13.3
Impact Factor
Theranostics 2021; 11(7):3348-3358. doi:10.7150/thno.45889 This issue Cite
Review
Roles of peptidyl-prolyl isomerase Pin1 in disease pathogenesis
1. The Second Affiliated Hospital of Chengdu Medical College, China National Nuclear Corporation 416 Hospital, Chengdu, Sichuan, China.
2. School of Biological Sciences and Technology, Chengdu Medical College, Chengdu, China.
3. Department of Immunology, School of Basic Medical Sciences, Chengdu Medical College, Chengdu, China.
*These authors contributed equally to this work.
Received 2020-3-11; Accepted 2020-12-2; Published 2021-1-19
Abstract
Pin1 belongs to the peptidyl-prolyl cis-trans isomerases (PPIases) superfamily and catalyzes the cis-trans conversion of proline in target substrates to modulate diverse cellular functions including cell cycle progression, cell motility, and apoptosis. Dysregulation of Pin1 has wide-ranging influences on the fate of cells; therefore, it is closely related to the occurrence and development of various diseases. This review summarizes the current knowledge of Pin1 in disease pathogenesis.
Keywords: peptidyl-prolyl cis-trans isomerases, Pin1, pathogenesis of diseases, viral infection, neurodegenerative diseases
Introduction
Some proteins exhibit either a cis or a trans state due to the presence of proline, which imparts distinct conformations and biological functions. The peptidyl-prolyl cis-trans isomerases (PPIases) superfamily [1] comprises four families according to their structural differences: cyclophilins, FK506-binding proteins (FKBPs), parvulins, and the protein phosphatase (PPase) 2A phosphatase activator (PTPA) [2-6].
In the human parvulin family, there are two genes: PIN1 and PIN4 [7-9]. The coded product of PIN1, PPIase NIMA-interacting 1 (Pin1) protein, was identified in 1996 by Lu et al. as a protein interacting with NIMA kinase [7]. PIN4 encodes the isoforms parvulin 14 (Par14) and parvulin 17 (Par17) [9, 10]. Parvulin 14 consists of 131 amino acids, while parvulin 17 is an N-terminal extended version of Par14 with an additional 25 amino acids. Among the three members of the human parvulin family, current research on the function of Pin1 and its role in disease pathogenesis is the most in-depth. Pin1 consists of 163 amino acid residues with a relative molecular mass of 18 kDa and contains 1 nuclear localization signal and 2 functional domains. The amino terminus (N-terminus) is the tryptophan-tryptophan central domain (WW domain), which is responsible for recognition and binding to the pSer/Thr-Pro motif of the substrate, while the C-terminal PPIase catalytic domain performs the function of cis-trans isomerization [7, 11]. The two domains are fastened by a flexible linker of 15 residues. Although they belong to the same family, Pin1 differs from parvulin-type PPIases in that Pin1 specifically catalyzes the isomerization of phosphorylated Ser-Pro or Thr-Pro (pSer-Pro or pThr-Pro) peptides, whereas Par14/Par17 show no preference for phosphorylated substrates [9, 12, 13].
Since Pin1 isomerizes phosphorylated substrates and phosphorylation and post-phosphorylation events play important roles in cell signaling pathways, Pin1 is involved in a variety of cellular processes such as cell cycle, cell proliferation, cell motility, and apoptosis [13-18]. In most cases, Pin1 functions as a molecular timer or switch that modulates proteins or entire signaling pathways. Dysregulation of Pin1 is closely related to the development of multiple diseases. In this review, we will discuss the role of Pin1 in the pathogenesis of various related diseases.
Pin1 and cancer
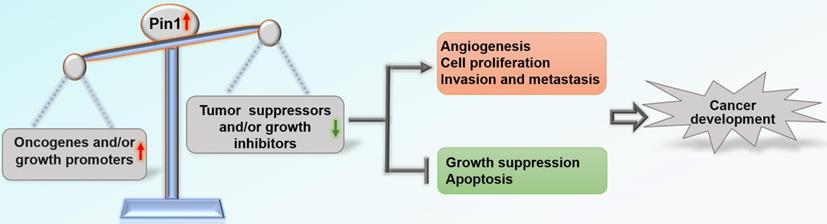
Overall, Pin1 drives tumor progression and is negatively associated with clinical outcome in patients with cancer [19-21]. Pin1 has been shown to activate more than 50 oncogenic proteins and growth promoters and/or shut down at least 20 tumor suppressors and growth inhibitors through positive and negative feedback mechanisms [12, 22] (Table 1). Most tumors exhibit overexpression and/or activation of Pin1 compared with corresponding normal tissues, including breast, prostate, lung, ovarian, gastric, esophageal, cervical, and brain tumors and melanoma [21, 23-25]. Expression of Pin1 in tumor cell lines cultured in vitro has also been found to be significantly higher than that in normal cell lines. Knockdown of the Pin1 gene inhibits cancer cell growth both in vitro and in vivo and results in cancer cell apoptosis [26, 27]. In addition, emerging evidence suggests that inhibitors targeting Pin1 have significant anti-cancer effects. These inhibitors include juglone [27, 28], all-trans retinoic acid (ATRA) [29, 30], 2-{[4-(4-tert-butylbenzenesulfonamido) -1-oxo-1,4-dihydronaphthalen-2-yl] sulfanyl} acetic acid (KPT-6566) [31], epigallocatechin-3-gallate (EGCG) [32], PiB [33], compound 20 [34], compound 23a [35], API-1 [36], arsenic trioxide (ATO) [37], and BJP-06-005-3 [38]. In a recent review [22], Chen et al. elaborated on how Pin1 contributes to all ten hallmarks of cancer [39] by dysregulating multiple cancer-driving pathways at various levels. Pin1 induces angiogenesis by facilitating expression of VEGF and inhibition of Pin1 by RNAi significantly suppresses angiogenesis [40, 41]; Pin1 sustains proliferative signaling and evades growth suppression by activating growth-promoting regulators and inactivating growth-inhibitory regulators [22]; Pin1 promotes migration and invasion by regulating NOTCH1 [42], TGF-β [43], β-catenin [44], and BRD4 [45]; Pin1 inhibits apoptosis of tumor cells by increasing the anti-apoptotic function of anti-apoptotic proteins and suppressing pro-apoptotic factors [46, 47]. This concept has recently been greatly expanded, demonstrating that overactivation of Pin1 disrupts the balance between carcinogenic proteins and tumor suppressors, which pushes cells towards carcinogenesis [12] (Figure 1).
Oncogenic proteins/growth-promoting regulators and tumor suppressors/growth-inhibitory regulators as Pin1 substrates
| Substrate | Function | Activity of substrate | Refs |
|---|---|---|---|
| AIB1 | Oncogenic protein | ↑ | [158] |
| AKT | Oncogenic protein | ↑ | [159] |
| BCL2 | Oncogenic protein | ↑ | [160] |
| JUN | Oncogenic protein | ↑ | [23] |
| COX2 | Oncogenic protein | ↑ | [161] |
| FOS | Oncogenic protein | ↑ | [162] |
| FOXM1 | Oncogenic protein | ↑ | [163] |
| HER2 | Oncogenic protein | ↑ | [164] |
| MYC | Oncogenic protein | ↑ | [165] |
| Survivin | Oncogenic protein | ↑ | [46] |
| Tax | Oncogenic protein | ↑ | [118] |
| XBP1 | Oncogenic protein | ↑ | [166] |
| AR | Growth-promoting regulator | ↑ | [167] |
| CDC25 | Growth-promoting regulator | ↑ | [168] |
| Cep55 | Growth-promoting regulator | ↑ | [169] |
| MYB | Growth-promoting regulator | ↑ | [170] |
| Cyclin D1 | Growth-promoting regulator | ↑ | [23] |
| ER | Growth-promoting regulator | ↑ | [164] |
| FAK | Growth-promoting regulator | ↑ | [171] |
| HBx | Growth-promoting regulator | ↑ | [113] |
| HIF1 | Growth-promoting regulator | ↑ | [172] |
| HSF1 | Growth-promoting regulator | ↑ | [173] |
| IRAK1 | Growth-promoting regulator | ↑ | [174] |
| MCL1 | Growth-promoting regulator | ↑ | [175] |
| Nanog | Growth-promoting regulator | ↑ | [176] |
| NF-κB | Growth-promoting regulator | ↑ | [177] |
| NOTCH1 | Growth-promoting regulator | ↑ | [178] |
| NOTCH3 | Growth-promoting regulator | ↑ | [179] |
| NUR77 | Growth-promoting regulator | ↑ | [180] |
| OCT4 | Growth-promoting regulator | ↑ | [181] |
| p47phox | Growth-promoting regulator | ↑ | [182] |
| p53M | Growth-promoting regulator | ↑ | [183] |
| PGK1 | Growth-promoting regulator | ↑ | [115] |
| PKM2 | Growth-promoting regulator | ↑ | [184] |
| PLK | Growth-promoting regulator | ↑ | [185] |
| PML-RARα | Growth-promoting regulator | ↑ | [186] |
| PTP | Growth-promoting regulator | ↑ | [187] |
| PTP-PEST | Growth-promoting regulator | ↑ | [171] |
| RAB2A | Growth-promoting regulator | ↑ | [188] |
| RAF1 | Growth-promoting regulator | ↑ | [189] |
| RSK2 | Growth-promoting regulator | ↑ | [190] |
| S642 | Growth-promoting regulator | ↑ | [189] |
| S6K | Growth-promoting regulator | ↑ | [191] |
| Separase | Growth-promoting regulator | ↑ | [192] |
| SEPT9 | Growth-promoting regulator | ↑ | [193] |
| BRD4 | Growth-promoting regulator | ↑ | [45] |
| STAT3 | Growth-promoting regulator | ↑ | [146] |
| v-Rel | Growth-promoting regulator | ↑ | [194] |
| β-catenin | Growth-promoting regulator | ↑ | [151] |
| BAX | Tumor suppressor | ↓ | [47] |
| CDK10 | Tumor suppressor | ↓ | [195] |
| CtIP | Tumor suppressor | ↓ | [107] |
| DAXX | Tumor suppressor | ↓ | [196] |
| FADD | Tumor suppressor | ↓ | [197] |
| FBXW7 | Tumor suppressor | ↓ | [198] |
| FOXO4 | Tumor suppressor | ↓ | [199] |
| IRF3 | Tumor suppressor | ↓ | [200] |
| KLF10 | Tumor suppressor | ↓ | [201] |
| PML | Tumor suppressor | ↓ | [202] |
| pRb | Tumor suppressor | ↓ | [203] |
| RARα | Tumor suppressor | ↓ | [186] |
| RUNX3 | Tumor suppressor | ↓ | [204] |
| SMRT | Tumor suppressor | ↓ | [205] |
| AMPK | Growth-inhibitory regulator | ↓ | [206] |
| ATR | Growth-inhibitory regulator | ↓ | [207] |
| AUF1 | Growth-inhibitory regulator | ↓ | [208] |
| BTK | Growth-inhibitory regulator | ↓ | [209] |
| Che1 | Growth-inhibitory regulator | ↓ | [210] |
| GRK2 | Growth-inhibitory regulator | ↓ | [211] |
| p27 | Growth-inhibitory regulator | ↓ | [212] |
| PIP4K | Growth-inhibitory regulator | ↓ | [213] |
| RBBP8 | Growth-inhibitory regulator | ↓ | [107] |
| SMAD | Growth-inhibitory regulator | ↓ | [55] |
| Smad3 | Growth-inhibitory regulator | ↓ | [214] |
| SUV39H1 | Growth-inhibitory regulator | ↓ | [215] |
| TRF1 | Growth-inhibitory regulator | ↓ | [216] |
| XPO5 | Growth-inhibitory regulator | ↓ | [217] |
AIB1: amplified in breast cancer 1; AKT: the serine/threonine protein kinase B; AMPK: AMP-activated protein kinase; AR: androgen receptor; ATR: ataxia telangiectasia and Rad3 related; BCL2: B-cell lymphoma 2; CDC25: cell division
cycle 25; CDK10: cyclin-dependent kinase 10; Cep55: centrosome protein 55; COX2: cyclooxygenase-2; ER: estrogen receptor; FAK: focal adhesion kinase; FBXW7 : F-box and WD40 repeat domain containing-7; FOXM1: forkhead box M1; FOXO4: forkhead box O4; HBX: hepatitis B virus X-protein; HER2: human epidermal growth factor receptor 2; HIF-1: hypoxia-inducible transcription factor-1; HSF1: heat shock transcription factor 1; IRAK1: interleukin-1 receptor-associated kinase 1; IRF3: interferon-regulatory factor 3; KLF10: kruppel-like factor 10; MCL1: myeloid cell leukemia-1; NF-κB: nuclear factor kappa-light-chain- enhancer of activated B cells; OCT4: octamer 4; PGK1: phosphoglycerate kinase 1; PKM2: pyruvate kinase M2; PLK: polo-like kinase; PML: promyelocytic leukemia protein; PML-RARα: promyelocytic leukemia- retinoic acid receptor alpha; pRb: retinoblastoma protein; PTP: protein tyrosine phosphatase; RARα: retinoic acid receptor alpha; RSK2: ribosomal protein S6 kinase 2; RUNX3: runt-related transcription factors 3; S6K: S6 kinase; SMRT: silencing mediator for retinoic acid and thyroid hormone receptor; STAT3: signal transducer and activator of transcription 3; XBP1: X-box-binding protein 1.
Roles of Pin1 in cancer development. Pin1 overactivation disrupts the balance between oncogenes and tumor suppressors, which affects biological behaviors related to tumor development.
Pin1 and cardiovascular diseases
Atherosclerosis (AS) is a chronic disease and the main cause of coronary heart disease, cerebral infarction, and peripheral vascular disease [48]. The early stage of AS is mainly caused by endothelial dysfunction. Endothelial nitric oxide synthetase (eNOS) plays a key role in the control of blood pressure and prevention of atherosclerosis by producing the vasodilator and vascular protective molecule nitric oxide (NO) [49]. eNOS interacts with Pin1 in a phosphorylation-dependent manner in endothelial cells. Phosphorylation of eNOS at Ser116 enhances this interaction, thus inhibiting eNOS activity and reducing NO release [50, 51]. Pin1 also drives diabetic vascular disease by causing mitochondrial oxidative stress and ROS production. Inhibition of Pin1 by gene silencing in human aortic endothelial cells (HAECs) or Pin1 knockout in mice was found to restore NO levels and relieve vascular dysfunction [52]. These results indicate that Pin1 reduces NO synthesis by inhibiting eNOS and, thus, exerts a negative effect in cardiovascular disease.
However, in some conditions, Pin1 may protect vascular endothelial homeostasis. TGF-β stimulates synthesis of proteoglycan in vascular smooth muscle cells (VSMC), especially expression of disaccharide chain protein and extension of glycosaminoglycan (GAG) chain on biglycan, which increases lipoprotein binding and promotes early inflammation of atherosclerosis [53, 54]. It has been shown that Pin1 enhances degradation of Smad2/3 ubiquitin proteasome induced by Smurf2 and inhibits TGF-β signal transduction [55], effectively preventing early occurrence of atherosclerosis [56]. Another study showed that Pin1 inhibition significantly suppresses NO production in human periodontal ligament cells (PDLCs) [57]. Taken together, Pin1 potentially plays a double-edged role in regulating the pathogenesis of cardiovascular diseases under different circumstances. Similarly, both overexpression and downregulation of Pin1 can reduce cardiac hypertrophy [58]. Further detailed investigations are needed to reveal the function of Pin1 in cardiovascular disease.
Pin1 and metabolic diseases
Insulin dysregulation is associated with various metabolic diseases including obesity, NASH, and type 2 diabetes. Pin1 promotes insulin secretion of islet β cells by enhancing the activity of SIK2, and also promotes cell proliferation and transformation by regulating activation of AP1 and ERK1/2 induced by insulin through interaction with p70S6K [59, 60]. Pin1 also positively regulates insulin-induced phosphorylation of IRS-1: Pin1 deletion inactivates IRS-1, thus leading to insulin resistance [61]. It can be concluded that Pin1 is involved in these metabolic diseases partially by controlling insulin signaling. However, Pin1 interacts with or regulates other key molecules involved in metabolic diseases, including obesity-related factors AMPK [62-65], PPARγ [66], and PRDM16 [67]; osteoporosis-related factors Runx2 [68-70] and BMP2 [71]; and Nash-related factors Smad2/Smad3 and the TGF-β1 pathway [72]. The detailed mechanisms by which Pin1 regulates metabolic diseases are summarized in other reviews [73, 74].
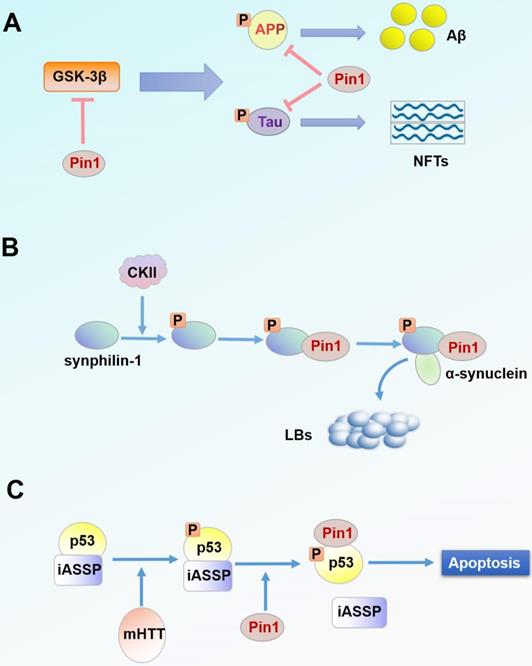
Pin1 in the pathogenesis of neurodegenerative diseases. (A) Accumulation of NFTs and Aβ is one of the pathogenic factors of AD. NFTs and Aβ are products of Tau and APP processing, respectively. Pin1 inhibits hyperphosphorylation of Tau protein and APP processing and suppresses upstream GSK-3β activity. (B) LBs are a characteristic protein polymer of PD. α-synuclein is the main component of LBs. Pin1 binds synphilin-1 phosphorylated by CKII and regulates its interaction with α-synuclein, thereby co-locating with α-synuclein intracellularly. (C) Pin1 binds and regulates p53 phosphorylated by mHTT. Subsequently, p53 is separated from the apoptosis inhibitor iASSP and is cascade activated, thus inducing neuronal apoptosis.
Pin1 and neurodegenerative diseases
Although emerging evidence has shown that Pin1 directly or indirectly regulates neuronal proteins such as Tau, amyloid precursor protein (APP), and α-synuclein, the physiological functions of Pin1 in neurodegenerative diseases remain to be elucidated. For example, in Parkinson's disease (PD) and Huntington's disease (HD), Pin1 is a pro-apoptotic factor in the process of neuronal degeneration, and high levels of Pin1 expression have been found in the brain tissue of patients [75-78]. In other studies, downregulation of Pin1 expression was found to increase the likelihood of developing Alzheimer's disease (AD), and low expression of Pin1 was found in patients with AD [74, 78-80].
Alzheimer's disease
Increased deposition of plaques and intracellular neurofibrillary tangles (NFTs) are the main mechanisms of AD pathogenesis. NFTs are microtubule aggregations produced by hyperphosphorylation of Tau protein [74]. Extracellular plaques are primarily composed of aggregates of amyloid-β-peptides (Aβ) derived from increased APP processing [74, 81, 82]. In the neuronal cells of patients with AD, Pin1 is usually underexpressed and exhibits a negative correlation with degeneration of neuronal fibers [83]. Pin1 catalyzes the conformational switch of GSK-3β-mediated phosphorylated Tau proteins from the dysfunctional cis structure to the functional trans structure, thus degrading Tau proteins [84-86]. Additionally, Pin1 catalyzes phosphorylation of APP Thr668-Pro from the cis to trans isomer and also transforms APP processing to healthy non-amyloid formation [84]. Pin1 can also directly inhibit activation of GSK-3β by binding to the phosphorylated Thr330-Pro motif of GSK-3β and catalyzing its isomerization [84, 87]. Evidence suggests that overexpression of Pin1 in mature neurons can prevent neurodegeneration caused by Tau hyperphosphorylation [79]. In general, events that decrease expression of Pin1 in the brain increase the likelihood of AD [88] (Figure 2A).
Parkinson's disease
Lewy bodies (LBs) are the characteristic protein aggregates in tissues of PD. LBs are mainly composed of α-synuclein [89, 90], which is an unfolded protein in the natural state but can be induced to form an insoluble α-synuclein aggregate in the pathological state [91, 92]. Synphilin-1 is a protein that can interact with α-synuclein; this interaction plays a very important role in the formation of LBs [93-95]. Co-expression of α-synuclein and synphilin-1 causes the formation of debris inclusion bodies in the cytoplasm [93, 96]. From immunohistochemical analysis of the brain tissue of patients with PD, Pin1 was found to be expressed in 50-60% of LBs and was co-located with α-synuclein in inclusion bodies [75]. Due to the absence of a pSer/Thr-Pro motif in α-synuclein, Pin1 cannot bind to free α-synuclein but affects α-synuclein through indirect effects [75, 97]. Under the phosphorylation mediated by casein kinase II (CKII), Pin1 binds to phosphorylated synphilin-1 through Ser211-Pro and Ser215-Pro motifs, thus indirectly interacting with α-synuclein [75]. Overexpression of Pin1 could inhibit degradation of α-synuclein, enhance the half-life and insolubility of α-synuclein, and contribute to the formation of debris inclusion bodies of α-synuclein [75] (Figure 2B). Therefore, it can be speculated that inhibitors targeting Pin1 may alleviate the process of PD.
Huntington's disease
HD is a neurodegenerative disease caused by repeated amplification of the gene encoding CAG in huntingtin protein (HTT) [98]. The mutant huntingtin protein (mHTT) forms an endonuclear inclusion by misfolding and aggregating [99, 100]. mHTT is toxic, and its aggregation causes glial proliferation of astrocytes and selective loss of striatal neurons [101, 102]. mHTT can also cause DNA damage in neurons (DDR) [103, 104], which is a significant pathological feature of HD. Studies have found that p53 mediates this cytotoxicity in HD cells and transgenic animal models, while p53 inhibitors block this process [105]. mHTT promotes phosphorylation of p53 at Ser46 through HIPK2 and PKCδ, making it a target for Pin1 binding and regulation [76]. Pin1-mediated p53 isolates from the apoptosis-inhibiting factor iASPP, thus promoting the activation cascade of p53 in striatal neurons and increasing neuronal apoptosis [76, 106] (Figure 2C). Conversely, when Pin1 is silenced, p53 binds to iASPP regardless of mHTT expression and p53 fails to induce apoptosis, thereby preventing mHTT-dependent neurodegeneration [76]. Pin1 is also associated with DDR in the regulation of DNA double-strand fracture repair [107]. In one study, DNA damage signal intensity in Pin1-knockout mice was significantly reduced by 20% compared with that of the wild-type HD mouse model [77]. However, another study revealed that Pin1 is a negative regulator of mHTT aggregation and that Pin1 overexpression reduces mHTT aggregates in HEK293 cells [108]. Nevertheless, experimental results from human neuronal cells and HD mice suggest that Pin1 is a potential therapeutic target for HD treatment.
Pin1 and viral infection
Viruses are common pathogens that cause infectious diseases. When a virus invades a host, the host activates its own immune system to fight or clear the infection [109]. But there are proteins in the host that help the virus reduce resistance from the host or promote the viral infection process. Some studies have found that Pin1 is one of these proteins and is closely related to viral infections [20, 110-119].
HIV
Acquired immunodeficiency syndrome (AIDS) is caused by human immunodeficiency virus (HIV) infection [120]. Host protein Pin1 promotes HIV infection by mediating three key processes in the HIV replication cycle [121-123] (Figure 3A).
First, the HIV core relies on Pin1 to remove capsid proteins: The HIV core is composed of ribonucleic acid (RNA) molecules and capsid protein. When HIV infects a host, it must remove the capsid and release RNA for subsequent reverse transcription, replication, and other processes [124]. The extracellular signal-regulated kinase 2 (ERK2) specifically phosphorylates Ser16-Pro17 residues on capsid proteins [124]. Pin1 then binds to the phosphorylated Ser16-Pro17 motif, which rearranges the structure of the capsid protein and removes the capsid from the HIV core [125].
Second, Pin1 facilitates reverse transcription of the HIV genome: Reverse transcription of the HIV genome is an important step in the life cycle of HIV, as DNA produced by reverse transcription can be incorporated into the host genome [126]. Host protein A3G (APOBEC3G) induces mutations in DNA during reverse transcription, which limits HIV replication [127]. But Pin1 downregulates A3G expression and prevents A3G from entering HIV [128]. HIV infection increases phosphorylation of Pin1 at Ser16 and enhances the inhibitory effect of Pin1 on A3G [111, 128].
Third, Pin1 helps integrate HIV cDNA into the host DNA: HIV needs to integrate its cDNA into the host genome to reliably transcribe its progeny RNA [129]. The cellular kinase JNK phosphorylates the pSer57 motif of HIV integrase [130]. Pin1 then binds to the phosphorylated pSer57-Pro motif to activate and stabilize HIV integrase activity, which helps it insert the HIV cDNA into the host genome [130, 131].
Roles of Pin1 in virus infection. (A) In HIV, the Ser16-Pro17 motif of the capsid protein is phosphorylated by ERK2. Pin1 specifically binds the motif and rearranges the capsid structure to release HIV RNA. Pin1 inhibits expression of catalytic polypeptide A3G to prevent incorrect coding of HIV cDNA during reverse transcription. Pin1 binds to the Ser57-Pro motif of integrase after its phosphorylation by the cellular kinase JNK, thus enhancing the stability of the integrase and promoting integration of HIV cDNA into the host cell DNA. (B) The viral protein NS5A/NS5B contains multiple phosphorylated Ser/Thr-Pro motifs. Interaction of overexpressed Pin1 with NS5A/NS5B increases HCV RNA replication and enhances HCV infection. (C) BALF5 is a key enzyme that regulates EBV DNA replication. Pin1 binds to the Thr178-Pro motif of BALF5 and actively regulates EBV DNA replication by regulating the conformation of the enzyme. Pin1 can also promote proliferation of EBV-infected nasopharyngeal carcinoma cells by upregulating expression of cyclin D1. (D) Viral oncoprotein Tax plays an important role in cell proliferation and viral replication. In HTLV-1-infected cells, Tax activates the RB/E2F pathway to increase expression of Pin1, which maintains the stability of Tax. Pin1 binds to the Ser160-Pro motif of Tax after its phosphorylation by mitotic kinase, which enhances the ability of Tax to directly bind IKKγ, activate NF-κB signaling, and finally promote cell proliferation and tumor occurrence. (E) In HPV-infected cells, overexpression of Pin1 causes NF-κB nuclear retention and activation of STAT3. Viral protein E2 can target and enhance the activity of Pin1 to increase the likelihood of cancer caused by HPV infection. (F) The viral protein HBx is a trans-activator of liver cancer. HBx activates cyclin D1 and the signaling pathway Wnt/β-catenin. Pin1 binds to the Ser41-Pro motif of HBx, which stabilizes the activity of HBx and induces overexpression of cyclin D1 and β-catenin, thus promoting liver cancer in HBV infection.
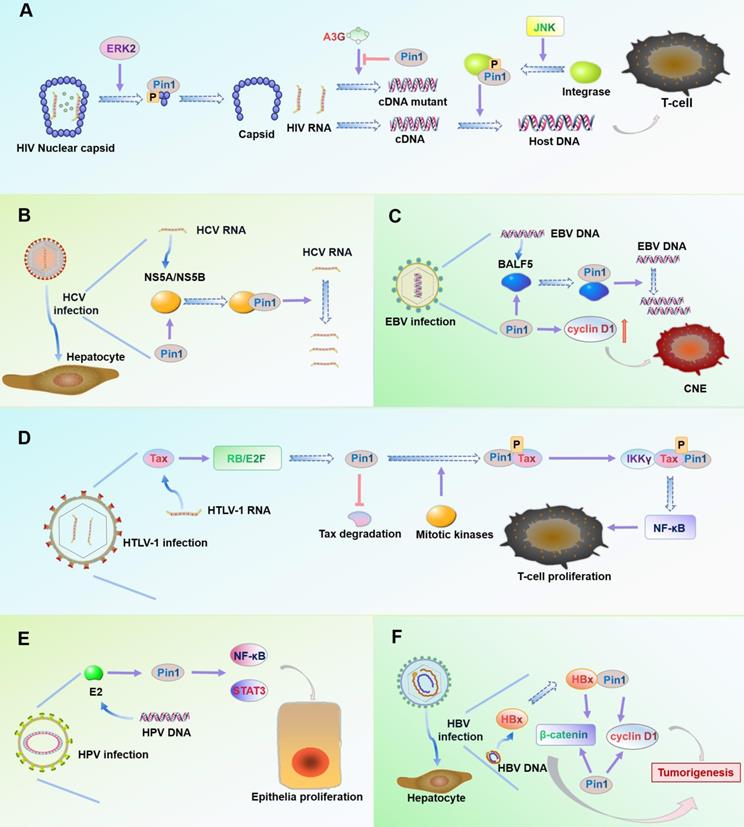
HCV
Hepatitis C virus (HCV) is the main pathogen of chronic hepatitis and hepatocellular carcinoma (HCC). HCV is an enveloped RNA virus [132]. The replication process of HCV depends mainly on the host cell cycle and requires participation of host proteins [133]. Pin1 has been shown to be a necessary cytokine for HCV replication and can increase HCV infection [114]. Overexpression of Pin1 increases intracellular HCV RNA and intracellular viral protein NS5A [114]. HCV proteins NS5A and NS5B contain phosphorylated Ser/Thr-Pro motifs and Pin1 specifically interacts with NS5A and NS5B in immunoprecipitation experiments. NS5B can also increase expression of Pin1 [114, 115]. In general, host protein Pin1 may be utilized to increase HCV replication and infection (Figure 3B).
EBV
Epstein-Barr virus (EBV) infection is associated with Burkitt lymphoma and production of T-cell malignancies [134]. BALF5, the EBV DNA polymerase subunit, is a key enzyme that affects replication during EBV cleavage [135]. Pin1 has been shown to be an important factor in regulating EBV replication. BALF5 interacts with Pin1 in a phosphorylation-dependent manner at Thr178-Pro of the BALF5 subunit. In one study, Pin1 knockdown by shRNA significantly inhibited EBV replication [119]. Another study showed that Pin1 is overexpressed in all EBV-associated nasopharyngeal carcinoma (NPC) cells, xenografts, and primary tumors [20]. Overexpression of Pin1 induces tumor cell growth by promoting production of cyclin D1 and activating the MAPK/JNK pathway (Figure 3C). The Pin1 inhibitor Juglone has been shown to inhibit growth of nasopharyngeal carcinoma cells and induce their apoptosis [20].
HTLV-1
Human T-cell leukemia virus type 1 (HTLV-1) is the pathogen that causes adult T-cell leukemia (ATL) [136]. The oncoprotein Tax encoded by HTLV-1 plays an important role in cell proliferation, viral gene replication, transformation, and tumor generation [137, 138]. Expression of Tax may cause overexpression of Pin1 in ATL [116, 118]. In cells infected by HTLV-1, Tax activates the E2F/RB pathway to increase transcription and expression of Pin1. Pin1 binds to the Tax phosphorylation motif pSer160-Pro in the presence of mitotic kinases. Pin1-regulated phosphorylated Tax then interacts with IKKγ to promote NF-κB activation [118, 139]. The activity of NF-κB plays an important role in cell transformation, cell proliferation and cancer development [140]. Pin1 can also inhibit both ubiquitination and lysosomal degradation of Tax, thus promoting its stability [116] (Figure 3D).
HR-HPV
High-risk human papillomavirus (HR-HPV) is closely related to cervical cancer, with HPV16 being the most common subtype [141]. E2 protein is a factor that regulates viral replication and transcription and can be a marker for early HPV infection [142]. Cancers caused by HR-HPV infection may be associated with activation of transcription factors NF-κB and STAT3 [143]. Overexpression of Pin1 in cervical cancer can increase nuclear retention of NF-κB and promote transactivation of STAT3, further promoting the occurrence of cancer [144-146]. In one study, increased Pin1 expression in E2-transfected HEK293 cells was found to be not significant (0.3-fold increase); however, E2 was found to enhance the activity of Pin1 [117]. This data indicates that E2 regulates activation of transcription factors NF-κB and STAT3 by targeting the activity of Pin1 [117], which further effects cancer progression (Figure 3E).
HBV
Hepatitis B virus (HBV) is a common pathogen in hepatocellular carcinoma (HCC) and HBV-encoded protein HBx is a trans-activator of liver cancer [147]. A study found that overexpression of Pin1 was most common in HBV-related HCC, and the majority of cases showed co-expression of Pin1 and HBx [113]. HBx contains two phosphorylated Ser-Pro motifs that are potential targets for Pin1 [148, 149]. Pin1 binds to the phosphorylated Ser41-Pro motif of HBx, which increases HBx stability and transactivation [113, 149]. HBx activates the oncogenic transcription factor cyclin D1 and the β-catenin signaling pathway associated with oncogenesis [150]. Overexpression of Pin1 not only increases expression of cyclin D1 but also promotes intracellular accumulation of β-catenin in the Wnt/β-catenin signaling pathway [151, 152]. These two aspects can increase expression of oncogenes and promote occurrence of HCC in HBV infection (Figure 3F).
In summary, Pin1 promotes most viral infectious diseases by two broad mechanisms: (1) Pin1 is directly involved in the life cycle of the virus to promote viral infection. For example, Pin1 is involved in core exuviation, reverse transcription, and integration of the virus in HIV infection [111]. In HCV infection, Pin1 is involved in viral RNA replication [114]. Pin1 is also involved in viral DNA replication in EBV infection [119]. (2) Pin1 enhances the stability and production of oncogenic proteins. For example, Pin1 increases the stability of Tax in HTLV-1 infection and mediates Tax transactivation of NF-κB factor [116, 118]. In HBV infection, Pin1 stabilizes the oncoprotein HBx and increases expression of oncogenic proteins cyclin D1 and catenin [112, 113]. In HPV infection, Pin1 is involved in increased activation of transcription factors STAT3 and NF-κB [117]. Pin1 also increases expression of the oncogenic protein cyclin D1 in EBV infection [20]. Tanaka et al. also found that dipentamethylene thiuram monosulfide, a specific inhibitor of Pin1, inhibited feline coronavirus (FCoV) replication [153], suggesting that targeting Pin1 may provide new insights for antiviral therapy.
Conclusions and future directions
In summary, our review illustrates the potential roles of Pin1 in several common diseases. Due to the overexpression of Pin1 in tumor tissues and its role in promoting tumor progression, drugs currently under development for targeting Pin1, including natural products, chemical compounds, and peptide drugs, are mainly focused on cancer treatment. Although Pin1 inhibitors have shown tumor suppressive effects in cell lines, animal models, and even clinical trials, some inhibitors reveal a Pin1-independent mechanism and the side effects have yet to be clarified. For example, Pin1 inhibitor KPT-6566 may exert anti-cancer effects through at least two simultaneously acting mechanisms: inhibition of Pin1 and ROS production [31]. API-1 shows significant anti-HCC activity, but its low water solubility and in vivo bioavailability limit its clinical application [154]. Researchers have also identified some compounds or peptides such as PEPTIDE [155] and benzothiophene [156, 157] that suppress Pin1 activity at nanomolar concentrations but are inactive in cell-based assays because of their poor membrane permeability. One potential countermeasure is to increase the membrane permeability of these compounds by optimizing their structure or looking for corresponding derivatives. Research on the application of Pin1 inhibitors and agonists in other related diseases is limited, and more detailed investigations need to be carried out for therapeutic potential, especially in diseases such as viral infection and AD in which the role of Pin1 is relatively clear. Studies on the upstream regulatory signals and downstream targets of Pin1 can also provide ideas for expanding treatment strategies for the above-mentioned diseases.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81772692), the Scientific Research Program for Young Talents of China National Nuclear Corporation (2020CNNC74), the Science & Technology Department of Sichuan Province (2018JY0368; 2019YJ0371), Health Commission of Sichuan Province (20PJ226), Chengdu Medical College (CYTD18-02), and Suzhou University (KJS1962).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fischer G, Bang H, Mech C. Determination of enzymatic catalysis for the cis-trans-isomerization of peptide binding in proline-containing peptides. Biomed Biochim Acta. 1984;43:1101-11
2. Fischer G, Tradler T, Zarnt T. The mode of action of peptidyl prolyl cis/trans isomerases in vivo: binding vs. catalysis. FEBS Lett. 1998;426:17-20
3. Fanghanel J, Fischer G. Insights into the catalytic mechanism of peptidyl prolyl cis/trans isomerases. Front Biosci. 2004;9:3453-78
4. Jordens J, Janssens V, Longin S, Stevens I, Martens E, Bultynck G. et al. The protein phosphatase 2A phosphatase activator is a novel peptidyl-prolyl cis/trans-isomerase. J Biol Chem. 2006;281:6349-57
5. Mueller JW, Bayer P. Small family with key contacts: par14 and par17 parvulin proteins, relatives of pin1, now emerge in biomedical research. Perspect Medicin Chem. 2008;2:11-20
6. Fischer G, Wittmann-Liebold B, Lang K, Kiefhaber T, Schmid FX. Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature. 1989;337:476-8
7. Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544-7
8. Rulten S, Thorpe J, Kay J. Identification of eukaryotic parvulin homologues: a new subfamily of peptidylprolyl cis-trans isomerases. Biochem Biophys Res Commun. 1999;259:557-62
9. Uchida T, Fujimori F, Tradler T, Fischer G, Rahfeld JU. Identification and characterization of a 14 kDa human protein as a novel parvulin-like peptidyl prolyl cis/trans isomerase. FEBS Lett. 1999;446:278-82
10. Mueller JW, Kessler D, Neumann D, Stratmann T, Papatheodorou P, Hartmann-Fatu C. et al. Characterization of novel elongated Parvulin isoforms that are ubiquitously expressed in human tissues and originate from alternative transcription initiation. BMC Mol Biol. 2006;7:9
11. Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283:1325-8
12. Zhou XZ, Lu KP. The isomerase PIN1 controls numerous cancer-driving pathways and is a unique drug target. Nat Rev Cancer. 2016;16:463-78
13. Ranganathan R, Lu KP, Hunter T, Noel JP. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell. 1997;89:875-86
14. Ryo A, Liou YC, Lu KP, Wulf G. Prolyl isomerase Pin1: a catalyst for oncogenesis and a potential therapeutic target in cancer. J Cell Sci. 2003;116:773-83
15. Wulf G, Finn G, Suizu F, Lu KP. Phosphorylation-specific prolyl isomerization: is there an underlying theme? Nat Cell Biol. 2005;7:435-41
16. Feng D, Yao J, Wang G, Li Z, Zu G, Li Y. et al. Inhibition of p66Shc-mediated mitochondrial apoptosis via targeting prolyl-isomerase Pin1 attenuates intestinal ischemia/reperfusion injury in rats. Clin Sci (Lond). 2017;131:759-73
17. Risal P, Shrestha N, Chand L, Sylvester KG, Jeong YJ. Involvement of prolyl isomerase PIN1 in the cell cycle progression and proliferation of hepatic oval cells. Pathol Res Pract. 2017;213:373-80
18. Lin CH, Li HY, Lee YC, Calkins MJ, Lee KH, Yang CN. et al. Landscape of Pin1 in the cell cycle. Exp Biol Med (Maywood). 2015;240:403-8
19. Ayala G, Wang D, Wulf G, Frolov A, Li R, Sowadski J. et al. The prolyl isomerase Pin1 is a novel prognostic marker in human prostate cancer. Cancer Res. 2003;63:6244-51
20. Xu M, Cheung CC, Chow C, Lun SW, Cheung ST, Lo KW. Overexpression of PIN1 Enhances Cancer Growth and Aggressiveness with Cyclin D1 Induction in EBV-Associated Nasopharyngeal Carcinoma. PLoS One. 2016;11:e0156833
21. Bao L, Kimzey A, Sauter G, Sowadski JM, Lu KP, Wang DG. Prevalent overexpression of prolyl isomerase Pin1 in human cancers. Am J Pathol. 2004;164:1727-37
22. Chen Y, Wu YR, Yang HY, Li XZ, Jie MM, Hu CJ. et al. Prolyl isomerase Pin1: a promoter of cancer and a target for therapy. Cell Death Dis. 2018;9:883
23. Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Petkova V. et al. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. 2001;20:3459-72
24. Shi M, Chen L, Ji J, Cai Q, Yu Y, Liu B. et al. Pin1 is overexpressed and correlates with poor prognosis in gastric cancer. Cell Biochem Biophys. 2015;71:857-64
25. Jin H, Jiang J, Sun L, Zheng F, Wu C, Peng L. et al. The prolyl isomerase Pin1 is overexpressed in human esophageal cancer. Oncol Lett. 2011;2:1191-6
26. Kim MR, Choi HK, Cho KB, Kim HS, Kang KW. Involvement of Pin1 induction in epithelial-mesenchymal transition of tamoxifen-resistant breast cancer cells. Cancer Sci. 2009;100:1834-41
27. Kanaoka R, Kushiyama A, Seno Y, Nakatsu Y, Matsunaga Y, Fukushima T. et al. Pin1 Inhibitor Juglone Exerts Anti-Oncogenic Effects on LNCaP and DU145 Cells despite the Patterns of Gene Regulation by Pin1 Differing between These Cell Lines. PLoS One. 2015;10:e0127467
28. Wang J, Liu K, Wang XF, Sun DJ. Juglone reduces growth and migration of U251 glioblastoma cells and disrupts angiogenesis. Oncol Rep. 2017;38:1959-66
29. Lian X, Lin YM, Kozono S, Herbert MK, Li X, Yuan X. et al. Pin1 inhibition exerts potent activity against acute myeloid leukemia through blocking multiple cancer-driving pathways. J Hematol Oncol. 2018;11:73
30. Liao XH, Zhang AL, Zheng M, Li MQ, Chen CP, Xu H. et al. Chemical or genetic Pin1 inhibition exerts potent anticancer activity against hepatocellular carcinoma by blocking multiple cancer-driving pathways. Sci Rep. 2017;7:43639
31. Campaner E, Rustighi A, Zannini A, Cristiani A, Piazza S, Ciani Y. et al. A covalent PIN1 inhibitor selectively targets cancer cells by a dual mechanism of action. Nat Commun. 2017;8:15772
32. Urusova DV, Shim JH, Kim DJ, Jung SK, Zykova TA, Carper A. et al. Epigallocatechin-gallate suppresses tumorigenesis by directly targeting Pin1. Cancer Prev Res (Phila). 2011;4:1366-77
33. Uchida T, Takamiya M, Takahashi M, Miyashita H, Ikeda H, Terada T. et al. Pin1 and Par14 peptidyl prolyl isomerase inhibitors block cell proliferation. Chem Biol. 2003;10:15-24
34. Potter A, Oldfield V, Nunns C, Fromont C, Ray S, Northfield CJ. et al. Discovery of cell-active phenyl-imidazole Pin1 inhibitors by structure-guided fragment evolution. Bioorg Med Chem Lett. 2010;20:6483-8
35. Potter AJ, Ray S, Gueritz L, Nunns CL, Bryant CJ, Scrace SF. et al. Structure-guided design of alpha-amino acid-derived Pin1 inhibitors. Bioorg Med Chem Lett. 2010;20:586-90
36. Pu W, Li J, Zheng Y, Shen X, Fan X, Zhou JK. et al. Targeting Pin1 by inhibitor API-1 regulates microRNA biogenesis and suppresses hepatocellular carcinoma development. Hepatology. 2018;68:547-60
37. Kozono S, Lin YM, Seo HS, Pinch B, Lian X, Qiu C. et al. Arsenic targets Pin1 and cooperates with retinoic acid to inhibit cancer-driving pathways and tumor-initiating cells. Nat Commun. 2018;9:3069
38. Pinch BJ, Doctor ZM, Nabet B, Browne CM, Seo HS, Mohardt ML. et al. Identification of a potent and selective covalent Pin1 inhibitor. Nat Chem Biol. 2020;16:979-87
39. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74
40. Shinoda K, Kuboki S, Shimizu H, Ohtsuka M, Kato A, Yoshitomi H. et al. Pin1 facilitates NF-kappaB activation and promotes tumour progression in human hepatocellular carcinoma. Br J Cancer. 2015;113:1323-31
41. Ryo A, Uemura H, Ishiguro H, Saitoh T, Yamaguchi A, Perrem K. et al. Stable suppression of tumorigenicity by Pin1-targeted RNA interference in prostate cancer. Clin Cancer Res. 2005;11:7523-31
42. Rustighi A, Tiberi L, Soldano A, Napoli M, Nuciforo P, Rosato A. et al. The prolyl-isomerase Pin1 is a Notch1 target that enhances Notch1 activation in cancer. Nat Cell Biol. 2009;11:133-42
43. Matsuura I, Chiang K N, Lai C Y, He D, Wang G, Ramkumar R. et al. Pin1 promotes transforming growth factor-beta-induced migration and invasion. J Biol Chem. 2010;285:1754-64
44. Zhu Z, Zhang H, Lang F, Liu G, Gao D, Li B. et al. Pin1 promotes prostate cancer cell proliferation and migration through activation of Wnt/beta-catenin signaling. Clin Transl Oncol. 2016;18:792-7
45. Hu X, Dong SH, Chen J, Zhou XZ, Chen R, Nair S. et al. Prolyl isomerase PIN1 regulates the stability, transcriptional activity and oncogenic potential of BRD4. Oncogene. 2017;36:5177-88
46. Cheng CW, Chow AK, Pang R, Fok EW, Kwong YL, Tse E. PIN1 inhibits apoptosis in hepatocellular carcinoma through modulation of the antiapoptotic function of survivin. Am J Pathol. 2013;182:765-75
47. Shen ZJ, Esnault S, Schinzel A, Borner C, Malter JS. The peptidyl-prolyl isomerase Pin1 facilitates cytokine-induced survival of eosinophils by suppressing Bax activation. Nat Immunol. 2009;10:257-65
48. Gimbrone MA Jr, Garcia-Cardena G. Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ Res. 2016;118:620-36
49. Forstermann U, Sessa WC. Nitric oxide synthases: regulation and function. Eur Heart J. 2012;33:829-37 37a-37d
50. Ruan L, Torres CM, Qian J, Chen F, Mintz JD, Stepp DW. et al. Pin1 prolyl isomerase regulates endothelial nitric oxide synthase. Arterioscler Thromb Vasc Biol. 2011;31:392-8
51. Li C, Ruan L, Sood SG, Papapetropoulos A, Fulton D, Venema RC. Role of eNOS phosphorylation at Ser-116 in regulation of eNOS activity in endothelial cells. Vascul Pharmacol. 2007;47:257-64
52. Paneni F, Costantino S, Castello L, Battista R, Capretti G, Chiandotto S. et al. Targeting prolyl-isomerase Pin1 prevents mitochondrial oxidative stress and vascular dysfunction: insights in patients with diabetes. Eur Heart J. 2015;36:817-28
53. Yang SN, Burch ML, Tannock LR, Evanko S, Osman N, Little PJ. Transforming growth factor-beta regulation of proteoglycan synthesis in vascular smooth muscle: contribution to lipid binding and accelerated atherosclerosis in diabetes. J Diabetes. 2010;2:233-42
54. Little PJ, Osman N, O'Brien KD. Hyperelongated biglycan: the surreptitious initiator of atherosclerosis. Curr Opin Lipidol. 2008;19:448-54
55. Nakano A, Koinuma D, Miyazawa K, Uchida T, Saitoh M, Kawabata M. et al. Pin1 down-regulates transforming growth factor-beta (TGF-beta) signaling by inducing degradation of Smad proteins. J Biol Chem. 2009;284:6109-15
56. Rostam MA, Piva TJ, Rezaei HB, Kamato D, Little PJ, Zheng W. et al. Peptidyl-prolyl isomerases: functionality and potential therapeutic targets in cardiovascular disease. Clin Exp Pharmacol Physiol. 2015;42:117-24
57. Cho YA, Jue SS, Bae WJ, Heo SH, Shin SI, Kwon IK. et al. PIN1 inhibition suppresses osteoclast differentiation and inflammatory responses. J Dent Res. 2015;94:371-80
58. Hariharan N, Sussman MA. Pin1: a molecular orchestrator in the heart. Trends Cardiovasc Med. 2014;24:256-62
59. Lee NY, Choi H-K, Shim J-H, Kang K-W, Dong Z, Choi HS. The prolyl isomerase Pin1 interacts with a ribosomal protein S6 kinase to enhance insulin-induced AP-1 activity and cellular transformation. Carcinogenesis. 2009;30:671-81
60. Nakatsu Y, Mori K, Matsunaga Y, Yamamotoya T, Ueda K, Inoue Y. et al. The prolyl isomerase Pin1 increases β-cell proliferation and enhances insulin secretion. J Biol Chem. 2017;292:11886-95
61. Nakatsu Y, Sakoda H, Kushiyama A, Zhang J, Ono H, Fujishiro M. et al. Peptidyl-prolyl cis/trans isomerase NIMA-interacting 1 associates with insulin receptor substrate-1 and enhances insulin actions and adipogenesis. J Biol Chem. 2011;286:20812-22
62. Nakatsu Y, Iwashita M, Sakoda H, Ono H, Nagata K, Matsunaga Y. et al. Prolyl isomerase Pin1 negatively regulates AMP-activated protein kinase (AMPK) by associating with the CBS domain in the γ subunit. J Biol Chem. 2015;290:24255-66
63. Steinberg GR, O'Neill HM, Dzamko NL, Galic S, Naim T, Koopman R. et al. Whole body deletion of AMP-activated protein kinase {beta}2 reduces muscle AMPK activity and exercise capacity. J Biol Chem. 2010;285:37198-209
64. O'Neill HM, Maarbjerg SJ, Crane JD, Jeppesen J, Jørgensen SB, Schertzer JD. et al. AMP-activated protein kinase (AMPK) beta1beta2 muscle null mice reveal an essential role for AMPK in maintaining mitochondrial content and glucose uptake during exercise. Proc Natl Acad Sci U S A. 2011;108:16092-7
65. Lessard SJ, Rivas DA, Chen Z-P, van Denderen BJ, Watt MJ, Koch LG. et al. Impaired skeletal muscle beta-adrenergic activation and lipolysis are associated with whole-body insulin resistance in rats bred for low intrinsic exercise capacity. Endocrinology. 2009;150:4883-91
66. Han Y, Lee SH, Bahn M, Yeo C-Y, Lee KY. Pin1 enhances adipocyte differentiation by positively regulating the transcriptional activity of PPARγ. Mol Cell Endocrinol. 2016;436:150-8
67. Nakatsu Y, Matsunaga Y, Yamamotoya T, Ueda K, Inoue M-K, Mizuno Y. et al. Prolyl Isomerase Pin1 Suppresses Thermogenic Programs in Adipocytes by Promoting Degradation of Transcriptional Co-activator PRDM16. Cell Rep. 2019;26:3221-30.e3
68. Yoon W-J, Cho Y-D, Kim W-J, Bae H-S, Islam R, Woo K-M. et al. Prolyl isomerase Pin1-mediated conformational change and subnuclear focal accumulation of Runx2 are crucial for fibroblast growth factor 2 (FGF2)-induced osteoblast differentiation. J Biol Chem. 2014;289:8828-38
69. Yoon W-J, Islam R, Cho Y-D, Woo K-M, Baek J-H, Uchida T. et al. Pin1-mediated Runx2 modification is critical for skeletal development. J Cell Physiol. 2013;228:2377-85
70. Fujita T, Azuma Y, Fukuyama R, Hattori Y, Yoshida C, Koida M. et al. Runx2 induces osteoblast and chondrocyte differentiation and enhances their migration by coupling with PI3K-Akt signaling. J Biol Chem. 2004;166:85-95
71. Yoon W-J, Islam R, Cho Y-D, Ryu K-M, Shin H-R, Woo K-M. et al. Pin1 plays a critical role as a molecular switch in canonical BMP signaling. J Cell Physiol. 2015;230:640-7
72. Yang JW, Hien TT, Lim SC, Jun DW, Choi HS, Yoon J-H. et al. Pin1 induction in the fibrotic liver and its roles in TGF-β1 expression and Smad2/3 phosphorylation. J Hepatol. 2014;60:1235-41
73. Nakatsu Y, Matsunaga Y, Yamamotoya T, Ueda K, Inoue Y, Mori K. et al. Physiological and Pathogenic Roles of Prolyl Isomerase Pin1 in Metabolic Regulations via Multiple Signal Transduction Pathway Modulations. Int J Mol Sci. 2016;17:1495
74. Bianchi M, Manco M. Pin1 Modulation in Physiological Status and Neurodegeneration. Any Contribution to the Pathogenesis of Type 3 Diabetes? Int J Mol Sci. 2018;19:2319
75. Ryo A, Togo T, Nakai T, Hirai A, Nishi M, Yamaguchi A. et al. Prolyl-isomerase Pin1 accumulates in lewy bodies of parkinson disease and facilitates formation of alpha-synuclein inclusions. J Biol Chem. 2006;281:4117-25
76. Grison A, Mantovani F, Comel A, Agostoni E, Gustincich S, Persichetti F. et al. Ser46 phosphorylation and prolyl-isomerase Pin1-mediated isomerization of p53 are key events in p53-dependent apoptosis induced by mutant huntingtin. Proc Natl Acad Sci U S A. 2011;108:17979-84
77. Agostoni E, Michelazzi S, Maurutto M, Carnemolla A, Ciani Y, Vatta P. et al. Effects of Pin1 Loss in Hdh(Q111) Knock-in Mice. Front Cell Neurosci. 2016;10:110
78. Matena A, Rehic E, Hönig D, Kamba B, Bayer P. Structure and function of the human parvulins Pin1 and Par14/17. Biol Chem. 2018;399:101-25
79. Angelucci F, Hort J. Prolyl isomerase Pin1 and neurotrophins: a loop that may determine the fate of cells in cancer and neurodegeneration. Ther Adv Med Oncol. 2017;9:59-62
80. Driver JA, Zhou XZ, Lu KP. Pin1 dysregulation helps to explain the inverse association between cancer and Alzheimer's disease. Biochim Biophys Acta. 2015;1850:2069-76
81. Harris RA, Tindale L, Cumming RC. Age-dependent metabolic dysregulation in cancer and Alzheimer's disease. Biogerontology. 2014;15:559-77
82. Zhang H, Ma Q, Zhang YW, Xu H. Proteolytic processing of Alzheimer's beta-amyloid precursor protein. J Neurochem. 2012;120(Suppl 1):9-21
83. Harris KD, Csicsvari J, Hirase H, Dragoi G, Buzsaki G. Role of the prolyl isomerase Pin1 in protecting against age-dependent neurodegeneration. Nature. 2003;424:552-6
84. Pastorino L, Sun A, Lu PJ, Zhou XZ, Balastik M, Finn G. et al. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-beta production. Nature. 2006;440:528-34
85. Lu PJ, Wulf G, Zhou XZ, Davies P, Lu KP. The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature. 1999;399:784-8
86. Driver JA, Zhou XZ, Lu KP. Regulation of protein conformation by Pin1 offers novel disease mechanisms and therapeutic approaches in Alzheimer's disease. Discov Med. 2014;17:93-9
87. Ma SL, Pastorino L, Zhou XZ, Lu KP. Prolyl isomerase Pin1 promotes amyloid precursor protein (APP) turnover by inhibiting glycogen synthase kinase-3beta (GSK3beta) activity: novel mechanism for Pin1 to protect against Alzheimer disease. J Biol Chem. 2012;287:6969-73
88. Butterfield DA, Abdul HM, Opii W, Newman SF, Joshi G, Ansari MA. et al. Pin1 in Alzheimer's disease. J Neurochem. 2006;98:1697-706
89. Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839-40
90. Lotharius J, Brundin P. Pathogenesis of Parkinson's disease: dopamine, vesicles and alpha-synuclein. Nat Rev Neurosci. 2002;3:932-42
91. Recchia A, Debetto P, Negro A, Guidolin D, Skaper SD, Giusti P. Alpha-synuclein and Parkinson's disease. FASEB J. 2004;18:617-26
92. Lundvig D, Lindersson E, Jensen PH. Pathogenic effects of alpha-synuclein aggregation. Brain Res Mol Brain Res. 2005;134:3-17
93. Engelender S, Kaminsky Z, Guo X, Sharp AH, Amaravi RK, Kleiderlein JJ. et al. Synphilin-1 associates with alpha-synuclein and promotes the formation of cytosolic inclusions. Nat Genet. 1999;22:110-4
94. Chung KK, Zhang Y, Lim KL, Tanaka Y, Huang H, Gao J. et al. Parkin ubiquitinates the alpha-synuclein-interacting protein, synphilin-1: implications for Lewy-body formation in Parkinson disease. Nat Med. 2001;7:1144-50
95. Lim KL, Chew KC, Tan JM, Wang C, Chung KK, Zhang Y. et al. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci. 2005;25:2002-9
96. O'Farrell C, Murphy DD, Petrucelli L, Singleton AB, Hussey J, Farrer M. et al. Transfected synphilin-1 forms cytoplasmic inclusions in HEK293 cells. Brain Res Mol Brain Res. 2001;97:94-102
97. Joseph JD, Yeh ES, Swenson KI. et al. The peptidyl-prolyl isomerase Pin1. Prog Cell Cycle Res. 2003;5:477-87
98. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971-83
99. Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R. et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549-58
100. Gutekunst CA, Li SH, Yi H, Mulroy JS, Kuemmerle S, Jones R. et al. Nuclear and neuropil aggregates in Huntington's disease: relationship to neuropathology. J Neurosci. 1999;19:2522-34
101. Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP Jr. Neuropathological classification of Huntington's disease. J Neuropathol Exp Neurol. 1985;44:559-77
102. Wheeler VC, Gutekunst CA, Vrbanac V, Lebel LA, Schilling G, Hersch S. et al. Early phenotypes that presage late-onset neurodegenerative disease allow testing of modifiers in Hdh CAG knock-in mice. Hum Mol Genet. 2002;11:633-40
103. Illuzzi J, Yerkes S, Parekh-Olmedo H, Kmiec EB. DNA breakage and induction of DNA damage response proteins precede the appearance of visible mutant huntingtin aggregates. J Neurosci Res. 2009;87:733-47
104. Anne SL, Saudou F, Humbert S. Phosphorylation of huntingtin by cyclin-dependent kinase 5 is induced by DNA damage and regulates wild-type and mutant huntingtin toxicity in neurons. J Neurosci. 2007;27:7318-28
105. Bae BI, Xu H, Igarashi S, Fujimuro M, Agrawal N, Taya Y. et al. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington's disease. Neuron. 2005;47:29-41
106. Mantovani F, Tocco F, Girardini J, Smith P, Gasco M, Lu X. et al. The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP. Nat Struct Mol Biol. 2007;14:912-20
107. Steger M, Murina O, Hühn D, Ferretti LP, Walser R, Hänggi K. et al. Prolyl isomerase PIN1 regulates DNA double-strand break repair by counteracting DNA end resection. Mol Cell. 2013;50:333-43
108. Carnemolla A, Michelazzi S, Agostoni E. PIN1 Modulates Huntingtin Levels and Aggregate Accumulation: An In vitro Model. Front Cell Neurosci. 2017;11:121
109. Finlay BB, McFadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124:767-82
110. Wang J, Liao XH, Zheng M, Yang D, Zhou XZ, Liu H. et al. The Roles of the Unique Prolyl Isomerase Pin1 in Cancer-Related Viral and Bacterial Infections. Curr Mol Med. 2016;16:793-802
111. Hou H, Wang JZ, Liu BG, Zhang T. Pin1 liberates the human immunodeficiency virus type-1 (HIV-1): Must we stop it? Gene. 2015;565:9-14
112. Huang L, Mo Z, Lao X, Zhang X, Liu Y, Sui J. et al. PIN1 genetic polymorphisms and the susceptibility of HBV-related hepatocellular carcinoma in a Guangxi population. Tumour Biol. 2016;37:6599-606
113. Pang R, Lee TK, Poon RT, Fan ST, Wong KB, Kwong YL. et al. Pin1 interacts with a specific serine-proline motif of hepatitis B virus X-protein to enhance hepatocarcinogenesis. Gastroenterology. 2007;132:1088-103
114. Lim YS, Tran HT, Park SJ, Yim SA, Hwang SB. Peptidyl-prolyl isomerase Pin1 is a cellular factor required for hepatitis C virus propagation. J Virol. 2011;85:8777-88
115. Li X, Jiang Y, Meisenhelder J, Yang W, Hawke DH, Zheng Y. et al. Mitochondria-Translocated PGK1 Functions as a Protein Kinase to Coordinate Glycolysis and the TCA Cycle in Tumorigenesis. Mol Cell. 2016;61:705-19
116. Jeong SJ, Ryo A, Yamamoto N. The prolyl isomerase Pin1 stabilizes the human T-cell leukemia virus type 1 (HTLV-1) Tax oncoprotein and promotes malignant transformation. Biochem Biophys Res Commun. 2009;381:294-9
117. Prabhavathy D, Vijayalakshmi R, Kanchana MP, Karunagaran D. HPV16 E2 enhances the expression of NF-kappaB and STAT3 target genes and potentiates NF-kappaB activation by inflammatory mediators. Cell Immunol. 2014;292:70-7
118. Peloponese JM Jr, Yasunaga J, Kinjo T, Watashi K, Jeang KT. Peptidylproline cis-trans-isomerase Pin1 interacts with human T-cell leukemia virus type 1 tax and modulates its activation of NF-kappaB. J Virol. 2009;83:3238-48
119. Narita Y, Murata T, Ryo A, Kawashima D, Sugimoto A, Kanda T. et al. Pin1 interacts with the Epstein-Barr virus DNA polymerase catalytic subunit and regulates viral DNA replication. J Virol. 2013;87:2120-7
120. Weiss RA. How does HIV cause AIDS? Science. 1993;260:1273-9
121. Dochi T, Nakano T, Inoue M, Takamune N, Shoji S, Sano K. et al. Phosphorylation of human immunodeficiency virus type 1 capsid protein at serine 16, required for peptidyl-prolyl isomerase-dependent uncoating, is mediated by virion-incorporated extracellular signal-regulated kinase 2. J Gen Virol. 2014;95:1156-66
122. Ott DE. Cellular proteins in HIV virions. Rev Med Virol. 1997;7:167-80
123. Ott DE, Coren LV, Johnson DG, Kane BP, Sowder RC 2nd, Kim YD. et al. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology. 2000;266:42-51
124. Fassati A. Multiple roles of the capsid protein in the early steps of HIV-1 infection. Virus Res. 2012;170:15-24
125. Misumi S, Inoue M, Dochi T, Kishimoto N, Hasegawa N, Takamune N. et al. Uncoating of human immunodeficiency virus type 1 requires prolyl isomerase Pin1. J Biol Chem. 2010;285:25185-95
126. Lehmann-Che J, Saib A. Early stages of HIV replication: how to hijack cellular functions for a successful infection. AIDS Rev. 2004;6:199-207
127. Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435:108-14
128. Watashi K, Khan M, Yedavalli VR, Yeung ML, Strebel K, Jeang KT. Human immunodeficiency virus type 1 replication and regulation of APOBEC3G by peptidyl prolyl isomerase Pin1. J Virol. 2008;82:9928-36
129. Warrilow D, Harrich D. HIV-1 replication from after cell entry to the nuclear periphery. Curr HIV Res. 2007;5:293-9
130. Manganaro L, Lusic M, Gutierrez MI, Cereseto A, Del Sal G, Giacca M. Concerted action of cellular JNK and Pin1 restricts HIV-1 genome integration to activated CD4+ T lymphocytes. Nat Med. 2010;16:329-33
131. Zhou Y, Zhang H, Siliciano JD, Siliciano RF. Kinetics of human immunodeficiency virus type 1 decay following entry into resting CD4+ T cells. J Virol. 2005;79:2199-210
132. Giannini C, Brechot C. Hepatitis C virus biology. Cell Death Differ. 2003;10(Suppl 1):S27-38
133. Scholle F, Li K, Bodola F, Ikeda M, Luxon BA, Lemon SM. Virus-host cell interactions during hepatitis C virus RNA replication: impact of polyprotein expression on the cellular transcriptome and cell cycle association with viral RNA synthesis. J Virol. 2004;78:1513-24
134. Ohga S. History of resaerch on Epstein-Barr virus-target cells of infection, and disease. Uirusu. 2014;64:67-74
135. Tsurumi T, Daikoku T, Nishiyama Y. Further characterization of the interaction between the Epstein-Barr virus DNA polymerase catalytic subunit and its accessory subunit with regard to the 3'-to-5' exonucleolytic activity and stability of initiation complex at primer terminus. J Virol. 1994;68:3354-63
136. Quaresma JA, Yoshikawa GT, Koyama RV, Dias GA, Fujihara S, Fuzii HT. HTLV-1, Immune Response and Autoimmunity. Viruses. 2015;8:5
137. Matsuoka M. Human T-cell leukemia virus type I and adult T-cell leukemia. Oncogene. 2003;22:5131-40
138. Akagi T, Ono H, Shimotohno K. Characterization of T cells immortalized by Tax1 of human T-cell leukemia virus type 1. Blood. 1995;86:4243-9
139. Chu ZL, Shin YA, Yang JM, DiDonato JA, Ballard DW. IKKgamma mediates the interaction of cellular IkappaB kinases with the tax transforming protein of human T cell leukemia virus type 1. J Biol Chem. 1999;274:15297-300
140. Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301-10
141. Bosch FX, Burchell AN, Schiffman M, Giuliano AR, de Sanjose S, Bruni L. et al. Epidemiology and natural history of human papillomavirus infections and type-specific implications in cervical neoplasia. Vaccine. 2008;26(Suppl 10):K1-16
142. Xue Y, Bellanger S, Zhang W, Lim D, Low J, Lunny D. et al. HPV16 E2 is an immediate early marker of viral infection, preceding E7 expression in precursor structures of cervical carcinoma. Cancer Res. 2010;70:5316-25
143. Tamim H, Finan RR, Sharida HE, Rashid M, Almawi WY. Cervicovaginal coinfections with human papillomavirus and Chlamydia trachomatis. Diagn Microbiol Infect Dis. 2002;43:277-81
144. Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G. et al. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell. 2003;12:1413-26
145. Shukla S, Shishodia G, Mahata S, Hedau S, Pandey A, Bhambhani S. et al. Aberrant expression and constitutive activation of STAT3 in cervical carcinogenesis: implications in high-risk human papillomavirus infection. Mol Cancer. 2010;9:282
146. Lufei C, Koh TH, Uchida T, Cao X. Pin1 is required for the Ser727 phosphorylation-dependent Stat3 activity. Oncogene. 2007;26:7656-64
147. Avila MA, Lu KP. Hepatitis B virus x protein and pin1 in liver cancer: “les liaisons dangereuses”. Gastroenterology. 2007;132:1180-3
148. Bouchard MJ, Schneider RJ. The enigmatic X gene of hepatitis B virus. J Virol. 2004;78:12725-34
149. Kim G, Kim JY, Choi HS. Peptidyl-Prolyl cis/trans Isomerase NIMA-Interacting 1 as a Therapeutic Target in Hepatocellular Carcinoma. Biol Pharm Bull. 2015;38:975-9
150. Shen L, Zhang X, Hu D, Feng T, Li H, Lu Y. et al. Hepatitis B virus X (HBx) play an anti-apoptosis role in hepatic progenitor cells by activating Wnt/beta-catenin pathway. Mol Cell Biochem. 2013;383:213-22
151. Ryo A, Nakamura M, Wulf G, Liou YC, Lu KP. Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nat Cell Biol. 2001;3:793-801
152. Liou YC, Ryo A, Huang HK, Lu PJ, Bronson R, Fujimori F. et al. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc Natl Acad Sci U S A. 2002;99:1335-40
153. Tanaka Y, Amano A, Morisaki M, Sato Y, Sasaki T. Cellular peptidyl-prolyl cis/trans isomerase Pin1 facilitates replication of feline coronavirus. Antiviral Res. 2016;126:1-7
154. Sun D, Tan S, Xiong Y, Pu W, Li J, Wei W. et al. MicroRNA Biogenesis is Enhanced by Liposome-Encapsulated Pin1 Inhibitor in Hepatocellular Carcinoma. Theranostics. 2019;9:4704-16
155. Zhang Y, Daum S, Wildemann D, Zhou XZ, Verdecia MA, Bowman ME. et al. Structural basis for high-affinity peptide inhibition of human Pin1. ACS Chem Biol. 2007;2:320-8
156. Guo C, Hou X, Dong L, Dagostino E, Greasley S, Ferre R. et al. Structure-based design of novel human Pin1 inhibitors (I). Bioorg Med Chem Lett. 2009;19:5613-6
157. Dong L, Marakovits J, Hou X, Guo C, Greasley S, Dagostino E. et al. Structure-based design of novel human Pin1 inhibitors (II). Bioorg Med Chem Lett. 2010;20:2210-4
158. Yi P, Wu RC, Sandquist J, Wong J, Tsai SY, Tsai MJ. et al. Peptidyl-prolyl isomerase 1 (Pin1) serves as a coactivator of steroid receptor by regulating the activity of phosphorylated steroid receptor coactivator 3 (SRC-3/AIB1). Mol Cell Biol. 2005;25:9687-99
159. Liao Y, Wei Y, Zhou X, Yang JY, Dai C, Chen YJ. et al. Peptidyl-prolyl cis/trans isomerase Pin1 is critical for the regulation of PKB/Akt stability and activation phosphorylation. Oncogene. 2009;28:2436-45
160. Nakatsu Y, Mori K, Matsunaga Y, Yamamotoya T, Ueda K, Inoue Y. et al. The prolyl isomerase Pin1 increases β-cell proliferation and enhances insulin secretion. J Biol Chem. 2017;292:11886-95
161. Liu T, Schneider RA, Shah V, Huang Y, Likhotvorik RI, Keshvara L. et al. Protein Never in Mitosis Gene A Interacting-1 regulates calpain activity and the degradation of cyclooxygenase-2 in endothelial cells. J Inflamm (Lond). 2009;6:20
162. Monje P, Hernández-Losa J, Lyons RJ, Castellone MD, Gutkind JS. Regulation of the transcriptional activity of c-Fos by ERK. A novel role for the prolyl isomerase PIN1. J Biol Chem. 2005;280:35081-4
163. Wang T, Liu Z, Shi F, Wang J. Pin1 modulates chemo-resistance by up-regulating FoxM1 and the involvements of Wnt/β-catenin signaling pathway in cervical cancer. Mol Cell Biochem. 2016;413:179-87
164. Ryo A, Wulf G, Lee TH, Lu KP. Pinning down HER2-ER crosstalk in SMRT regulation. Trends Biochem Sci. 2009;34:162-5
165. Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN. et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A. 2004;101:9085-90
166. Chae U, Park SJ, Kim B, Wei S, Min JS, Lee JH. et al. Critical role of XBP1 in cancer signalling is regulated by PIN1. Biochem J. 2016;473:2603-10
167. La Montagna R, Caligiuri I, Maranta P, Lucchetti C, Esposito L, Paggi MG. et al. Androgen receptor serine 81 mediates Pin1 interaction and activity. Cell Cycle. 2012;11:3415-20
168. Stukenberg PT, Kirschner MW. Pin1 acts catalytically to promote a conformational change in Cdc25. Mol Cell. 2001;7:1071-83
169. van der Horst A, Khanna KK. The peptidyl-prolyl isomerase Pin1 regulates cytokinesis through Cep55. Cancer Res. 2009;69:6651-9
170. Pani E, Menigatti M, Schubert S, Hess D, Gerrits B, Klempnauer KH. et al. Pin1 interacts with in a phosphorylation-dependent manner and regulates its transactivation activity. Biochim Biophys Acta. 2008;1783:1121-8
171. Zheng Y, Xia Y, Hawke D, Halle M, Tremblay ML, Gao X. et al. FAK phosphorylation by ERK primes ras-induced tyrosine dephosphorylation of FAK mediated by PIN1 and PTP-PEST. Mol Cell. 2009;35:11-25
172. Lonati E, Brambilla A, Milani C, Masserini M, Palestini P, Bulbarelli A. Pin1, a new player in the fate of HIF-1α degradation: an hypothetical mechanism inside vascular damage as Alzheimer's disease risk factor. Front Cell Neurosci. 2014;8:1
173. Wang HY, Fu JC, Lee YC, Lu PJ. Hyperthermia stress activates heat shock protein expression via propyl isomerase 1 regulation with heat shock factor 1. Mol Cell Biol. 2013;33:4889-99
174. Tun-Kyi A, Finn G, Greenwood A, Nowak M, Lee TH, Asara JM. et al. Essential role for the prolyl isomerase Pin1 in Toll-like receptor signaling and type I interferon-mediated immunity. Nat Immunol. 2011;12:733-41
175. Ding Q, Huo L, Yang JY, Xia W, Wei Y, Liao Y. et al. Down-regulation of myeloid cell leukemia-1 through inhibiting Erk/Pin 1 pathway by sorafenib facilitates chemosensitization in breast cancer. Cancer Res. 2008;68:6109-17
176. Moretto-Zita M, Jin H, Shen Z, Zhao T, Briggs SP, Xu Y. Phosphorylation stabilizes Nanog by promoting its interaction with Pin1. Proc Natl Acad Sci U S A. 2010;107:13312-7
177. Atkinson GP, Nozell SE, Harrison DK, Stonecypher MS, Chen D, Benveniste EN. The prolyl isomerase Pin1 regulates the NF-kappaB signaling pathway and interleukin-8 expression in glioblastoma. Oncogene. 2009;28:3735-45
178. Takahashi K, Akiyama H, Shimazaki K, Uchida C, Akiyama-Okunuki H, Tomita M. et al. Ablation of a peptidyl prolyl isomerase Pin1 from p53-null mice accelerated thymic hyperplasia by increasing the level of the intracellular form of Notch1. Oncogene. 2007;26:3835-45
179. Franciosa G, Diluvio G, Gaudio FD, Giuli MV, Palermo R, Grazioli P. et al. Prolyl-isomerase Pin1 controls Notch3 protein expression and regulates T-ALL progression. Oncogene. 2016;35:4741-51
180. van Tiel CM, Kurakula K, Koenis DS, van der Wal E, de Vries CJ. Dual function of Pin1 in NR4A nuclear receptor activation: enhanced activity of NR4As and increased Nur77 protein stability. Biochim Biophys Acta. 2012;1823:1894-904
181. Nishi M, Akutsu H, Masui S, Kondo A, Nagashima Y, Kimura H. et al. A distinct role for Pin1 in the induction and maintenance of pluripotency. J Biol Chem. 2011;286:11593-603
182. Makni-Maalej K, Boussetta T, Hurtado-Nedelec M, Belambri SA, Gougerot-Pocidalo MA, El-Benna J. The TLR7/8 agonist CL097 primes N-formyl-methionyl-leucyl-phenylalanine-stimulated NADPH oxidase activation in human neutrophils: critical role of p47phox phosphorylation and the proline isomerase Pin1. J Immunol. 2012;189:4657-65
183. Girardini JE, Napoli M, Piazza S, Rustighi A, Marotta C, Radaelli E. et al. A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell. 2011;20:79-91
184. Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F. et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14:1295-304
185. Eckerdt F, Yuan J, Saxena K, Martin B, Kappel S, Lindenau C. et al. Polo-like kinase 1-mediated phosphorylation stabilizes Pin1 by inhibiting its ubiquitination in human cells. J Biol Chem. 2005;280:36575-83
186. Gianni M, Boldetti A, Guarnaccia V, Rambaldi A, Parrella E, Raska I Jr. et al. Inhibition of the peptidyl-prolyl-isomerase Pin1 enhances the responses of acute myeloid leukemia cells to retinoic acid via stabilization of RARalpha and PML-RARalpha. Cancer Res. 2009;69:1016-26
187. Zheng Y, Yang W, Xia Y, Hawke D, Liu DX, Lu Z. Ras-induced and extracellular signal-regulated kinase 1 and 2 phosphorylation-dependent isomerization of protein tyrosine phosphatase (PTP)-PEST by PIN1 promotes FAK dephosphorylation by PTP-PEST. Mol Cell Biol. 2011;31:4258-69
188. Luo ML, Gong C, Chen CH, Hu H, Huang P, Zheng M. et al. The Rab2A GTPase promotes breast cancer stem cells and tumorigenesis via Erk signaling activation. Cell Rep. 2015;11:111-24
189. Dougherty MK, Müller J, Ritt DA, Zhou M, Zhou XZ, Copeland TD. et al. Regulation of Raf-1 by direct feedback phosphorylation. Mol Cell. 2005;17:215-24
190. Cho YS, Park SY, Kim DJ, Lee SH, Woo KM, Lee KA. et al. TPA-induced cell transformation provokes a complex formation between Pin1 and 90 kDa ribosomal protein S6 kinase 2. Mol Cell Biochem. 2012;367:85-92
191. Lee NY, Choi HK, Shim JH, Kang KW, Dong Z, Choi HS. The prolyl isomerase Pin1 interacts with a ribosomal protein S6 kinase to enhance insulin-induced AP-1 activity and cellular transformation. Carcinogenesis. 2009;30:671-81
192. Hellmuth S, Rata S, Brown A, Heidmann S, Novak B, Stemmann O. Human chromosome segregation involves multi-layered regulation of separase by the peptidyl-prolyl-isomerase Pin1. Mol Cell. 2015;58:495-506
193. Estey MP, Di Ciano-Oliveira C, Froese CD, Fung KY, Steels JD, Litchfield DW. et al. Mitotic regulation of SEPT9 protein by cyclin-dependent kinase 1 (Cdk1) and Pin1 protein is important for the completion of cytokinesis. J Biol Chem. 2013;288:30075-86
194. Fan G, Fan Y, Gupta N, Matsuura I, Liu F, Zhou XZ. et al. Peptidyl-prolyl isomerase Pin1 markedly enhances the oncogenic activity of the rel proteins in the nuclear factor-kappaB family. Cancer Res. 2009;69:4589-97
195. Khanal P, Yun HJ, Lim SC, Ahn SG, Yoon HE, Kang KW. et al. Proyl isomerase Pin1 facilitates ubiquitin-mediated degradation of cyclin-dependent kinase 10 to induce tamoxifen resistance in breast cancer cells. Oncogene. 2012;31:3845-56
196. Ryo A, Hirai A, Nishi M, Liou YC, Perrem K, Lin SC. et al. A suppressive role of the prolyl isomerase Pin1 in cellular apoptosis mediated by the death-associated protein Daxx. J Biol Chem. 2007;282:36671-81
197. Oh J, Malter JS. Pin1-FADD interactions regulate Fas-mediated apoptosis in activated eosinophils. J Immunol. 2013;190:4937-45
198. Zheng M, Xu H, Liao XH, Chen CP, Zhang AL, Lu W. et al. Inhibition of the prolyl isomerase Pin1 enhances the ability of sorafenib to induce cell death and inhibit tumor growth in hepatocellular carcinoma. Oncotarget. 2017;8:29771-84
199. Brenkman AB, de Keizer PL, van den Broek NJ, van der Groep P, van Diest PJ, van der Horst A. et al. The peptidyl-isomerase Pin1 regulates p27kip1 expression through inhibition of Forkhead box O tumor suppressors. Cancer Res. 2008;68:7597-605
200. Saitoh T, Tun-Kyi A, Ryo A, Yamamoto M, Finn G, Fujita T. et al. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat Immunol. 2006;7:598-605
201. Hwang YC, Yang CH, Lin CH, Ch'ang HJ, Chang VHS, Yu WCY. Destabilization of KLF10, a tumor suppressor, relies on thr93 phosphorylation and isomerase association. Biochim Biophys Acta. 2013;1833:3035-45
202. Reineke EL, Lam M, Liu Q, Liu Y, Stanya KJ, Chang KS. et al. Degradation of the tumor suppressor PML by Pin1 contributes to the cancer phenotype of breast cancer MDA-MB-231 cells. Mol Cell Biol. 2008;28:997-1006
203. Rizzolio F, Lucchetti C, Caligiuri I, Marchesi I, Caputo M, Klein-Szanto AJ. et al. Retinoblastoma tumor-suppressor protein phosphorylation and inactivation depend on direct interaction with Pin1. Cell Death Differ. 2012;19:1152-61
204. Nicole Tsang YH, Wu XW, Lim JS, Wee Ong C, Salto-Tellez M, Ito K. et al. Prolyl isomerase Pin1 downregulates tumor suppressor RUNX3 in breast cancer. Oncogene. 2013;32:1488-96
205. Stanya KJ, Liu Y, Means AR, Kao HY. Cdk2 and Pin1 negatively regulate the transcriptional corepressor SMRT. J Cell Biol. 2008;183:49-61
206. Ki SH, Lee JW, Lim SC, Hien TT, Im JH, Oh WK. et al. Protective effect of nectandrin B, a potent AMPK activator on neointima formation: inhibition of Pin1 expression through AMPK activation. Br J Pharmacol. 2013;168:932-45
207. Hilton BA, Li Z, Musich PR, Wang H, Cartwright BM, Serrano M. et al. ATR Plays a Direct Antiapoptotic Role at Mitochondria, which Is Regulated by Prolyl Isomerase Pin1. Mol Cell. 2016;61:487
208. Shen ZJ, Esnault S, Malter JS. The peptidyl-prolyl isomerase Pin1 regulates the stability of granulocyte-macrophage colony-stimulating factor mRNA in activated eosinophils. Nat Immunol. 2005;6:1280-7
209. Mohamed AJ, Yu L, Bäckesjö CM, Vargas L, Faryal R, Aints A. et al. Bruton's tyrosine kinase (Btk): function, regulation, and transformation with special emphasis on the PH domain. Immunol Rev. 2009;228:58-73
210. Suenaga M, Schirripa M, Cao S, Zhang W, Yang D, Murgioni S. et al. Genetic variants of DNA repair-related genes predict efficacy of TAS-102 in patients with refractory metastatic colorectal cancer. Ann Oncol. 2017;28:1015-22
211. Penela P, Rivas V, Salcedo A, Mayor F Jr. G protein-coupled receptor kinase 2 (GRK2) modulation and cell cycle progression. Proc Natl Acad Sci U S A. 2010;107:1118-23
212. Cheng CW, Leong KW, Ng YM, Kwong YL, Tse E. The peptidyl-prolyl isomerase PIN1 relieves cyclin-dependent kinase 2 (CDK2) inhibition by the CDK inhibitor p27. J Biol Chem. 2017;292:21431-41
213. Keune WJ, Jones DR, Bultsma Y, Sommer L, Zhou XZ, Lu KP. et al. Regulation of phosphatidylinositol-5-phosphate signaling by Pin1 determines sensitivity to oxidative stress. Sci Signal. 2012;5:ra86
214. Tarasewicz E, Rivas L, Hamdan R, Dokic D, Parimi V, Bernabe BP. et al. Inhibition of CDK-mediated phosphorylation of Smad3 results in decreased oncogenesis in triple negative breast cancer cells. Cell Cycle. 2014;13:3191-201
215. Khanal P, Kim G, Lim SC, Yun HJ, Lee KY, Choi HK. et al. Prolyl isomerase Pin1 negatively regulates the stability of SUV39H1 to promote tumorigenesis in breast cancer. FASEB J. 2013;27:4606-18
216. Lee TH, Tun-Kyi A, Shi R, Lim J, Soohoo C, Finn G. et al. Essential role of Pin1 in the regulation of TRF1 stability and telomere maintenance. Nat Cell Biol. 2009;11:97-105
217. Li J, Pu W, Sun HL, Zhou JK, Fan X, Zheng Y. et al. Pin1 impairs microRNA biogenesis by mediating conformation change of XPO5 in hepatocellular carcinoma. Cell Death Differ. 2018;25:1612-24
Author contact
![]() Corresponding authors: Jingyi Li, Jichun Shao and Dan He, The Second Affiliated Hospital of Chengdu Medical College, China National Nuclear Corporation 416 Hospital, Chengdu, Sichuan, China. E-mail: lijingyiedu.cn; shaoji93com; hd_baby81com.
Corresponding authors: Jingyi Li, Jichun Shao and Dan He, The Second Affiliated Hospital of Chengdu Medical College, China National Nuclear Corporation 416 Hospital, Chengdu, Sichuan, China. E-mail: lijingyiedu.cn; shaoji93com; hd_baby81com.
 Global reach, higher impact
Global reach, higher impact