13.3
Impact Factor
Theranostics 2021; 11(11):5553-5568. doi:10.7150/thno.52670 This issue Cite
Research Paper
Computational methods for cancer driver discovery: A survey
1. UniSA STEM, University of South Australia, Mawson Lakes, SA 5095, AU.
2. Centre for Cancer Biology, SA Pathology, Adelaide, SA 5000, AU.
3. Department of Medicine, The University of Adelaide, Adelaide, SA 5005, AU.
Abstract
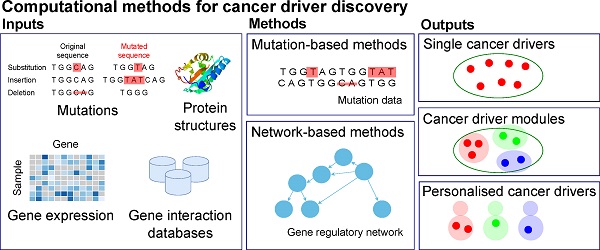
Identifying the genes responsible for driving cancer is of critical importance for directing treatment. Accordingly, multiple computational tools have been developed to facilitate this task. Due to the different methods employed by these tools, different data considered by the tools, and the rapidly evolving nature of the field, the selection of an appropriate tool for cancer driver discovery is not straightforward. This survey seeks to provide a comprehensive review of the different computational methods for discovering cancer drivers. We categorise the methods into three groups; methods for single driver identification, methods for driver module identification, and methods for identifying personalised cancer drivers. In addition to providing a “one-stop” reference of these methods, by evaluating and comparing their performance, we also provide readers the information about the different capabilities of the methods in identifying biologically significant cancer drivers. The biologically relevant information identified by these tools can be seen through the enrichment of discovered cancer drivers in GO biological processes and KEGG pathways and through our identification of a small cancer-driver cohort that is capable of stratifying patient survival.
Keywords: cancer driver, cancer driver discovery, computational method, coding gene, microRNA
 Global reach, higher impact
Global reach, higher impact