13.3
Impact Factor
Theranostics 2021; 11(6):2755-2769. doi:10.7150/thno.56174 This issue Cite
Review
Circular RNA in pancreatic cancer: a novel avenue for the roles of diagnosis and treatment
1. Department of Pancreatic Surgery, Fudan University Shanghai Cancer Center, Shanghai, 200032, China.
2. Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, 200032, China.
3. Pancreatic Cancer Institute, Fudan University, Shanghai, 200032, China.
4. Shanghai Pancreatic Cancer Institute, Shanghai, 200032, China.
#These authors contributed equally to this work.
Received 2020-11-19; Accepted 2020-12-12; Published 2021-1-1
Abstract
Pancreatic cancer (PC), an important cause of cancer-related deaths worldwide, is one of the most malignant cancers characterized by a dismal prognosis. Circular RNAs (circRNAs), a class of endogenous ncRNAs with unique covalently closed loops, have attracted great attention in regard to various diseases, especially cancers. Compelling studies have suggested that circRNAs are aberrantly expressed in different cancer tissues and cell types, including PC. More specifically, circRNAs can modify the proliferation, progression, tumorigenesis and chemosensitivity of PC, and some circRNAs could serve as biomarkers for diagnosis and prognosis. Herein, we summarize what is currently known to be related to the biogenesis, functions and potential roles of human circRNAs in PC and their application prospects for PC clinical treatments.
Keywords: Circular RNA, pancreatic cancer, sponge function, biomarker, treatment
Introduction
As one of the most malignant tumors with a dismal prognosis, pancreatic cancer (PC), ranks fourth in cancer mortality in the United States, and accounts for 8% of all estimated cancer-related deaths [1, 2, 3]. Even worse, it is emerging as the second primary cause of cancer-related deaths by 2030 [1, 2, 3]. PC is characterized by rapid progression, high metastasis and recurrence, and fast development of drug resistance. Despite numerous efforts to improve the efficacy of surgery and chemoradiotherapy over the last several years, there are still few reliable biomarkers or notably better therapeutic strategies for daily clinical practice in PC [4]. Consequently, it is absolutely essential to determine the potential mechanisms of cancer initiation and development for the diagnosis and treatment of pancreatic cancer.
CircRNAs are a group of endogenous noncoding RNAs that were originally misinterpreted as the products of splicing errors [5]. Currently, it has been clarified that circRNAs are generated from introns or exons through back-splicing and possess a covalently closed circular loop without 5' end caps and 3' polyadenylated tails [6]. Most of them are widespread, abundant, conserved, and stable and have tissue/developmental-stage-specific characteristics in eukaryotes [7, 8]. CircRNAs are more resistant to exonuclease-mediated degradation and regular mechanisms of linear RNA decay than linear RNAs because of their unique single-stranded closed circle loop, and are not sensitive to regular mechanisms of linear RNA decay because of their unique single-stranded closed circular loop, which contributes to their abundance in tissues, serum, and urine and makes them promising biomarkers for aging and human cancers [9, 10]. Currently, circRNAs have garnered much scientific attention because of their aberrant expression and pivotal impacts on the regulation and pathogenesis of different diseases, particularly human tumors [11]. Increasing evidence indicates that circRNAs have considerable functional potential to alter proliferation, invasion, apoptosis, metastasis, angiogenesis, and the response to chemotherapy, indicating that circRNAs may function as novel potential therapeutic targets for the treatment of various tumors [12, 13, 14, 15, 16, 17], including pancreatic cancer [18, 19, 20, 21]. For instance, a landmark discovery confirmed the exonic circRNA ciRS-7 (CDR1as), which contains over seventy binding sites for miR-7 [9], as the first functional circRNA. CiRS-7 plays an oncogenic role by binding with miR-7 and elevating the expression of its downstream oncogenes in pancreatic cancer [18].
In this review, we summarize the current understanding of the biogenesis and functions of circRNAs and their roles in pancreatic cancer and discuss the application prospects of circRNAs for pancreatic cancer clinical treatments.
Classification and biogenesis of circRNAs
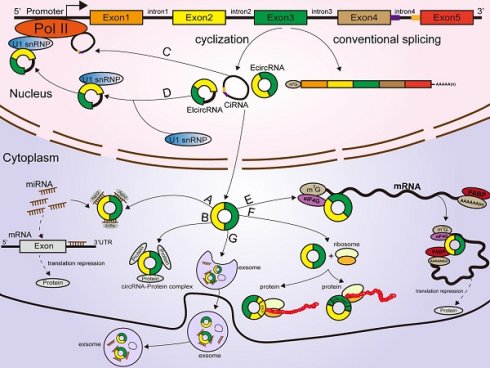
According to their origin and genomic organization, circRNAs are classified into 3 groups: exonic circRNAs (EcircRNAs, produced from exons), exon-intron circRNAs (EIcircRNAs, produced from exons and introns), and intronic circRNAs (ciRNAs, produced from introns) [22, 23, 24]. Unlike the mechanisms of linear RNA generation (Figure 1A), the specific formation mechanism of circRNAs is not yet fully clarified. As the most abundant circRNAs, EcircRNAs are derived from a specific splicing mechanism known as exon back-splicing. An upstream 5' splice donor attacks the downstream 3' splice acceptor, leading to the formation of a covalently closed circRNA [25]. When the introns located in the 5' donor and the 3' acceptor on the precursor mRNA (premRNA)are retained [25], the resultant circRNAs are called EIcircRNAs and are composed of both exons and introns. Existing studies have proposed three biogenesis models of the synthesis of circRNAs: intron-pairing circularization, lariat- induced circularization, and RNA-binding protein (RBP)-induced circularization [26]. First, for intron-pairing circularization, the complementary sequences from the introns in premRNAs generally facilitate the circularization of EcircRNAs [25]. By means of base pairing between intronic repeats, the 3' splice acceptor site on the exon combines with the 5' splice donor site, leading to spatial proximity of the downstream and upstream splice exons [27], which facilitates exon circularization to form EcircRNAs (Figure 1B). A previous study demonstrated that very short introns with lengths of 30-40 nucleotides comprising repeat complementary interdependent sequences located in upstream and downstream introns (such as Alu repeats) could be used to interfere with exon circularization after extensive mutagenesis of expression plasmids in human cells [25, 28]. In lariat-driven circularization, the splice sites of skipped exons are connected to generate a lariat during the transcription of premRNA [25, 29, 30, 31]. Then, the intronic sequences are removed from the lariats, and EcircRNAs are formed (Figure 1C). In addition, RBPs are considered critical regulators of EcircRNA biogenesis [32, 33]. They can specifically bind to the introns near splice sites and bring flanking introns closer together to facilitate the production of circRNAs (Figure 1D) [34]. For example, the Drosophila Muscleblind (Mbl)-binding protein, derived from its second exon, can recognize and bind to particular motifs that are located in flanking introns on its own premRNA to increase circMbl production [35, 36]. The flanking introns of circMbl contain highly conserved Mbl-binding elements [37], which can be recognized and precisely bound by the Mbl protein, thereby influencing the biogenesis of circMbl [35, 36]. Quaking (QKI), an alternative splicing factor [38], can induce the production of hundreds of circRNAs by binding to recognition elements within introns and forming dimers, which can promote the efficiency of back-splicing during human epithelial-mesenchymal transition (EMT) [33]. A study from Errichelli L et al. illustrated that the RBP fused in sarcoma (FUS) participated in circRNA biosynthetic processes by associating with the introns near splice junctions [39, 40]. Under some circumstances, the end of the 2'-OH group of the intron and the 5' splice site form a branchpoint 2'-5' concatenation, after releasing the 3' exon [25].The ciRNAs are formed. Their structure depends on conserved elements, including seven-nucleotide GU abundant motifs near the 5' splice site and eleven-nucleotide C abundant motifs near the branchpoint site [41, 42, 43]. These motifs can protect ciRNAs from debranching and degradation (Figure 1E) [22, 42].
Online databases related to circRNAs
To advance the research of the multiple applications of circRNAs, many online databases related to circRNAs have been established, such as CircRNADb [44], TransCirc [45], CircBase [46], Circ2Traits [47], CIRCpedia v2 [48], CircInteractome [49], CircNet [50], MiOncoCirc [51], TSCD (tissue-specific circRNA) [52], cancer-specific circRNA (CSCD) [53], exoRBase [54], and circR2Disease [55] (Table 1). Combined with the advancement of biotechnology [42], these circRNA-related databases could help discover meaningful circRNAs, forecast the interactions between circRNAs and target molecules and translation potency, and investigate their functions in the processes of physiological and pathological development in different diseases.
Functions of CircRNAs
CircRNAs act as sponges of miRNAs
Mechanistically, most identified circRNAs are mainly localized in the cytoplasm of the cell [56], indicative of their roles in posttranscriptional regulation [57]. MiRNAs are a group of ubiquitous, conserved small noncoding RNAs with lengths of 19-25 nucleotides that can affect the expression of genes and a broad range of biogenesis functions in tumors [58]. The ceRNA hypothesis indicates that circRNAs harbor MREs that bind miRNAs to reversely regulate the activity of the miRNAs [59], thus attenuating the inhibitory effect on their target molecules. Mounting evidence has confirmed that some circRNAs can repress miRNA function and modulate target gene expression to play a tumor suppressor or oncogenic role by acting as miRNA sponges in different cancers [15, 60, 61, 62]. Furthermore, some circRNAs can also target multiple miRNAs and carry out opposite effects in different diseases. For instance, circ-ITCH suppresses the proliferation and progression of bladder cancer by absorbing miR-17 and miR-224 [63]. Moreover, circ-ITCH can also target miR-7 to elevate the level of EGFR and promote the migration and invasion of osteosarcoma [64]. Recently, circBFAR was found to function for miR-34b-5p to promote the proliferation and metastasis of pancreatic ductal adenocarcinoma (PDAC) [65]. In contrast, circNFIB1 directly sponge miR-486-5p to inhibit lymphangiogenesis and lymphatic metastasis of pancreatic cancer [66]. Overall, acting as miRNA sponges may be a general function of circRNAs in tumors [67] (Figure 2A).
Classification and biogenesis of circRNAs (A) Canonical splicing related to linear RNA generation. (B) Intron-pairing circularization: the 3' splicing receptor site in the exon combines with the 5' splicing donor site, with the assistance of base pairing between intronic repeats, resulting in spatial proximity of the donor and acceptor splice exons. (C) Lariat-induced circularization: the 5' splice site attacks the 3' splice site, and skipped exons 2 and 3 are connected by producing a lariat. (D) RBP-induced circularization: RBPs bind to introns near splice sites, which can bridge flanking introns together, facilitating the production of circRNAs. (E) CiRNA formation: ciRNAs are produced from lariat introns. The GU-rich motif close to the 5' splice site (purple box) and the C-rich motif close to the branchpoint site (yellow box) prevent the intron from debranching and form a stable circRNA.
Online databases related to circRNAs
| Database | Website | Description | References |
|---|---|---|---|
| CircBase | http://www.circbase.org/ | A comprehensive database that provides published circRNAs in different species (human, mouse, C. elegans, and Latimeria organisms) and identification of circRNAs. | [46] |
| Circ2Traits | http://gyanxet-beta.com/circdb/ | A comprehensive knowledgebase of the potential association of circRNAs with diseases and traits in humans. | [47] |
| CIRCpedia v2 | http://www.picb.ac.cn/rnomics/circpedia | A database for comprehensive circRNA annotations and expression comparisons from over 180 RNA-seq datasets across six different species. | [48] |
| CircInteractome | http://circinteractome.nia.nih.gov | A database to explore the possible interactions of circRNAs with miRNAs and RBPs. | [49] |
| CircNet | http://circnet.mbc.nctu.edu.tw | A useful tool to investigate the regulatory relationship between circRNAs, miRNAs and genes. | [50] |
| CircRNADb | http://reprod.njmu.edu.cn/circrnadb | A database to provide detailed information on circRNAs including genome sequence, exon splicing, ORF, IRES, and references to predict the translation potential of certain circRNAs. | [44] |
| TransCirc | https://www.biosino.org/transcirc/ | A database used to predict the potential of all circRNAs to encode functional peptides. | [45] |
| MiOncoCirc | mioncocirc.github.io | A database compiled from clinical cancer samples that provides the expression of a certain circRNA in different cancer clinical samples. | [51] |
| TSCD | http://gb.whu.edu.cn/TSCD | A database used to characterize the features of human and mouse tissue-specific circRNAs. | [52] |
| CSCD | http://gb.whu.edu.cn/CSCD | A cancer-specific circRNA database contributes to the study of the function and regulation of cancer-related circRNAs. | [53] |
| exoRBase | http://www.exoRBase.org | A database containing more than 58000 circRNAs from 87 human blood exosomal RNA-seq datasets that provides circRNA annotation and expression levels and can assist researchers to discover new exosomal biomarkers for human diseases. | [54] |
| CircR2Disease | http://cgga.org.cn:9091/circRNADisease/ | CircR2Disease is a comprehensive database for circRNAs dysregulated in different diseases and contains indicators showing that circRNAs participate in gene posttranscriptional regulation. | [55] |
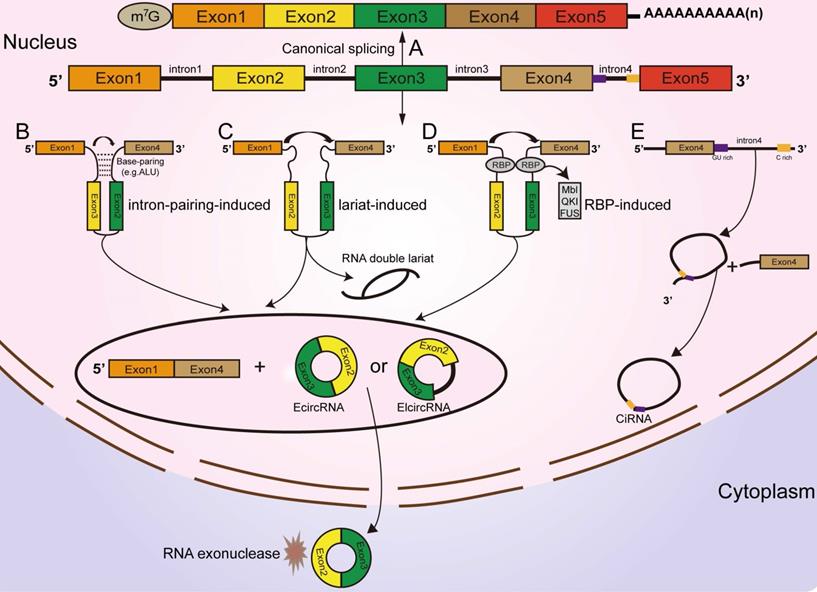
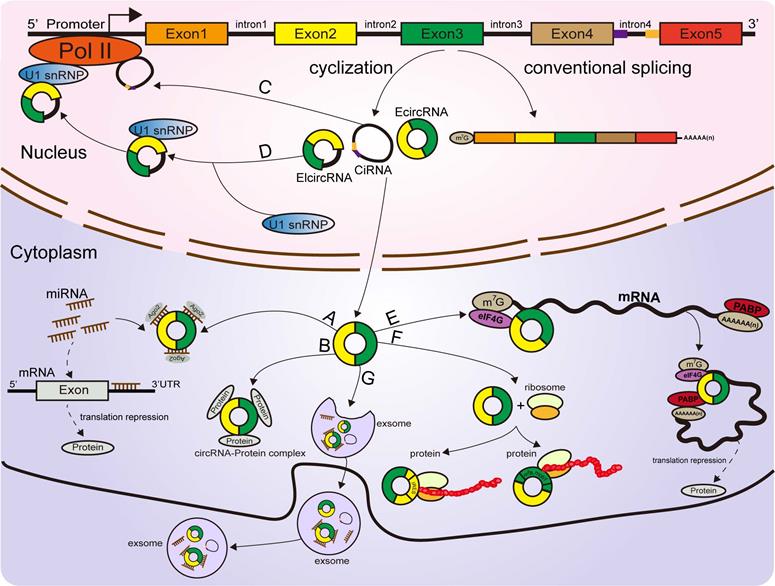
Functions of circRNAs. (A) CircRNAs function as miRNA sponges to influence the expression of downstream gene. (B, C, D) CircRNAs regulate parental gene expression at the transcriptional and translational levels. (E) CircRNAs bind with proteins to establish circRNA-protein complexes and alter the functions of some proteins. (F) CircRNAs encode proteins based on IRES-driven and m6A-driven models. (G) Some circRNAs carried by exosomes are derived from cancer cells.
CircRNAs bind with proteins
Several circRNAs were revealed to bind with proteins to promote their functions. A ciRNA (circAGO2), produced from the premRNA of AGO2, physically interacts with the HuR protein and facilitates its transfer from the nucleus into the cytoplasm, leading to a decrease in AGO2 binding and suppression of the function of AGO2-miRNA complexes [68]. Circ-Amotl1, derived from the Amotl1 gene, is predominantly expressed in the nucleus, where it colocalizes and interacts with the c-Myc protein. Upregulation of circ-Amotl1 in the nucleus facilitated translocation of the c-Myc protein into the nucleus and increased its binding affinity for some promoters [69], but did not alter the level of total c-Myc in breast cancer cells [69]. CircECE1 binds to c-Myc and inhibits the degradation process mediated by SPOP, thereby reducing the efficiency of c-Myc modification and degradation by ubiquitination and promoting the stabilization of the c-Myc protein in osteosarcoma [70]. Typically, circFOXK2 interacts with YBX1 and hnRNPK to increase the expression of the oncogenic proteins NUF2 and PDXK in PDAC [71].
Conversely, some circRNAs were confirmed to bind with proteins to suppress their functions [67]. Recently, a study revealed that circSTAG1 binds with the well-known m6A demethylase ALKBH5, resulting in elevated m6A methylation of the mRNA of FAAH and its degradation in astrocytes [72]. Circ-Foxo3, P21 and CDK2 form the ternary complex circ-Foxo3-P21-CDK2, which can suppress the progression of the cell cycle by inhibiting CDK2 function [73, 74, 75]. All these findings suggest that circRNAs can act as decoys or scaffolds to affect the expression or function of some proteins [76] (Figure 2B).
CircRNAs affect parental gene expression
Emerging reports have shown that some circRNAs are retained in the cell nucleus to establish a large quantity of posttranscriptional regulatory factors that affect their parental gene expression [9]. Zhang et al discovered that ci-ANKRD52 specifically interacts with the elongation RNA Pol II complex and directly promotes ANKRD52 transcription (Figure 2C). Moreover, ci-ANKRD52 silencing reduced the transcription rate of ANKRD52 [22]. Another novel study discovered that circ-EIF3J and circ-PAIP2, identified as EIcircRNAs, can combine with RNA Pol II and facilitate their parental gene transcription in interaction with U1 snRNP in the nucleus of 293T and HeLa cells [77] (Figure 2D). These findings suggest that circRNAs affect parental gene expression at the transcriptional level. Moreover, circRNAs can regulate gene expression at the translational level. For example, a complex is formed by the direct interaction of circYap (generated from the Yap gene), Yap mRNA, eIF4G and PABP, in which eIF4G and PABP are translation initiation-associated proteins. Overexpression of circYap in this complex inhibited the interaction of PABP with eIF4G and consequently suppressed the translation initiation of Yap mRNA (Figure 2E) [78]. CircPABPN1 (hsa_circ_0031288), produced from the PABPN1 gene, suppressed HuR binding to PABPN1 mRNA and subsequently decreased the translation of PABPN1 [79]. All these discoveries indicated that circRNAs affect their parental gene expression at both the transcriptional and translational levels.
CircRNAs encode proteins
Similar to most noncoding RNAs, circRNAs were originally considered untranslatable due to the lack of distinct ORFs. Nonetheless, emerging evidence corroborated that these so-called “noncoding RNAs” are translatable and can directly encode functional proteins (Figure 2F) [80, 81, 82, 83, 84, 85]. Some EcircRNAs containing an IRES in the ORF have the ability to encode functional proteins or peptides in the cytoplasm [25, 44, 49, 86]. For example, circFNDC3B translated a 218-amino acid novel protein (circFNDC3B-218aa) driven by an IRES [87]. The junction-spanning ORF of circ-FBXW7 driven by an IRES encodes a novel 21-kDa protein, termed FBXW7-185aa, in glioblastoma [84, 88]. Circβ-catenin has a putative IRES sequence and encodes a novel 370 aa β-catenin isoform that can stabilize β-catenin by inhibiting β-catenin phosphorylation and degradation induced by GSK3β in hepatocellular carcinoma [89, 90]. Yang and coworkers revealed that in addition to IRES-mediated translation, a circRNA containing consensus N6-methyladenosine (m6A) motifs is translated [81]. Moreover, this m6A-driven translation from the circRNA was decreased by FTO (m6A demethylase) and enhanced by METTL3/14 (adenosine methyltransferase) [81, 91]. To date, circRNAs have been confirmed to be directly translated into proteins. However, more information is required to clarify the mechanism of circRNA translation to better understand other aspects of gene regulation.
CircRNAs and pancreatic cancer
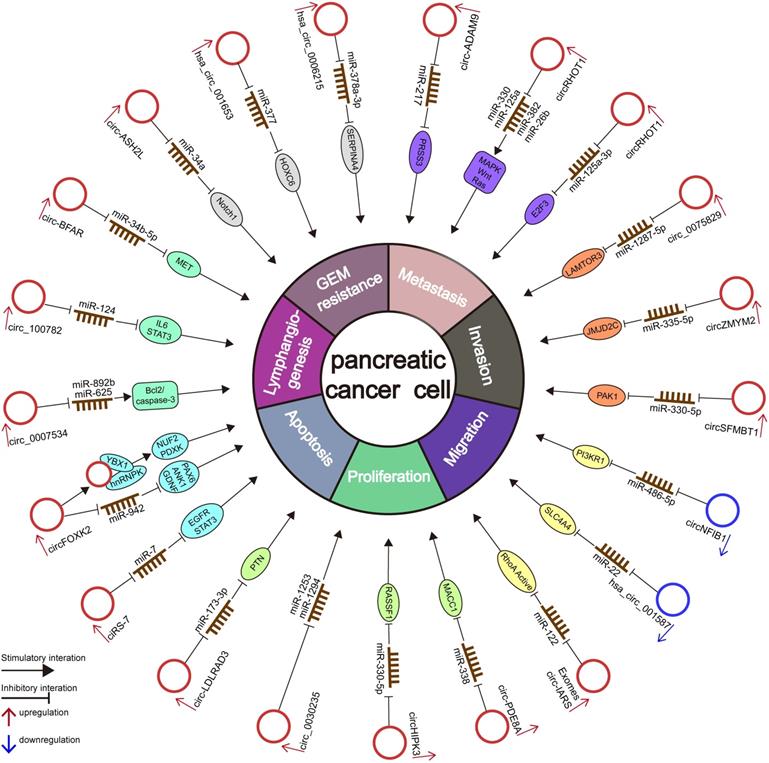
Numerous circRNAs are abnormally expressed in several cancer types and exhibit a high degree of tissue- or disease-specificity differences [92], indicating that circRNAs may be used for diagnostic and therapeutic applications [51, 93]. Although considerable studies have confirmed that circRNAs play essential roles in the occurrence and progression of PC, the study of the potential correlation and potential mechanism between circRNAs and PC is still in its early stage [51, 93]. Here, we summarize the current circRNA-related studies in PC and list the deregulated circRNAs in Table 2 and Figure 3.
Deregulated circRNAs in pancreatic cancer
| CircRNAs | Expression change | Roles in PC | Putative function | Possible mechanism | Relationships with clinical features | Clinical association | Reference |
|---|---|---|---|---|---|---|---|
| circPDAC | Up | Unknown | Unknown | Unknown | LNM, TNM stage | A noninvasive biomarker | [96] |
| ciRS-7 | Up | Oncogene | Promotes proliferation, invasion, metastasis | miRNA sponge (ciRS-7/miR-7 EGFR/STAT3 pathway) | LNM, tumor venous invasion | Unknown | [18] |
| circFOXK2 | Up | Oncogene | Promotes growth, migration, invasion, liver metastasis | miRNA sponge (circFOXK2/miR-942/(ANK1, GDNF, PAX6); interaction with YBX1 and hnRNPK | Unknown | Unknown | [71] |
| hsa_circ_0007534 | Up | Oncogene | Promotes proliferation, migration, invasion. Inhibits apoptosis. | miRNA sponges (circ_000753/miR-625 and miR-892b | Tumor stage, lymphatic invasion | OS | [100] |
| hsa_circ_100782 | Up | Oncogene | Promotes proliferation | miRNA sponge (hsa_circ_100782/miR-124/IL6-STAT3 pathway) | Unknown | Unknown | [101] |
| circ-BFAR | Up | Oncogene | Promotes proliferation, migration, invasion, metastasis | miRNA sponges (circ-BFAR/miR-34b-5p/MET/Akt axis) | TNM stage | OS DFS | [65] |
| circ-ASH2L | Up | Oncogene | Promotes invasion, proliferation, angiogenesis | miRNA sponge (circ-ASH2L/miR-34a/Notch1 axis) | Lymphatic invasion, TNM stage | OS | [103] |
| hsa_circ_001653 | Up | Oncogene | Promotes proliferation, invasion, tumorigenesis angiogenesis. Inhibits apoptosis. | miRNA sponge (hsa_circ_001653/miR-377/HOXC6 axis) | Unknown | OS | [104] |
| hsa_circ_0006215 | Up | Oncogene | Promotes growth, migration. Inhibits apoptosis. | miRNA sponge (hsa_circ_0006215/miR-378a 3p/SERPINA4 axis) | Unknown | Unknown | [106] |
| circ-ADAM9 | Up | Oncogene | Promotes proliferation, migration and invasion | miRNA sponge (circADAM9/miR-217/PRSS3 axis) | Lymphatic metastasis, TNM stage | OS | [112] |
| circRHOT1 | Up | Oncogene | Promotes proliferation, invasion, migration | miRNA sponge (circRHOT1/miR-26b, miR-125a, miR-330 and miR-382) | Unknown | Unknown | [107] |
| circRHOT1 | Up | Oncogene | Promotes proliferation, migration, invasion. Inhibits apoptosis | miRNA sponge (circRHOT1/miR-125a-3p/E2F3 axis) | Lymphatic metastasis | Unknown | [108] |
| circ_0075829 | Up | Oncogene | Promotes proliferation, migration and invasion, tumorigenicity and metastasis | miRNA sponge (circ_0075829/miR-1287-5p/LAMTOR3 axis) | Tumor size, lymphatic metastasis | Unknown | [109] |
| circZMYM2 | Up | Oncogene | Promotes proliferation and invasion. Inhibits apoptosis | miRNA sponge (circZMYM2/miR-335-5p/JMJD2C axis) | LNM | Unknown | [110] |
| circSFMBT1 | Up | Oncogene | Promotes proliferation, migration, invasion, EMT, metastasis. Inhibits apoptosis. | miRNA sponge (circSFMBT1/miR-330-5p/PAK1 axis) | Unknown | Unknown | [113] |
| circNFIB1 | Down | Tumor Suppressor | Inhibits lymphangiogenesis, LNM, tumorigenesis | miRNA sponge (circNFIB1/miR-486-5p/PIK3R1/VEGF-C axis) | LNM | OS DFS | [66] |
| hsa_circ_001587 | Down | Tumor Suppressor | Inhibits proliferation, migration, invasion, angiogenesis, tumorigenesis | miRNA sponge (hsa_circ_001587/miR-22/SLC4A4 axis) | Unknown | Unknown | [116] |
| hsa_circ_0001649 | Down | Tumor Suppressor, Biomarker | Inhibits proliferation, colony-forming ability. Promotes apoptosis. | Unknown | TNM stage, differentiation grade | OS | [117] |
| circ-LDLRAD3 | Up | Oncogene | Promotes proliferation, migration, invasion | miRNA sponge (circ-LDLRAD3/miR-137-3p/PTN axis) | Unknown | OS | [124] |
| circ-LDLRAD3 | Up | Biomarker | Unknown | Unknown | Tissue samples (venous invasion, lymphatic invasion); plasma samples (CA19-9 level, N stage, venous invasion, lymphatic metastasis) | Combination with CA19-9 increased the diagnostic value in PC | [122] |
| hsa_circ_0030235 | Up | Oncogene | Promotes growth, migration, invasion. Inhibits apoptosis. | miRNA sponge (circ_0030235/miR-1253 and miR-1294) | Lymphatic invasion, TNM stage | OS | [121] |
| circ-IARS | Up | Oncogene | Promotes metastasis | miRNA sponge (circ-IARS/miR-122/RhoA/F-actin and ZO-1 axis) | Differentiation grade, vascular invasion, liver metastasis, TNM stage | OS; circ-IARS in exosomes as a marker for the early diagnosis and prognostic prediction in PC | [129] |
| circ-PDE8A | Up | Oncogene, Biomarker | Promotes invasion, growth, liver metastasis | miRNA sponge (circ-PDE8A/miR-338/MACC1/MET axis) | Lymphatic invasion, T factor, TNM stage | OS (both in PC patient tissues and plasma exosomes) | [130] |
| circRNA_101672, circRNA_004077 | Up | Unknown | Enhances GEM resistance | Unknown | Unknown | Unknown | [135] |
| chr14:101402109-101464448C, chr4:52729603-52780244C | Up | Unknown | Enhances GEM resistance | Possibly related to the ErbB and VEGF pathways | Unknown | Unknown | [13] |
| circHIPK3 | Up | Oncogene, Biomarker | Promotes proliferation, invasion, migration, EMT. Inhibits apoptosis. Enhances GEM resistance | miRNA sponge (circHIPK3/miR-330-5p/RASSF1 axis) | Unknown | OS | [132] |
The diagram illustrates the mechanism underlying circRNAs in the regulation of cellular properties and miRNA-related gene regulation networks in PC.
Profiles of circRNA expression in PC
Extensive reports have confirmed that many circRNAs are deregulated in pancreatic ductal adenocarcinoma (PDAC). By analyzing the data from the GEO (No: GSE79634) database, 256 differentially expressed circRNAs (DECs) were identified, among which 115 and 141 were upregulated and downregulated, respectively, in PDAC tissues in comparison with matched normal tissues [94]. Utilizing Arraystar Human CircRNA Array Analysis, Guo et al. identified 289 DECs between 20 PC tissues and corresponding paracancerous tissues, of which 128 were upregulated and 161 were downregulated [95]. Subsequently, the qRT-PCR results were consistent with the microarray data [95]. A recent study identified more than 40,000 unknown circRNAs that have not been previously described in the circBase database via RNA sequencing analyses [96]. More importantly, the authors also discovered a novel circRNA, “circPDAC”, that was produced from the 5' end of exon 3 of the noncoding RNA LOC107987178 and the 3' end of exon 3 of the adjacent noncoding RNA LOC100507377 [96]. This circRNA was significantly overexpressed in PC tissues and cells, whereas it was barely detected in normal pancreas tissues [96]. Similarly, Li and colleagues identified 351 DECs between six paired PDAC tissues and normal tissues by utilizing circRNA microarray analysis, of which 209 and 142 circRNAs were upregulated and downregulated, respectively, in tumor samples [97]. Based on two GEO microarray datasets, Xiao et al identified 289 and 170 DECs in GSE79634 and GSE69362, respectively; analysis using publicly available circRNA and miRNA databases of the top ten DECs revealed that hsa_circ_0007767 and hsa_circ_0092367 play essential roles in PDAC by acting as miRNA sponges [98]. Additionally, Wong et al. performed circRNA sequencing to identify DECs between PANC-1 and SW1990 PC cells and nontumor human pancreatic ductal epithelial (HPDE) cells. Overall, 17,158 circRNAs were identified, of which 84% were EcircRNAs (GEO; No: GSE135731). Among these circRNAs, 83 upregulated and 86 downregulated circRNAs were found in PANC-1 and SW1990 cells compared with HPDE cells [71].
However, only a very small proportion of the DECs in these expression profiles have been verified to regulate carcinogenesis in PDAC. Although numerous efforts have been made to discover more circRNAs in PDAC, the study of PDAC-related circRNAs still faces great challenges.
CircRNAs alter the proliferation and progression of PC
Numerous reports have highlighted that circRNAs alter proliferation, migration, invasion, metastasis, apoptosis, and the cell cycle by acting as oncogenes or tumor suppressors in PC. Multiple mechanisms, including miRNA sponge activity, cancer-related signaling pathway regulation, and interaction with proteins, are related to these functions. Therefore, miRNA sponges are still the leading mechanism related to circRNAs in PC.
Oncogene
A previous study showed upregulated ciRS-7 in PC tissues, which was positively associated with LNM and venous invasion. In general, ciRS-7 enhanced the proliferation and invasion of PC by sponging miR-7 and inhibiting its activity, subsequently activating the EGFR and STAT3 signaling pathways [18]. CircFOXK2 was markedly overexpressed in both PDAC cells and tissues. Silencing circFOXK2 significantly inhibited migration, invasion, liver metastasis and tumor growth in PDAC [71]. CircFOXK2 competitively sponges miR-942 to increase the expression of its target genes ANK1, GDNF, and PAX6 [71, 99]. Notably, circFOXK2 interacted with YBX1 and hnRNPK to enhance the expression of the oncogenic proteins NUF2 and PDXK in PDAC [71]. Moreover, the increase in NUF2 and PDXK expression caused by the overexpression of circFOXK2 was attenuated by knockdown of YBX1 and hnRNPK [71].
Another novel study found that overexpression of circ_0007534 promoted the proliferation, migration, and invasion while inhibiting the apoptosis of PDAC cells by sponging miR‐625 and miR‐892b [19, 100]. Circ_100782 was overexpressed in PC tissues. Circ_100782 knockdown effectively inhibited proliferation through the IL6-STAT3 pathway by directly sponging miR-124 in PC cells [101]. Circ-BFAR is overexpressed in PDAC patients [65], and its knockdown significantly inhibits the proliferation, migration, invasion, tumor growth and metastasis of PDAC cells [65]. Circ-BFAR enhanced the expression of its target gene mesenchymal-epithelial transition factor (MET) by absorbing miR-34b-5p in PC cells. MET is frequently overexpressed and acts as an oncoprotein in PC [102]. Collectively, these findings highlighted the significance of the circ-BFAR/miR-34b-5p/MET axis in the progression of PDAC and demonstrated that circ-BFAR performs an oncogenic function and might function as a possible diagnostic biomarker and therapeutic target in PDAC [65].
Circ-ASH2L was overexpressed in PDAC tissues and cells [103]. Based on bioinformatic analysis and experiments, circ-ASH2L was confirmed to promote invasion, proliferation and angiogenesis by sponging miR-34a to enhance the level of Notch 1 [103]. A recent study found that hsa_circ_001653 was upregulated in PDAC tissues and cells. The knockdown of hsa_circ_001653 inhibited miR-377-targeted HOXC6 and suppressed PDAC cell proliferation, invasion, angiogenesis, and tumorigenesis whereas promoting cell apoptosis [104]. The upregulation of human HOXC6 in PCa is related to tumor progression and serves as an independent prognostic biomarker [105]. Zhu et al. revealed that overexpression of hsa_circ_0006215 enhanced SERPINA4 expression by absorbing miR-378a-3p, therefore initiating and promoting the occurrence and progression of PC [106]. Hsa_circ_0005397 (termed as circRHOT1) knockdown inhibited the cell proliferation, migration and invasion of PDAC cells. Bioinformatic analysis revealed that circRHOT1 can function as a miRNA sponge for miR-26b, miR-125a, miR-330 and miR-382 to affect various cancer-related pathways in PDAC [107]. Interestingly, another study confirmed that circRHOT1 promoted proliferation, apoptosis and invasion by downregulating the expression of miR-125a-3p to increase the expression of its target gene E2F3 in PC [108]. Zhang et al found that the expression of circ_0075829 was significantly overexpressed in PC tissues compared with adjacent noncancerous tissues and was associated with tumor size and lymphatic metastasis [109]. Circ_0075829 significantly promoted the proliferation and metastasis of PC cells by directly sponging miR-1287-5p and elevating the expression of LAMTOR3 [109].
Microarray analysis and qRT-PCR results confirmed that circZMYM2 (hsa_circ_0099999) was markedly upregulated in human PC tissues and cells [110]. The overexpression of circZMYM2 promoted PC cell proliferation and invasion and inhibited apoptosis, while the knockdown of circZMYM2 had opposite effects. Moreover, circZMYM2 knockdown attenuated PC cell tumor formation and growth in vivo [110]. Importantly, circZMYM2 downregulates the expression of miR-335-5p, which is a crucial factor that suppresses PC progression by inhibiting JMJD2C [110]. MJD2/KDM4, which is a member of the JMJ family, is involved in the proliferation and progression of PC and transforms abnormal cells into invasive and metastatic forms by enhancing cell invasive and migratory abilities [111]. Briefly, circZMYM2 knockdown can increase the expression of miR-335-5p to subsequently attenuate JMJD2C and inhibit the progression of PC [110]. Xing et al. detected low levels of miR-217 and high levels of circ-ADAM9 in PC tissues and cells [112]. Of note, the knockdown of circ-ADAM9 dramatically suppressed the cell growth, migration, and invasion of PC in vitro and inhibited tumor growth in vivo [112]. Mechanistically, circ-ADAM9 directly sponges miR-217 to suppress its effect on its target serine protease 3 (PRSS3) and then indirectly activates the ERK/VEGF pathway. PRSS3 is an oncogene that is significantly overexpressed in PC [112]. It was suggested that circ-ADAM9 may be an oncogene influencing cancer growth and progression through the miR-217/PRSS3 axis [112]. Xu et al. discovered that elevated circSFMBT1 (hsa_circ_0066147) enhanced the proliferation, invasion, migration, and EMT process of PC cells and inhibited the apoptosis of PC cells in vitro by regulating the miR-330-5p/PAK1 pathway by sponging miR-330-5p [113]. Moreover, circSFMBT1 knockdown suppressed the growth of tumor and lung metastases in vivo through the miR-330-5p/PAK1 axis [113].
Tumor suppressors
Hsa_circ_0086375, generated from the NFIB1 gene (circNFIB1), was obviously downregulated in PC tissues in comparation with adjacent normal tissues and negatively correlated with LNM in PC patients [66]. CircNFIB1 depletion increased the expression and secretion of VEGF-C in PC cells [66]. VEGF-C is a VEGFR3 ligand that participate in lymphangiogenesis and is considered the upstream regulator of the PI3K/Akt signaling pathway [114, 115]. Furthermore, in vitro experiments showed that conditioned medium from circNFIB1-knockdown PC cells dramatically enhanced HLEC tube formation and migration, whereas conditioned medium from circNFIB1-overexpressing PC cells exerted the opposite effects [66]. In vivo experiments showed that circNFIB1 knockdown markedly enhanced LNM in PC cells [66]. Importantly, circ-NFIB1 directly absorbs miR-486-5p to attenuate the oncogenic function of miR-486-5p to some degree and subsequently upregulates the expression of the miR-486-5p target PI3K p85α, a regulatory subunit of PI3K (PIK3R1) [66]. Circ-NFIB1-induced VEGF-C attenuated the activation of the PI3K/Akt signaling pathway and suppressed the lymphangiogenesis and LN metastasis of PC [66]. Hsa_circ_001587 expression was markedly lower in PDAC cells and tissues. Hsa_circRNA_001587 overexpression inhibited proliferation, angiogenesis, tumorigenesis, migration and invasion abilities by decreasing the expression of MMP-2, MMP-9, MCM2 and VEGF in PC [116]. Mechanistic studies suggested that hsa_circRNA_001587 directly sponges miR-223 to enhance the level of its target gene SLC4A4, a cancer-promoting gene [116]. Jiang et al discovered that hsa_circ_0001649 is also aberrantly downregulated in both PDAC tissues and cells [117]. Moreover, hsa_circ_0001649 overexpression obviously suppressed the proliferation and colony formation abilities and enhanced the apoptosis rate of PDAC cells [117].
CircRNAs function as diagnostic and prognostic biomarkers in pancreatic cancer
Most PC patients present with symptomatic, surgically unresectable disease due to a lack of dependable and valid early diagnostic techniques. Therefore, achieving early detection of PC is important and would result in a significant improvement in overall survival (OS) [4]. Unlike their linear counterparts, circRNAs have a unique stable closed loop structure, which contributes to their stable expression in tissues, saliva, plasma, and exosomes [118]. Additionally, circRNAs may function as special molecular markers in cancers because of their abundance, conservation and specificity in tissues and cells [25, 119, 120].
To date, several investigations have confirmed the roles of circRNAs in different cancers, including PC [107, 121, 122, 123]. As mentioned above, it was found that high expression of circ_0007534 was correlated with poor prognosis in PC patients [104]. Patients with lower expression of circ-ADAM9 had a better OS rate than those with higher expression of circ-ADAM9 (p=0.001) [112]. High expression of circ-ASH2L was positively correlated with tumor malignancy, lymphatic invasion and TNM stage [103].
Yao's study revealed that PC patients whose tumors expressed high levels of circ-LDLRAD3 (hsa_circ_0006988) had a worse prognosis (p=0.0476) [124]. Circ-LDLRAD3 overexpression could act as an oncogene by sponging miR-137-3p to promote the cell proliferation, migration and invasion of PC [124]. Interestingly, another study also confirmed that circ-LDLRAD3 was significantly upregulated in PC tissues and plasma [122]. A high level of circ-LDLRAD3 was positively associated with tumor venous invasion (p=0.025) and lymphatic metastasis (p=0.014) [122], and its expression in plasma was significantly related to CA19-9 levels (p=0.03), N stage (p=0.049), venous invasion (p=0.005), and lymphatic metastasis (p=0.014) in PC tissues [122]. Additionally, circ-LDLRAD3 coupled with CA19-9 was confirmed to have higher sensitivity and specificity for the diagnosis of PC [122]. Therefore, these findings suggest that circ-LDLRAD3 may function as a novel biomarker for the diagnosis of PC [122]. A high-throughput circRNA microarray showed that circ_0030235 is highly expressed in PDAC tissue samples [97]. qRT-PCR further confirmed that circ_0030235 was also markedly elevated in PDAC tissues and cells compared to paired nontumorous tissue specimens and HPDE cells, respectively [121]. In PDAC tissues, high expression of circ_0030235 was confirmed as a possible biomarker for poor prognosis by Kaplan-Meier (KM) analysis (p=0.001) [121]. Moreover, high expression of circ_0030235 is an independent prognostic indicator of unfavorable OS for PDAC patients according to a multivariate Cox analysis [121].
Conversely, as mentioned above, PDAC patients with a high level of hsa_circ_0001649 had a higher OS rate (p=0.002) [117]. It was also found that PDAC patients with low levels of hsa_circ_0001649 presented with more advanced tumor stage (p=0.038) and lower differentiation grade (p=0.018) [117]. The univariate analysis of OS verified that high hsa_circ_0001649 expression (p=0.003) and high differentiation grade (p=0.006) were all good prognostic indicators [117]. Furthermore, the Cox proportional hazards model demonstrated that hsa_circ_0001649 may serve as an independent prognostic predictor of OS in PDAC patients (p=0.039) [117].
Exosomes are a type of nanosized (30-150 nm) extracellular vesicle with a lipid bilayer membrane released by multiple cell types and can be detected in various bodily fluids, such as plasma, saliva, and urine [125, 126]. As crucial mediators of intercellular communication, exosomes participate in carcinogenesis and cancer progression [125, 127]. Recent studies have identified the abundance and stability of circRNAs in exosomes (Figure 2G) [128]. Hence, exosomal circRNAs might be potential biomarkers for the detection of some cancers [128]. For instance, circ-IARS located within exosomes, generated and released by PC cells, was obviously overexpressed in the plasma exosomes of patients with metastasis and in PC patient tissue [129]. A high level of circ-IARS was positively associated with liver metastasis (p=0.000), vascular invasion (p=0.038), and TNM stage (p=0.011) [129]. PC patients with higher circ-IARS expression showed a lower OS rate than those with lower circ-IARS expression (p=0.01) [129]. Furthermore, it was found that circ-IARS enters HUVECs via exosomes to enhance cancer metastasis [129]. Collectively, these findings show that circ-IARS carried by exosomes from PC cells was taken up by HUVECs, specifically sponging miR-122 in HUVECs to relieve its inhibition of the target gene RhoA [129]. This further elevated the expression and activity of RhoA, thereby reducing the expression of the tight junction protein ZO-1 and increasing the expression of F-actin and endothelial monolayer permeability [129]. Another study identified the tumor-released exosomal circ-PDE8A (hsa_circ_0036627) in the exosomes of PC cells from patients with liver metastasis by using human circRNA microarrays [130]. Circ-PDE8A levels were significantly higher in PC tissues than in adjacent normal tissues. It was also discovered that the level of circ-PDE8A was related to lymphatic invasion (p=0.014), T factor (p=0.049) and TNM stage (p=0.005) in PC patients [130]. PC patients with higher levels of circ-PDE8A had a significantly poorer OS than those with lower expression of circ-PDE8A (p=0.016) [130]. Further investigations verified that circ-PDE8A absorbed miR-338 to enhance the growth and invasion of PC cells by upregulating the expression level of MET [130, 131]. In addition, KM survival curves demonstrated that the high expression of circ-PDE8A in plasma exosomes predicted worse OS in PC patients (p=0.011) [130]. Based on these findings, exosomal circRNAs may function as potential biomarkers for the diagnosis, prognostic prediction and progression of PC [129, 130].
CircRNAs as therapeutic targets in pancreatic cancer
Recent studies have determined that circRNAs affect the sensitivity of PC to chemotherapy [13, 132, 133]. Gemcitabine (GEM) is currently an effective monotherapy for the treatment of advanced PC. Nevertheless, acquired GEM resistance has resulted in treatment failures in a large number of PC patients [134]. Xu et al. detected 12,866 circRNAs and identified 81 DECs between GEM-resistant SW1990 cells and parental SW1990 cells by using circRNA microarrays [135]. Of the 81 DECs, 26 were upregulated and 55 were downregulated [135]. Moreover, circRNA_101672 and circRNA_004077 were the top two upregulated circRNAs and were both located on chromosome 16, which may make a difference in GEM resistance in PC. RNA sequencing analysis demonstrated that 68 and 58 circRNAs were upregulated and downregulated, respectively, in PANC-1-GR cells in comparation with control PANC-1 cells [13]. qRT-PCR experiments demonstrated that although the expression trends of the top 10 DECs were consistent with the microarray data, only two of them (chr14:101402109-101464448C and chr4:52729603-52780244C) were verified to be the most markedly upregulated in PANC-1-GR cells [13]. Similarly, it was confirmed that these two most significantly upregulated circRNAs were also upregulated in plasma samples of GEM-nonresponsive PDAC patients but not in GEM-responsive PDAC patients [13]. More importantly, knockdown and overexpression of these two circRNAs enhanced the GEM sensitivity of PANC-1-GR cells and GEM resistance of PANC-1 and MIA PACA-2 cells [13]. In addition, GO and pathway analyses of the parental genes related to the DECs revealed eight significantly enriched pathways, among which the VEGF and ErbB signaling pathways were previously confirmed to participate in GEM resistance [136] and PDAC progression [137]. All these findings suggest that in addition to the two most significantly upregulated circRNAs that have been identified, more circRNAs might take part in the GEM resistance of PANC-1-GR cells [13]. CircHIPK3 (hsa_circ_0000284) was proposed to be involved in tumorigenesis and chemotherapy resistance in different cancers [138, 139, 140, 141]. In a recent study, Zhu's team discovered that circHIPK3 was elevated in PC tissues and cells with GEM resistance [132]. Briefly, circHIPK3 absorbed miR-330-5p to enhance the resistance of GEM and regulate proliferation and progression by upregulating the expression of RASSF1 in PC [132].
Conclusions and Prospects
Due to late diagnosis and low response to chemotherapy, PC patients have poor prognosis, and the specific pathogenesis of PC is still unclear. Hence, it is of paramount importance to identify earlier diagnostic and more efficient therapeutic approaches for the clinical management of PC. Initially, considered to be RNA splicing errors, circRNAs have generated increasing attention due to their strong correlation with different physiological and pathological processes, their important roles in various diseases, and their high degree of tissue and development specificity [142]. Currently, many studies have revealed the value of circRNAs in clinical practice in various tumors, including PC. In this review, circRNAs were shown to participate in different biological processes of PC and serve as promising biomarkers for the diagnosis, prognosis, response to chemotherapy, and risk evaluation of PC. At present, m6A-modified circRNAs can promote the transport of circRNAs to cytoplasm [143], protein translation [81], and degradation processes [144]. However, there has not been a study involving m6A modification of circRNA in PC, so it is worth exploring in later circRNA research on PC. In fact, research on the field of circRNAs in PC is still in its early stage compared with miRNA and long noncoding RNA (lncRNA) research, and only a tiny proportion of important circRNAs in PC have been identified and characterized. Herein, some suggestions are put forward for future research on circRNAs in PC. First, although many scholars have initially proposed models related to circRNA formation, the detailed mechanism of circRNA formation is far from understood. More scientific explorations and efforts are imperatively needed to fully illustrate the mechanisms of biogenesis, turnover and degradation of circRNAs. A thorough annotation of circRNA biogenesis and regulation will undoubtedly strengthen our understanding of circRNA functions. Second, in PC, circRNAs exert their effects mainly by serving as miRNA sponges. Nonetheless, because a large number of circRNAs have much lower abundance than miRNAs and there are only a few circRNAs with many MREs [145] , the miRNA sponge mechanism for circRNA is faced with a great dilemma. Therefore, it is imperative to further elucidate other mechanisms by which circRNAs function, including gene transcription regulation, interaction with RBPs and translation potential. Third, the detection of circRNAs is currently mainly performed in clinical tissue specimens. In future research, the expression of circRNAs should be detected in more extensive clinical samples related to the disease, such as blood, cerebrospinal fluid, urine, and saliva. Studies can also utilize combined detection methods to obtain better diagnostic value, including combining the detection methods for various circRNAs with traditional diagnostic markers related to tumors. Nevertheless, whether circRNAs can successfully function as effective biomarkers for the diagnosis and prognosis of cancers is still far from clinical application. Fourth, future studies may regard circRNAs as potential targets for the treatment of tumors. How to deliver circRNAs to the corresponding parts of the body, how to ensure longstanding and effective function, and how to avoid the occurrence of immune rejection are all difficult problems that need to be resolved. Last, the application of specific circRNAs related to human diseases in the treatment of tumors is the ultimate goal of circRNA-related research. Therefore, more controlled clinical studies and experiments need to be conducted on a large scale in tumor patients.
In summary, the current understanding of circRNA functions in PC is still very limited. Fortunately, following the accelerated advancement of high-throughput RNA sequencing and biotechnology [42] and the numerous available online databases, more circRNAs will be identified and validated. In the near future, we believe that an in-depth understanding of the characteristics of circRNAs and correct application of circRNAs in clinical practice will represent a giant leap in the treatment of PC.
Abbreviations
ceRNA: competing endogenous RNA; miR: miRNA; MREs: miRNA response elements; EGFR: epidermal growth factor receptor; HuR: human antigen R; Amotl1: angiomotin-like1; YBX1: Y-box binding protein; hnRNPK: heterogeneous nuclear ribonucleoprotein K; PDXK: pyridoxal kinase; NUF2: NUF2 component of NDC80 kinetochore complex; ALKBH5: alkB homologue 5; FAAH: fatty acid amide hydrolase; CDK2: cyclin-dependent kinase 2; P21: cyclin-dependent kinase inhibitor 1; ANKRD52: ankyrin repeat domain 52; RNA Pol II: RNA polymerase II; U1 snRNP: U1 small nuclear ribonucleoprotein; PABPN1: poly (A)-binding protein nuclear 1; ORFs: open reading frames; IRES: internal ribosome entry site element; FTO: obesity-associated protein; METTL3/14: methyltransferase-like 3/14; GEO: Gene Expression Omnibus; LNM: lymph node metastasis; ANK1: ankyrin 1; LAMTOR3: lysosomal adaptor, MAPK and MTOR activator 3; GDNF: glial cell-derived neurotrophic factor; PAX6: paired box 6; HOXC6: homeobox C6; PCa: prostate cancer; SERPINA4: serpin family A member 4; PAK1: p21-activated kinase 1; VEGF-C: vascular endothelial growth factor C; VEGFR3: vascular endothelial growth factor receptor-3; HLEC: lymphatic endothelial cell; SLC4A4: solute carrier family 4 member 4; HUVECs: human microvascular vein endothelial cells; PANC-1-GR: GEM-resistant PANC-1; GO: Gene Ontology; RASSF1: ras association domain family member 1.
Acknowledgements
We are grateful to the editors of AJE (American Journal Experts) for their help in editing the manuscript. This study was supported by the National Natural Science Foundation of China (No. 81902428, 81802352 and 81772555), the Shanghai Sailing Program (No. 19YF1409400), the National Science Foundation for Distinguished Young Scholars of China (No. 81625016), Clinical and Scientific Innovation Project of Shanghai Hospital Development Center (SHDC12018109) and Scientific Innovation Project of Shanghai Education Committee (2019-01-07-00-07-E00057).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Crawford HC, Pasca di Magliano M, Banerjee S. Signaling Networks That Control Cellular Plasticity in Pancreatic Tumorigenesis, Progression, and Metastasis. Gastroenterology. 2019;156:2073-84
2. Neoptolemos JP, Kleeff J, Michl P, Costello E, Greenhalf W, Palmer DH. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat Rev Gastroenterol Hepatol. 2018;15:333-48
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30
4. Singhi AD, Koay EJ, Chari ST, Maitra A. Early Detection of Pancreatic Cancer: Opportunities and Challenges. Gastroenterology. 2019;156:2024-40
5. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852-6
6. Wang M, Yu F, Wu W, Zhang Y, Chang W, Ponnusamy M. et al. Circular RNAs: A novel type of non-coding RNA and their potential implications in antiviral immunity. Int J Biol Sci. 2017;13:1497-506
7. Wang Y, Mo Y, Gong Z, Yang X, Yang M, Zhang S. et al. Circular RNAs in human cancer. Mol Cancer. 2017;16:25
8. Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y. et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61:221-30
9. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-8
10. Fang X, Wen J, Sun M, Yuan Y, Xu Q. CircRNAs and its relationship with gastric cancer. J Cancer. 2019;10:6105-13
11. Li J, Yang J, Zhou P, Le Y, Zhou C, Wang S. et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5:472-80
12. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205-11
13. Shao F, Huang M, Meng F, Huang Q. Circular RNA Signature Predicts Gemcitabine Resistance of Pancreatic Ductal Adenocarcinoma. Front Pharmacol. 2018;9:584
14. Rong D, Tang W, Li Z, Zhou J, Shi J, Wang H. et al. Novel insights into circular RNAs in clinical application of carcinomas. Onco Targets Ther. 2017;10:2183-8
15. Luo Z, Rong Z, Zhang J, Zhu Z, Yu Z, Li T. et al. Circular RNA circCCDC9 acts as a miR-6792-3p sponge to suppress the progression of gastric cancer through regulating CAV1 expression. Mol Cancer. 2020;19:86
16. Li J, Ma M, Yang X, Zhang M, Luo J, Zhou H. et al. Circular HER2 RNA positive triple negative breast cancer is sensitive to Pertuzumab. Mol Cancer. 2020;19:142
17. Drula R, Braicu C, Harangus A, Nabavi SM, Trif M, Slaby O. et al. Critical function of circular RNAs in lung cancer. Wiley Interdiscip Rev RNA. 2020;11:e1592
18. Liu L, Liu FB, Huang M, Xie K, Xie QS, Liu CH. et al. Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat Dis Int. 2019;18:580-6
19. Wang YZ, An Y, Li BQ, Lu J, Guo JC. Research progress on circularRNAs in pancreatic cancer: emerging but promising. Cancer Biol Ther. 2019;20:1163-71
20. Limb C, Liu DSK, Veno MT, Rees E, Krell J, Bagwan IN. et al. The Role of Circular RNAs in Pancreatic Ductal Adenocarcinoma and Biliary-Tract Cancers. Cancers (Basel). 2020;12:11
21. Sharma GG, Okada Y, Von Hoff D, Goel A. Non-coding RNA biomarkers in pancreatic ductal adenocarcinoma. Semin Cancer Biol. 2020 doi: 10.1016/j.semcancer.2020.10.001
22. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH. et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792-806
23. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675-91
24. Wang F, Nazarali AJ, Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res. 2016;6:1167-76
25. Guo Y, Yang J, Huang Q, Hsueh C, Zheng J, Wu C. et al. Circular RNAs and their roles in head and neck cancers. Mol Cancer. 2019;18:44
26. Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18:6
27. Ma S, Kong S, Wang F, Ju S. CircRNAs: biogenesis, functions, and role in drug-resistant Tumours. Mol Cancer. 2020;19:119
28. Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10:3503-17
29. Kelly S, Greenman C, Cook PR, Papantonis A. Exon Skipping Is Correlated with Exon Circularization. J Mol Biol. 2015;427:2414-7
30. Wilusz JE. A 360° view of circular RNAs: From biogenesis to functions. Wiley Interdiscip Rev RNA. 2018;9:e1478
31. Guo Y, Yang J, Huang Q, Hsueh C, Zheng J, Wu C. et al. Circular RNAs and their roles in head and neck cancers. Mol Cancer. 2019;18:44
32. Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF. et al. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol Cell. 2017;67:214-27.e7
33. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125-34
34. Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J Neurosci Res. 2020;98:87-97
35. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M. et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55-66
36. Qian L, Yu S, Chen Z, Meng Z, Huang S, Wang P. The emerging role of circRNAs and their clinical significance in human cancers. Biochim Biophys Acta Rev Cancer. 2018;1870:247-60
37. Cui X, Wang J, Guo Z, Li M, Li M, Liu S. et al. Emerging function and potential diagnostic value of circular RNAs in cancer. Mol Cancer. 2018;17:123
38. Han J, Meng J, Chen S, Wang X, Yin S, Zhang Q. et al. YY1 Complex Promotes Quaking Expression via Super-Enhancer Binding during EMT of Hepatocellular Carcinoma. Cancer Res. 2019;79:1451-64
39. Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D. et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8:14741
40. Sun J, Li B, Shu C, Ma Q, Wang J. Functions and clinical significance of circular RNAs in glioma. Mol Cancer. 2020;19:34
41. Lasda E, Parker R. Circular RNAs: diversity of form and function. Rna. 2014;20:1829-42
42. Wang M, Yu F, Li P. Circular RNAs: Characteristics, Function and Clinical Significance in Hepatocellular Carcinoma. Cancers (Basel). 2018;10:8
43. Zhao W, Dong M, Pan J, Wang Y, Zhou J, Ma J. et al. Circular RNAs: A novel target among non-coding RNAs with potential roles in malignant tumors (Review). Mol Med Rep. 2019;20:3463-74
44. Chen X, Han P, Zhou T, Guo X, Song X, Li Y. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985
45. Huang W, Ling Y, Zhang S, Xia Q, Cao R, Fan X. et al. TransCirc: an interactive database for translatable circular RNAs based on multi-omics evidence. Nucleic Acids Res. 2020 doi: 10.1093/nar/gkaa823
46. Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. Rna. 2014;20:1666-70
47. Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283
48. Dong R, Ma XK, Li GW, Yang L. CIRCpedia v2: An Updated Database for Comprehensive Circular RNA Annotation and Expression Comparison. Genomics Proteomics Bioinformatics. 2018;16:226-33
49. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34-42
50. Liu YC, Li JR, Sun CH, Andrews E, Chao RF, Lin FM. et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44:D209-15
51. Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y. et al. The Landscape of Circular RNA in Cancer. Cell. 2019;176:869-81.e13
52. Xia S, Feng J, Lei L, Hu J, Xia L, Wang J. et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform. 2017;18:984-92
53. Xia S, Feng J, Chen K, Ma Y, Gong J, Cai F. et al. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res. 2018;46:D925-d9
54. Li S, Li Y, Chen B, Zhao J, Yu S, Tang Y. et al. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018;46:D106-d12
55. Yao D, Zhang L, Zheng M, Sun X, Lu Y, Liu P. Circ2Disease: a manually curated database of experimentally validated circRNAs in human disease. Sci Rep. 2018;8:11018
56. Noh JH, Kim KM, McClusky WG, Abdelmohsen K, Gorospe M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA. 2018;9:e1471
57. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733
58. Xiong G, Feng M, Yang G, Zheng S, Song X, Cao Z. et al. The underlying mechanisms of non-coding RNAs in the chemoresistance of pancreatic cancer. Cancer Lett. 2017;397:94-102
59. Liu J, Liu T, Wang X, He A. Circles reshaping the RNA world: from waste to treasure. Mol Cancer. 2017;16:58
60. Zhu Z, Rong Z, Luo Z, Yu Z, Zhang J, Qiu Z. et al. Circular RNA circNHSL1 promotes gastric cancer progression through the miR-1306-3p/SIX1/vimentin axis. Mol Cancer. 2019;18:126
61. Song T, Xu A, Zhang Z, Gao F, Zhao L, Chen X. et al. CircRNA hsa_circRNA_101996 increases cervical cancer proliferation and invasion through activating TPX2 expression by restraining miR-8075. J Cell Physiol. 2019;234:14296-305
62. Sang Y, Chen B, Song X, Li Y, Liang Y, Han D. et al. circRNA_0025202 Regulates Tamoxifen Sensitivity and Tumor Progression via Regulating the miR-182-5p/FOXO3a Axis in Breast Cancer. Mol Ther. 2019;27:1638-52
63. Yang C, Yuan W, Yang X, Li P, Wang J, Han J. et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17:19
64. Li H, Lan M, Liao X, Tang Z, Yang C. Circular RNA cir-ITCH Promotes Osteosarcoma Migration and Invasion through cir-ITCH/miR-7/EGFR Pathway. Technol Cancer Res Treat. 2020;19:1533033819898728
65. Guo X, Zhou Q, Su D, Luo Y, Fu Z, Huang L. et al. Circular RNA circBFAR promotes the progression of pancreatic ductal adenocarcinoma via the miR-34b-5p/MET/Akt axis. Mol Cancer. 2020;19:83
66. Kong Y, Li Y, Luo Y, Zhu J, Zheng H, Gao B. et al. circNFIB1 inhibits lymphangiogenesis and lymphatic metastasis via the miR-486-5p/PIK3R1/VEGF-C axis in pancreatic cancer. Mol Cancer. 2020;19:82
67. Li R, Jiang J, Shi H, Qian H, Zhang X, Xu W. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci. 2020;77:1661-80
68. Chen Y, Yang F, Fang E, Xiao W, Mei H, Li H. et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26:1346-64
69. Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L. et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24:1609-20
70. Shen S, Yao T, Xu Y, Zhang D, Fan S, Ma J. CircECE1 activates energy metabolism in osteosarcoma by stabilizing c-Myc. Mol Cancer. 2020;19:151
71. Wong CH, Lou UK, Li Y, Chan SL, Tong JH, To KF. et al. CircFOXK2 Promotes Growth and Metastasis of Pancreatic Ductal Adenocarcinoma by Complexing with RNA-Binding Proteins and Sponging MiR-942. Cancer Res. 2020;80:2138-49
72. Huang R, Zhang Y, Bai Y, Han B, Ju M, Chen B. et al. N(6)-Methyladenosine Modification of Fatty Acid Amide Hydrolase Messenger RNA in Circular RNA STAG1-Regulated Astrocyte Dysfunction and Depressive-like Behaviors. Biol Psychiatry. 2020;88:392-404
73. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846-58
74. Zhang H, Shen Y, Li Z, Ruan Y, Li T, Xiao B. et al. The biogenesis and biological functions of circular RNAs and their molecular diagnostic values in cancers. J Clin Lab Anal. 2020;34:e23049
75. Chen B, Huang S. Circular RNA: An emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett. 2018;418:41-50
76. Shan C, Zhang Y, Hao X, Gao J, Chen X, Wang K. Biogenesis, functions and clinical significance of circRNAs in gastric cancer. Mol Cancer. 2019;18:136
77. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256-64
78. Wu N, Yuan Z, Du KY, Fang L, Lyu J, Zhang C. et al. Translation of yes-associated protein (YAP) was antagonized by its circular RNA via suppressing the assembly of the translation initiation machinery. Cell Death Differ. 2019;26:2758-73
79. Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S. et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361-9
80. Schneider T, Bindereif A. Circular RNAs: Coding or noncoding? Cell Res. 2017;27:724-5
81. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y. et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626-41
82. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L. et al. Translation of CircRNAs. Mol Cell. 2017;66:9-21.e7
83. Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F. et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805-14
84. Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F. et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst. 2018;110:304-15
85. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O. et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017;66:22-37.e9
86. Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415-7
87. Pan Z, Cai J, Lin J, Zhou H, Peng J, Liang J. et al. A novel protein encoded by circFNDC3B inhibits tumor progression and EMT through regulating Snail in colon cancer. Mol Cancer. 2020;19:71
88. Zhou B, Yang H, Yang C, Bao YL, Yang SM, Liu J. et al. Translation of noncoding RNAs and cancer. Cancer Lett. 2021;497:89-99
89. Liang WC, Wong CW, Liang PP, Shi M, Cao Y, Rao ST. et al. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20:84
90. Zhao X, Cai Y, Xu J. Circular RNAs: Biogenesis, Mechanism, and Function in Human Cancers. Int J Mol Sci. 2019;20:16
91. Xu Z, Yan Y, Zeng S, Dai S, Chen X, Wei J. et al. Circular RNAs: clinical relevance in cancer. Oncotarget. 2018;9:1444-60
92. Zhang M, Xin Y. Circular RNAs: a new frontier for cancer diagnosis and therapy. J Hematol Oncol. 2018;11:21
93. Wang X, Fang L. Advances in circular RNAs and their roles in breast Cancer. J Exp Clin Cancer Res. 2018;37:206
94. Zhang Q, Wang JY, Zhou SY, Yang SJ, Zhong SL. Circular RNA expression in pancreatic ductal adenocarcinoma. Oncol Lett. 2019;18:2923-30
95. Guo S, Xu X, Ouyang Y, Wang Y, Yang J, Yin L. et al. Microarray expression profile analysis of circular RNAs in pancreatic cancer. Mol Med Rep. 2018;17:7661-71
96. Seimiya T, Otsuka M, Iwata T, Tanaka E, Sekiba K, Shibata C. et al. Aberrant expression of a novel circular RNA in pancreatic cancer. J Hum Genet. 2020 doi: 10.1038/s10038-020-00826-5
97. Li H, Hao X, Wang H, Liu Z, He Y, Pu M. et al. Circular RNA Expression Profile of Pancreatic Ductal Adenocarcinoma Revealed by Microarray. Cell Physiol Biochem. 2016;40:1334-44
98. Xiao Y. Construction of a circRNA-miRNA-mRNA network to explore the pathogenesis and treatment of pancreatic ductal adenocarcinoma. J Cell Biochem. 2020;121:394-406
99. Visci G, Tolomeo D, Agostini A, Traversa D, Macchia G, Storlazzi CT. CircRNAs and Fusion-circRNAs in cancer: New players in an old game. Cell Signal. 2020;75:109747
100. Hao L, Rong W, Bai L, Cui H, Zhang S, Li Y. et al. Upregulated circular RNA circ_0007534 indicates an unfavorable prognosis in pancreatic ductal adenocarcinoma and regulates cell proliferation, apoptosis, and invasion by sponging miR-625 and miR-892b. J Cell Biochem. 2019;120:3780-9
101. Chen G, Shi Y, Zhang Y, Sun J. CircRNA_100782 regulates pancreatic carcinoma proliferation through the IL6-STAT3 pathway. Onco Targets Ther. 2017;10:5783-94
102. Neesse A, Bauer CA, Öhlund D, Lauth M, Buchholz M, Michl P. et al. Stromal biology and therapy in pancreatic cancer: ready for clinical translation? Gut. 2019;68:159-71
103. Chen Y, Li Z, Zhang M, Wang B, Ye J, Zhang Y. et al. Circ-ASH2L promotes tumor progression by sponging miR-34a to regulate Notch1 in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res. 2019;38:466
104. Shi H, Li H, Zhen T, Dong Y, Pei X, Zhang X. hsa_circ_001653 Implicates in the Development of Pancreatic Ductal Adenocarcinoma by Regulating MicroRNA-377-Mediated HOXC6 Axis. Mol Ther Nucleic Acids. 2020;20:252-64
105. Zhou J, Yang X, Song P, Wang H, Wang X. HOXC6 in the prognosis of prostate cancer. Artif Cells Nanomed Biotechnol. 2019;47:2715-20
106. Zhu P, Ge N, Liu D, Yang F, Zhang K, Guo J. et al. Preliminary investigation of the function of hsa_circ_0006215 in pancreatic cancer. Oncol Lett. 2018;16:603-11
107. Qu S, Hao X, Song W, Niu K, Yang X, Zhang X. et al. Circular RNA circRHOT1 is upregulated and promotes cell proliferation and invasion in pancreatic cancer. Epigenomics. 2019;11:53-63
108. Ling S, He Y, Li X, Hu M, Ma Y, Li Y. et al. CircRHOT1 mediated cell proliferation, apoptosis and invasion of pancreatic cancer cells by sponging miR-125a-3p. J Cell Mol Med. 2020;24:9881-9
109. Zhang X, Xue C, Cui X, Zhou Z, Fu Y, Yin X. et al. Circ_0075829 facilitates the progression of pancreatic carcinoma by sponging miR-1287-5p and activating LAMTOR3 signalling. J Cell Mol Med. 2020 doi: 10.1111/jcmm.16089
110. An Y, Cai H, Zhang Y, Liu S, Duan Y, Sun D. et al. circZMYM2 Competed Endogenously with miR-335-5p to Regulate JMJD2C in Pancreatic Cancer. Cell Physiol Biochem. 2018;51:2224-36
111. Garcia J, Lizcano F. KDM4C Activity Modulates Cell Proliferation and Chromosome Segregation in Triple-Negative Breast Cancer. Breast Cancer (Auckl). 2016;10:169-75
112. Xing C, Ye H, Wang W, Sun M, Zhang J, Zhao Z. et al. Circular RNA ADAM9 facilitates the malignant behaviours of pancreatic cancer by sponging miR-217 and upregulating PRSS3 expression. Artif Cells Nanomed Biotechnol. 2019;47:3920-8
113. Xu S, Lei S-L, Liu K-J, Yi S-G, Yang Z-L, Yao H-L. circSFMBT1 promotes pancreatic cancer growth and metastasis via targeting miR-330-5p/PAK1 axis. Cancer Gene Therapy. 2020 doi: 10.1038/s41417-020-00215-2
114. Jussila L, Alitalo K. Vascular growth factors and lymphangiogenesis. Physiol Rev. 2002;82:673-700
115. Brouillard P, Boon L, Vikkula M. Genetics of lymphatic anomalies. J Clin Invest. 2014;124:898-904
116. Zhang X, Tan P, Zhuang Y, Du L. Hsa_circRNA_001587 upregulates SLC4A4 expression to inhibit migration, invasion and angiogenesis of pancreatic cancer cells via binding to microRNA-223. Am J Physiol Gastrointest Liver Physiol. 2020;319:G703-g17
117. Jiang Y, Wang T, Yan L, Qu L. A novel prognostic biomarker for pancreatic ductal adenocarcinoma: hsa_circ_0001649. Gene. 2018;675:88-93
118. Han YN, Xia SQ, Zhang YY, Zheng JH, Li W. Circular RNAs: A novel type of biomarker and genetic tools in cancer. Oncotarget. 2017;8:64551-63
119. Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S. et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58:870-85
120. Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P. et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94
121. Xu Y, Yao Y, Gao P, Cui Y. Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294. Biochem Biophys Res Commun. 2019;509:138-42
122. Yang F, Liu DY, Guo JT, Ge N, Zhu P, Liu X. et al. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. 2017;23:8345-54
123. Fathizadeh H, Hallajzadeh J, Asemi Z. Circular RNAs as diagnostic biomarker in pancreatic cancer. Pathol Res Pract. 2020;216:153075
124. Yao J, Zhang C, Chen Y, Gao S. Downregulation of circular RNA circ-LDLRAD3 suppresses pancreatic cancer progression through miR-137-3p/PTN axis. Life Sci. 2019;239:116871
125. Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W. et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18:116
126. Boriachek K, Islam MN, Möller A, Salomon C, Nguyen NT, Hossain MSA. et al. Biological Functions and Current Advances in Isolation and Detection Strategies for Exosome Nanovesicles. Small. 2018;14:16
127. Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B. et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer. 2020;19:117
128. Xie Y, Li J, Li P, Li N, Zhang Y, Binang H. et al. RNA-Seq Profiling of Serum Exosomal Circular RNAs Reveals Circ-PNN as a Potential Biomarker for Human Colorectal Cancer. Front Oncol. 2020;10:982
129. Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K. et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37:177
130. Li Z, Yanfang W, Li J, Jiang P, Peng T, Chen K. et al. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237-50
131. Luna J, Boni J, Cuatrecasas M, Bofill-De Ros X, Núñez-Manchón E, Gironella M. et al. DYRK1A modulates c-MET in pancreatic ductal adenocarcinoma to drive tumour growth. Gut. 2019;68:1465-76
132. Liu Y, Xia L, Dong L, Wang J, Xiao Q, Yu X. et al. CircHIPK3 Promotes Gemcitabine (GEM) Resistance in Pancreatic Cancer Cells by Sponging miR-330-5p and Targets RASSF1. Cancer Manag Res. 2020;12:921-9
133. Xie W, Chu M, Song G, Zuo Z, Han Z, Chen C. et al. Emerging roles of long noncoding RNAs in chemoresistance of pancreatic cancer. Semin Cancer Biol. 2020 doi: 10.1016/j.semcancer
134. Amrutkar M, Gladhaug IP. Pancreatic Cancer Chemoresistance to Gemcitabine. Cancers (Basel). 2017;9:11
135. Xu C, Yu Y, Ding F. Microarray analysis of circular RNA expression profiles associated with gemcitabine resistance in pancreatic cancer cells. Oncol Rep. 2018;40:395-404
136. Skrypek N, Vasseur R, Vincent A, Duchêne B, Van Seuningen I, Jonckheere N. The oncogenic receptor ErbB2 modulates gemcitabine and irinotecan/SN-38 chemoresistance of human pancreatic cancer cells via hCNT1 transporter and multidrug-resistance associated protein MRP-2. Oncotarget. 2015;6:10853-67
137. Zhou R, Curry JM, Roy LD, Grover P, Haider J, Moore LJ. et al. A novel association of neuropilin-1 and MUC1 in pancreatic ductal adenocarcinoma: role in induction of VEGF signaling and angiogenesis. Oncogene. 2016;35:5608-18
138. Zhang Y, Li C, Liu X, Wang Y, Zhao R, Yang Y. et al. circHIPK3 promotes oxaliplatin-resistance in colorectal cancer through autophagy by sponging miR-637. EBioMedicine. 2019;48:277-88
139. Xie F, Zhao N, Zhang H, Xie D. Circular RNA CircHIPK3 Promotes Gemcitabine Sensitivity in Bladder Cancer. J Cancer. 2020;11:1907-12
140. Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C. et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646-59
141. Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T. et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417
142. Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11:98
143. Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD. et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695
144. Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L. et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol Cell. 2019;76:96-109.e9
145. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409
Author contact
![]() Corresponding authors: Chen Liang, Department of Pancreatic Surgery, Fudan University Shanghai Cancer Center, 270 Dong'An Road, Shanghai 200032, P. R. China. Tel.:+86-021-64175590; Fax: +86-021-64031446; E-mail: liangchenorg. Xianjun Yu, Department of Pancreatic Surgery, Fudan University Shanghai Cancer Center, 270 Dong'An Road, Shanghai 200032, P.R. China. Tel.: +86-021-64175590; Fax: +86-021-64031446; E-mail:yuxianjunorg.
Corresponding authors: Chen Liang, Department of Pancreatic Surgery, Fudan University Shanghai Cancer Center, 270 Dong'An Road, Shanghai 200032, P. R. China. Tel.:+86-021-64175590; Fax: +86-021-64031446; E-mail: liangchenorg. Xianjun Yu, Department of Pancreatic Surgery, Fudan University Shanghai Cancer Center, 270 Dong'An Road, Shanghai 200032, P.R. China. Tel.: +86-021-64175590; Fax: +86-021-64031446; E-mail:yuxianjunorg.
 Global reach, higher impact
Global reach, higher impact