13.3
Impact Factor
Theranostics 2021; 11(18):9038-9053. doi:10.7150/thno.63416 This issue Cite
Research Paper
Mapping the landscape of synthetic lethal interactions in liver cancer
1. Department of Clinical Medicine, School of Medicine, Zhejiang University City College, Hangzhou, China.
2. State Key Laboratory of Oncogenes and Related Genes, Shanghai Cancer Institute, Renji Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China.
3. Department of Clinical Medicine, Fujian Medical University, Fuzhou, China.
4. Division of Gastroenterology and Hepatology, Key Laboratory of Gastroenterology and Hepatology, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China.
5. Hepatic Surgery Center, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China.
*These authors contributed equally to this work.
Abstract
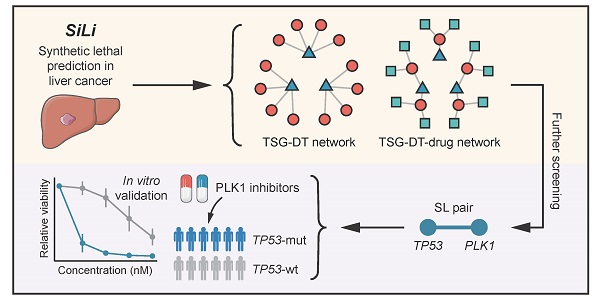
Almost all the current therapies against liver cancer are based on the “one size fits all” principle and offer only limited survival benefit. Fortunately, synthetic lethality (SL) may provide an alternate route towards individualized therapy in liver cancer. The concept that simultaneous losses of two genes are lethal to a cell while a single loss is non-lethal can be utilized to selectively eliminate tumors with genetic aberrations.
Methods: To infer liver cancer-specific SL interactions, we propose a computational pipeline termed SiLi (statistical inference-based synthetic lethality identification) that incorporates five inference procedures. Based on large-scale sequencing datasets, SiLi analysis was performed to identify SL interactions in liver cancer.
Results: By SiLi analysis, a total of 272 SL pairs were discerned, which included 209 unique target candidates. Among these, polo-like kinase 1 (PLK1) was considered to have considerable therapeutic potential. Further computational and experimental validation of the SL pair TP53-PLK1 demonstrated that inhibition of PLK1 could be a novel therapeutic strategy specifically targeting those patients with TP53-mutant liver tumors.
Conclusions: In this study, we report a comprehensive analysis of synthetic lethal interactions of liver cancer. Our findings may open new possibilities for patient-tailored therapeutic interventions in liver cancer.
Keywords: liver cancer, synthetic lethality, precision medicine, TP53, PLK1
 Global reach, higher impact
Global reach, higher impact