xu X, Jia J, Liu Z. FBXO31 Suppresses Gastric Cancer EMT by Targeting Snail1 for Proteasomal Degradation. Mol Cancer Res. 2018;16:286-95
154. Malonia SK, Dutta P, Santra MK, Green MR. F-box protein FBXO31 directs degradation of MDM2 to facilitate p53-mediated growth arrest following genotoxic stress. Proc Natl Acad Sci U S A. 2015;112:8632-7
155. Liu J, Han L, Li B, Yang J, Huen MS, Pan X. et al. F-box only protein 31 (FBXO31) negatively regulates p38 mitogen-activated protein kinase (MAPK) signaling by mediating lysine 48-linked ubiquitination and degradation of mitogen-activated protein kinase kinase 6 (MKK6). J Biol Chem. 2014;289:21508-18
156. Zhou H, Liu Y, Zhu R, Ding F, Wan Y, Li Y. et al. FBXO32 suppresses breast cancer tumorigenesis through targeting KLF4 to proteasomal degradation. Oncogene. 2017;36:3312-21
157. Meshram SN, Paul D, Manne R, Choppara S, Sankaran G, Agrawal Y. et al. FBXO32 activates NF-kappaB through IkappaBalpha degradation in inflammatory and genotoxic stress. Int J Biochem Cell Biol. 2017;92:134-40
158. Sahu SK, Tiwari N, Pataskar A, Zhuang Y, Borisova M, Diken M. et al. FBXO32 promotes microenvironment underlying epithelial-mesenchymal transition via CtBP1 during tumour metastasis and brain development. Nat Commun. 2017;8:1523
159. Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. P Natl Acad Sci USA. 2001;98:14440-5
160. Kogure N, Yokobori T, Ogata K, Altan B, Mochiki E, Ohno T. et al. Low Expression of FBXO45 Is Associated with Gastric Cancer Progression and Poor Prognosis. Anticancer Res. 2017;37:191-6
161. Chen X, Sahasrabuddhe AA, Szankasi P, Chung F, Basrur V, Rangnekar VM. et al. Fbxo45-mediated degradation of the tumor-suppressor Par-4 regulates cancer cell survival. Cell Death Differ. 2014;21:1535-45
162. Peschiaroli A, Scialpi F, Bernassola F, Pagano M, Melino G. The F-box protein FBXO45 promotes the proteasome-dependent degradation of p73. Oncogene. 2009;28:3157-66
163. Chung FZ, Sahasrabuddhe AA, Ma K, Chen X, Basrur V, Lim MS. et al. Fbxo45 inhibits calcium-sensitive proteolysis of N-cadherin and promotes neuronal differentiation. J Biol Chem. 2014;289:28448-59
164. Watanabe K, Yumimoto K, Nakayama KI. FBXO21 mediates the ubiquitylation and proteasomal degradation of EID1. Genes Cells. 2015;20:667-74
165. Spaich S, Will RD, Just S, Spaich S, Kuhn C, Frank D. et al. F-Box and Leucine-Rich Repeat Protein 22 Is a Cardiac-Enriched F-Box Protein That Regulates Sarcomeric Protein Turnover and Is Essential for Maintenance of Contractile Function In Vivo. Circ Res. 2012;111:1504 -+
166. Meng X, Liu X, Guo X, Jiang S, Chen T, Hu Z. et al. FBXO38 mediates PD-1 ubiquitination and regulates anti-tumour immunity of T cells. Nature. 2018;564:130-5
167. Clement E, Inuzuka H, Nihira NT, Wei W, Toker A. Skp2-dependent reactivation of AKT drives resistance to PI3K inhibitors. Sci Signal. 2018:11
168. Johnson NB, Deck KM, Nizzi CP, Eisenstein RS. A synergistic role of IRP1 and FBXL5 proteins in coordinating iron metabolism during cell proliferation. J Biol Chem. 2017;292:15976-89
169. Fell VL, Schild-Poulter C. The Ku heterodimer: function in DNA repair and beyond. Mutat Res Rev Mutat Res. 2015;763:15-29
170. Chiorazzi M, Rui L, Yang Y, Ceribelli M, Tishbi N, Maurer CW. et al. Related F-box proteins control cell death in Caenorhabditis elegans and human lymphoma. Proc Natl Acad Sci U S A. 2013;110:3943-8
171. Jackson SJ, Zhang Z, Feng D, Flagg M, O'Loughlin E, Wang D. et al. Rapid and widespread suppression of self-renewal by microRNA-203 during epidermal differentiation. Development. 2013;140:1882-91
172. Guo X, Zhu Y, Hong X, Zhang M, Qiu X, Wang Z. et al. miR-181d and c-myc-mediated inhibition of CRY2 and FBXL3 reprograms metabolism in colorectal cancer. Cell Death Dis. 2017;8:e2958
173. Wang D, Han X, Li C, Bai W. FBXL3 is regulated by miRNA-4735-3p and suppresses cell proliferation and migration in non-small cell lung cancer. Pathol Res Pract. 2019;215:358-65
174. He ZJ, Li W, Chen H, Wen J, Gao YF, Liu YJ. miR-1306-3p targets FBXL5 to promote metastasis of hepatocellular carcinoma through suppressing snail degradation. Biochem Biophys Res Commun. 2018;504:820-6
175. Gong J, Cui Z, Li L, Ma Q, Wang Q, Gao Y. et al. MicroRNA-25 promotes gastric cancer proliferation, invasion, and migration by directly targeting F-box and WD-40 Domain Protein 7, FBXW7. Tumour Biol. 2015;36:7831-40
176. Zhou C, Shen L, Mao L, Wang B, Li Y, Yu H. miR-92a is upregulated in cervical cancer and promotes cell proliferation and invasion by targeting FBXW7. Biochem Biophys Res Commun. 2015;458:63-9
177. Mavrakis KJ, Van Der Meulen J, Wolfe AL, Liu X, Mets E, Taghon T. et al. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL). Nat Genet. 2011;43:673-8
178. Kurashige J, Watanabe M, Iwatsuki M, Kinoshita K, Saito S, Hiyoshi Y. et al. Overexpression of microRNA-223 regulates the ubiquitin ligase FBXW7 in oesophageal squamous cell carcinoma. Br J Cancer. 2012;106:182-8
179. Chen Y, Wei H, Liu Y, Zheng S. Promotional effect of microRNA-194 on breast cancer cells via targeting F-box/WD repeat-containing protein 7. Oncol Lett. 2018;15:4439-44
180. Yang CH, Pfeffer SR, Sims M, Yue J, Wang Y, Linga VG. et al. The oncogenic microRNA-21 inhibits the tumor suppressive activity of FBXO11 to promote tumorigenesis. J Biol Chem. 2015;290:6037-46
181. Pfeffer SR, Yang CH, Pfeffer LM. The Role of miR-21 in Cancer. Drug Dev Res. 2015;76:270-7
182. Shi D, Tan Z, Lu R, Yang W, Zhang Y. MicroRNA-218 inhibits the proliferation of human choriocarcinoma JEG-3 cell line by targeting Fbxw8. Biochem Biophys Res Commun. 2014;450:1241-6
183. Zhang X, Kong Y, Xu X, Xing H, Zhang Y, Han F. et al. F-box protein FBXO31 is down-regulated in gastric cancer and negatively regulated by miR-17 and miR-20a. Oncotarget. 2014;5:6178-90
184. Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D. et al. Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. 2008;111:4690-9
185. Chan CH, Morrow JK, Li CF, Gao Y, Jin G, Moten A. et al. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell. 2013;154:556-68
186. Rico-Bautista E, Wolf DA. Skipping cancer: small molecule inhibitors of SKP2-mediated p27 degradation. Chem Biol. 2012;19:1497-8
187. Wu L, Grigoryan AV, Li Y, Hao B, Pagano M, Cardozo TJ. Specific small molecule inhibitors of Skp2-mediated p27 degradation. Chem Biol. 2012;19:1515-24
188. Huang HC, Way TD, Lin CL, Lin JK. EGCG stabilizes p27kip1 in E2-stimulated MCF-7 cells through down-regulation of the Skp2 protein. Endocrinology. 2008;149:5972-83
189. Huang HC, Lin CL, Lin JK. 1,2,3,4,6-penta-O-galloyl-beta-D-glucose, quercetin, curcumin and lycopene induce cell-cycle arrest in MDA-MB-231 and BT474 cells through downregulation of Skp2 protein. J Agric Food Chem. 2011;59:6765-75
190. Yang ES, Burnstein KL. Vitamin D inhibits G1 to S progression in LNCaP prostate cancer cells through p27Kip1 stabilization and Cdk2 mislocalization to the cytoplasm. J Biol Chem. 2003;278:46862-8
191. Nakajima H, Fujiwara H, Furuichi Y, Tanaka K, Shimbara N. A novel small-molecule inhibitor of NF-kappaB signaling. Biochem Biophys Res Commun. 2008;368:1007-13
192. Blees JS, Bokesch HR, Rubsamen D, Schulz K, Milke L, Bajer MM. et al. Erioflorin stabilizes the tumor suppressor Pdcd4 by inhibiting its interaction with the E3-ligase beta-TrCP1. PLoS One. 2012;7:e46567
193. Wei S, Lin LF, Yang CC, Wang YC, Chang GD, Chen H. et al. Thiazolidinediones modulate the expression of beta-catenin and other cell-cycle regulatory proteins by targeting the F-box proteins of Skp1-Cul1-F-box protein E3 ubiquitin ligase independently of peroxisome proliferator-activated receptor gamma. Mol Pharmacol. 2007;72:725-33
194. Gao J, Azmi AS, Aboukameel A, Kauffman M, Shacham S, Abou-Samra AB. et al. Nuclear retention of Fbw7 by specific inhibitors of nuclear export leads to Notch1 degradation in pancreatic cancer. Oncotarget. 2014;5:3444-54
195. Huang HL, Weng HY, Wang LQ, Yu CH, Huang QJ, Zhao PP. et al. Triggering Fbw7-mediated proteasomal degradation of c-Myc by oridonin induces cell growth inhibition and apoptosis. Mol Cancer Ther. 2012;11:1155-65
196. Hirota T, Lee JW, St John PC, Sawa M, Iwaisako K, Noguchi T. et al. Identification of small molecule activators of cryptochrome. Science. 2012;337:1094-7
197. Pao KC, Wood NT, Knebel A, Rafie K, Stanley M, Mabbitt PD. et al. Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity. Nature. 2018;556:381-5
198. Ungermannova D, Lee J, Zhang G, Dallmann HG, McHenry CS, Liu X. High-throughput screening AlphaScreen assay for identification of small-molecule inhibitors of ubiquitin E3 ligase SCFSkp2-Cks1. J Biomol Screen. 2013;18:910-20
199. Ma J, Cheng L, Liu H, Zhang J, Shi Y, Zeng F. et al. Genistein down-regulates miR-223 expression in pancreatic cancer cells. Curr Drug Targets. 2013;14:1150-6
200. Nangle S, Xing W, Zheng N. Crystal structure of mammalian cryptochrome in complex with a small molecule competitor of its ubiquitin ligase. Cell Res. 2013;23:1417-9
Author contact
![]() Corresponding authors: Hai-long Piao, hpiaoac.cn; Hongxu Liu, hxliuedu.cn
Corresponding authors: Hai-long Piao, hpiaoac.cn; Hongxu Liu, hxliuedu.cn
 Global reach, higher impact
Global reach, higher impact