13.3
Impact Factor
Theranostics 2022; 12(1):87-104. doi:10.7150/thno.64096 This issue Cite
Review
The potential role of exosomal circRNAs in the tumor microenvironment: insights into cancer diagnosis and therapy
1. Key Laboratory of Environmental Health, Ministry of Education, Department of Toxicology, School of Public Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei 430030, China.
2. School of Basic Medicine, Health Science Center, Yangtze University, 1 Nanhuan Road, Jingzhou, Hubei 434023, China.
3. Department of Clinical Pharmacy and Pharmacy Administration, School of Pharmacy, Fudan University, Shanghai 201203, China.
4. Department of Pharmacology, Yong Loo Lin School of Medicine, National University of Singapore, 117600 Singapore.
5. Cancer Science Institute of Singapore, National University of Singapore, 117599 Singapore.
6. Department of Haematology-Oncology, National University Cancer Institute, Singapore 119228, Singapore.
* Juan Li and Guo Zhang contributed equally to this work.
Received 2021-6-20; Accepted 2021-10-21; Published 2022-1-1
Abstract
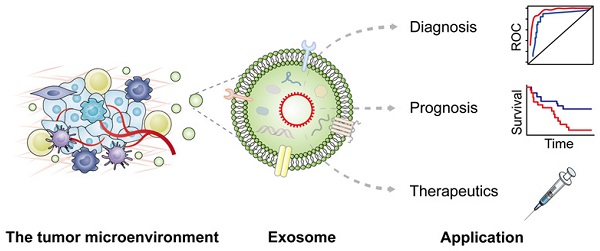
Exosomes are multifunctional regulators of intercellular communication by carrying various messages under both physiological and pathological status of cancer patients. Accumulating studies have identified the presence of circular RNAs (circRNAs) in exosomes with crucial regulatory roles in diverse pathophysiological processes. Exosomal circRNAs derived from donor cells can modulate crosstalk with recipient cells locally or remotely to enhance cancer development and propagation, and play crucial roles in the tumor microenvironment (TME), leading to significant enhancement of tumor immunity, metabolism, angiogenesis, drug resistance, epithelial mesenchymal transition (EMT), invasion and metastasis. In this review, we describe the advances of exosomal circRNAs and their roles in modulating cancer hallmarks, especially those in the TME. Moreover, clinical application potential of exosomal circRNAs in cancer diagnosis and therapy are highlighted, bridging the gap between basic knowledge and clinical practice.
Keywords: Exosome, circular RNAs, tumor microenvironment, diagnosis, cancer therapy.
Background
Recently, exosome biology and its clinical significance in oncology have attracted increasing attention. Exosomes are small extracellular vesicles (sEVs) that originate from intraluminal vesicles (ILVs) in the endosomal system, and are secreted from various cells upon fusion of multivesicular bodies with the plasma membrane [1]. As such, exosomes can be detected in various bodily fluids, including blood, urine, saliva, and breast milk, thereby carrying a wide repertoire of cellular components from their parental cells, including proteins, lipids, DNAs, mRNAs, and noncoding RNAs (ncRNAs) [2]. These exosomal cargoes can be delivered from their parental cells to recipient cells [3, 4]. Exosome-mediated interplays between cancer cells and surrounding cells remodel the tumor microenvironment (TME), thereby creating favorable conditions for cancer progression and metastasis [5, 6].
Circular RNAs (circRNAs), which are abundant and become more stable in exosomes, can be transferred to neighboring or distant cells where they exert their functions [7]. Exosomal circRNAs originating from cancer cells can be delivered to the recipient cells where they play vital roles in cancer cell initiation, progression, and proliferation, invasion and metastasis, as well as drug resistance [8]. The relative stability, high abundance, and conserved nature across species of exosomal circRNAs make them potential biomarkers for various diseases, including cancers. Previous reports have illustrated the functions of various circRNAs in the TME, a complicated ecosystem incorporating the coevolution of both tumor cells and the surrounding stroma [9, 10]. Dynamic alterations in the oncogenic components, including circRNAs and other TME features such as acidosis and hypoxia, play a crucial role in tumorigenesis and metastasis [11]. In recent years, the focus on cancer therapy has shifted away from targeting malignant cancer cells to mediating the TME to interfere their complicated crosstalk. Hence, the TME has been considered a promising therapeutic target for cancer treatment [12].
Several reviews have discussed the roles of exosomal circRNAs in human diseases, including cancer [13-15], which focused on the functions and potential applications in different cancer types. Nevertheless, the role of exosome-derived circRNAs in regulating the TME has not been systematically summarized. Currently, targeting vital components of the TME is very attractive research areas for cancer therapy [10]. Understanding the exosomal circRNA-mediated TME network may boost the development of therapies for diverse cancers. Herein, we conducted a comprehensive literature search to focus on latest research progress in the role of exosomal circRNAs in regulating the TME through affecting tumor immunity, tumor progression and metastasis, angiogenesis, drug resistance, and tumor metabolism. Furthermore, potential clinical applications of exosomal circRNAs as cancer biomarkers or therapeutic targets in cancer therapy have been discussed.
Emerging roles of exosomes and circRNAs in cancer biology
Characteristics of exosomes
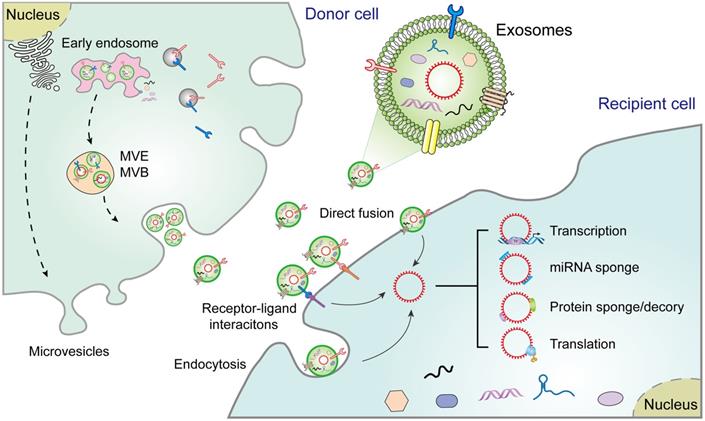
The biogenesis of EVs, including exosomes, microvesicles, and apoptotic bodies, is relatively well documented [16]. Exosomes, the sEVs in diameter (40-150 nm), are secreted from diverse cell types [17]. Their biogenesis begins with the formation of endosomes via endocytosis of the plasma membrane, followed by maturation of early endosomes into multivesicular endosomes/bodies (MVEs and MVBs). Exosomes are intraluminal vesicles (ILVs) produced within the endolysosomal system and shaped by the inward budding of the limiting membrane of MVEs/MVBs. Exosome formation involves a range of ordered molecular machineries, the most important of which is the endosomal sorting complexes required for transport (ESCRT), which is a key driver of membrane formation and scission, thus playing a crucial role in the formation of ILVs and MVEs (Figure 1) [3]. Although exosomes were originally considered to be "garbage bags", functioning to extrude obsolete components from the cells, evidence suggests that they are in fact crucial regulators of the physiological or pathological processes by transferring biomolecules to recipient cells [3]. For instance, once released, exosomes may act directly on neighboring, or distant recipient cells via various mechanisms, including ligand/receptor interaction, direct membrane fusion, and endocytosis, thus orchestrating multiple cellular processes in recipient cells [4]. Alternatively, exosomes may merge with the membrane of a recipient cell and transfer their constituents into its cytosol [16]. However, although exosomes share common characteristics with progenitor cells, they also contain distinct content that makes them unique [18].
Exosomal circRNAs: emerging players in the TME
CircRNAs, a type of endogenous ncRNAs, are produced by a particular form of alternative splicing called back-splicing [19]. CircRNAs were first discovered in viroids in 1976, and later considered as by-products of splicing errors [20]. However, recent studies have highlighted the underlying importance of circRNAs while also characterizing their biogenesis and functions. CircRNAs have high abundance, conservation stability, and prevalence. Interestingly, unlike other RNAs, the absence of 5′ caps and 3′ tails allows for circRNAs to resist RNases and be more stable than linear RNAs. Hence, circRNAs may aggregate in cells to cause pathologies such as cancer progression [21]. Therefore, circRNAs may become valuable diagnostic biomarkers or potential therapeutic targets for cancer treatment. Exosomes have been found to contain abundant diverse RNA species (e.g., circRNAs), and exosomal circRNAs contribute to tumorigenesis and progression via diverse mechanisms, including transcription, miRNA sponge, protein sponge/decoy, and translation (Figure 1). Therefore, studies involved in exosomal circRNAs have become a hotspot in intercellular communication in the TME, which is a complicated scaffold of stromal cells, extracellular matrix (ECM) components, and exosomes [22]. The stromal cells include endothelial cells, pericytes, cancer-associated fibroblasts (CAFs), and immune cells, such as various types of lymphocytes, regulatory T cells (Treg), natural killer (NK) cells, tumor-associated macrophages (TAMs), and matrix metalloproteinases (MMPS), myeloid-derived suppressor cells chemokines, integrins, and other secreted molecules [22, 23]. The tumor-derived exosomes (TDEs) contain various signaling molecules that mediate intercellular crosstalk and reprogramming of the TME [24]. Thus, a thorough understanding of the phenotypic and functional characteristics in the TME may provide insights into novel treatment strategies for a diverse range of cancers.
The TME orchestrates cell-cell interactions through a various signaling networks, such as paracrine and juxtracrine interactions [23]. Exosomes are excreted by nearly all paracrine cell types commonly detected in the TME [25, 26], and transfer diverse biomolecules including circRNAs, which participate in intercellular interactions when they are released and delivered into recipient cells [4]. Some studies have illustrated the potential role of exosomal circRNAs in cancer progression, angiogenesis, chemoresistance and metabolism through regulating the TME [27-29].
Functions of exosomal circRNAs in the TME
Certain innovative methods for investigating the biological foundations of exosome transport have been developed to monitor the dynamic communication of exosomes in different models [30-33]. However, current isolation techniques of exosomes, including centrifugation, filtration, polymeric precipitation, and immunoaffinity separation [34, 35], often cause variations in downstream analysis results. Hence, there remains an urgent need to develop effective strategies for functional identification of exosomal cargoes (e.g., circRNAs) in different cancers.
In the TME, exosomes mediate the interplay between immune cells and tumor cells [36, 37]. The TDEs play pivotal roles in the establishment of pre-metastatic niches, immune escape, tumor immune microenvironment, immune suppression, and immune surveillance [38]. Notably, circRNAs can specifically interact with tumor-specific miRNAs or mRNAs in exosomes, functioning as new tumor antigens for mediating immune responses [39, 40]. In the following sections, we focus on the expanding landscape of exosomal circRNAs in the TME, including tumor immunity, angiogenesis, drug resistance, EMT, tumor cell migration, tumor metastasis, as well as tumor metabolism (Table 1 and Figure 2).
Extracellular vesicle biogenesis and secretion in donor cells and their functions in recipient cells. Exosomes participate in cell-to-cell communication via transferring diverse molecules, including exosomal circRNAs, new players in cancer diagnosis and therapy. Exosomes contribute to the interplay between donor cells and recipient cells through a variety of pathways, including cell-cell interactions, receptor-ligand interactions, and endocytosis. MVE: multivesicular endosome, MVB: Multivesicular body.
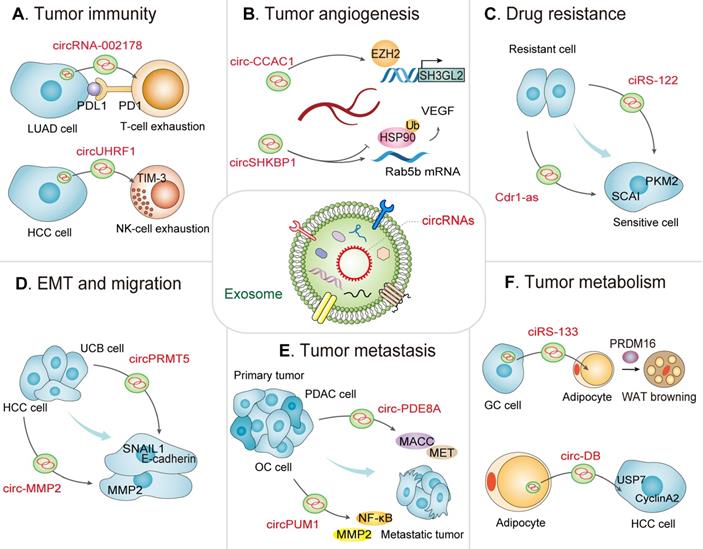
Exosomal circRNA-mediated intercellular crosstalk in the TME. A view of the molecular crosstalk pathways involving exosomal circRNAs in the TME, including (A) tumor immunity, (B) tumor angiogenesis, (C) drug resistance, (D) EMT and migration, (E) tumor metastasis, and (F) tumor metabolism.
The emerging roles of exosomal circRNAs in the TME.
| Exosomal circRNA | Source cell | Cancer types | Expression | Molecular axis | Functions | Ref. |
|---|---|---|---|---|---|---|
| Exosomal circRNAs and tumor immunity | ||||||
| circRNA-002178 | Lung adenocarcinoma cells | Lung adenocarcinoma | Up | circRNA-002178/miR-34/PDL1/PD1 | Immune escape, T-cell exhaustion | [47] |
| circ-0001068 | Ovarian cancer | Ovarian cancer | Up | circ-0001068/miR-28-5p /PD1 | Immune escape, induce PD1 expression | [48] |
| circ-CPA4 | Non-small cell lung cancer cells | Non-small cell lung cancer | Up | circ-CPA4/let-7 miRNA/PD-L1 | Immune escape | [49] |
| circUHRF1 | Hepatocellular carcinoma cells | Hepatocellular carcinoma | Up | circUHRF1/miR-449c-5p/TIM-3/anti-PD1 | NK cell exhaustion | [27] |
| circ-PDE8A (hsa_circ_0036627) | Pancreatic ductal adenocarcinoma | Pancreatic ductal adenocarcinoma | Up | circ-PDE8A/miR-338/MACC1/MET | Lymphatic invasion and tumor progression, tumor infiltrating lymphocytes | [60] |
| circPACRGL | Colorectal cancer | Colorectal cancer | Up | circPACRGL/miR-142-3p/miR-506-3p-TGF-β1 | Promote proliferation, migration and invasion, differentiation of N1 to N2 neutrophils. | [61] |
| hsa-circ-0048117 | Hypoxia pre-challenged esophageal squamous cell carcinoma cells | Esophageal squamous cell carcinoma | Up | hsa-circ-0048117/miR-140 | Promote M2 macrophage polarization | [52] |
| circFARSA | Non-small cell lung cancer | Non-small cell lung cancer | Up | circFARSA/PTEN/PI3K/AKT/eIF4A3 | Promote M2 macrophage polarization | [53] |
| Exosomal circRNAs and tumor angiogenesis | ||||||
| circ-CCAC1 | Cholangiocarcinoma | Cholangiocarcinoma | Up | circ-CCAC1/miR-514a-5p/ YY1 | Tumor angiogenesis by interacting with epigenetic regulators | [28] |
| circSHKBP1(hsa_circ_0000936) | Gastric cancer | Gastric cancer | Up | circSHKBP1/miR-582-3p/HUR/VEGF | Angiogenesis by acting as RBPs and miRNA "sponges" | [67] |
| circ-RanGAP1 | Gastric cancer | Gastric cancer | Up | circ-RanGAP1/miR-877-3p/VEGFA | Regulate the expression of VEGFA by acting as specific miRNA “sponges” | [68] |
| circFNDC3B | Colorectal cancer | Colorectal cancer | Down | circFNDC3B/miR-97-5p/TIMP3 | Inhibit angiogenesis and progression. | [69] |
| circ-IARS | Pancreatic cancer | Pancreatic cancer | Up | circRNA IARS /miR-122/RhoA /F-actin | Elevate endothelial cell permeability, facilitate tumor metastasis and invasion. | [71] |
| circRNA-100,338 | Hepatocellular carcinoma cells | Hepatocellular carcinoma | Up | circRNA-100,338/ NOVA2 | Influence the permeability, angiogenesis and proliferation | [72] |
| Exosomal circRNAs and therapeutic resistance | ||||||
| ciRS-122 | Oxaliplatin-resistant colorectal cancer cells | Colorectal cancer | Up | ciRS-122/miR-122/PKM2 | Promote glycolysis to induce chemoresistance | [77] |
| circ_0000338 | FOLFOX-resistant colorectal cancer cells | Colorectal cancer | Up | —— | Induce chemoresistance | [78] |
| circNFIX | Temozolomide-Resistant glioma cells | Glioma | Up | circNFIX/miR-132. | Enhance temozolomide resistance, diagnostic biomarker (AUC 0.885) and prognostic biomarker | [29] |
| circRNA-SORE | Sorafenib-resistant hepatocellular carcinoma cells | Hepatocellular carcinoma | Up | circRNA-SORE/YBX1/PRP19 | Spread sorafenib resistance | [79] |
| Cdr1as | Cisplatin-resistant ovarian cancer | Ovarian cancer | Down | Cdr1as/miR-1270/SCAI | Suppress cisplatin resistance | [80]. |
| hsa_circ_0014235 | Non-small cell lung cancer | Non-small cell lung cancer | Up | circ_0014235/miR-520a-5p/CDK4 | Promote cisplatin resistance | [81] |
| mc-COX2 | Chronic lymphocytic leukemia | Chronic lymphocytic leukemia | Up | —— | Strengthen drug resistance | [82] |
| Exosomal circRNAs and migration and epithelial mesenchymal transition (EMT) | ||||||
| circPRMT5 | Urothelial carcinoma of the bladder | Urothelial carcinoma of the bladder | Up | circPRMT5/miR-30c/SNAIL1/E-cadherin | Epithelial mesenchymal transition (EMT), biomarker of metastasis | [85] |
| circ_MMP2 (hsa_circ_0039411) | Hepatocellular carcinoma cells | Hepatocellular carcinoma | Up | circ_MMP2/miR-136-5p/MMP2 | Enhance the EMT process and invasion | [86] |
| circIFT80 | colorectal cancer | Colorectal cancer | Up | circIFT80/miR-1236-3p/HOXB7 | Promote growth, proliferation, migration, and invasion via EMT | [87] |
| circRNA_100284 | Arsenite-transformed human hepatic epithelial cells | Carcinogenesis induced by Arsenic | Up | circRNA_100284/microRNA-217 | Accelerate the cell cycle and promote cell proliferation | [88] |
| circ-0000284 | Cholangiocarcinoma cells | Cholangiocarcinoma | Up | circ-0000284/miR-637/LY6E | Promote proliferation and migration, suppress apoptosis | [89] |
| circRASSF2 | Laryngeal squamous cell carcinoma | Laryngeal squamous cell carcinoma | Up | circRASSF2 /miR-302b-3p/IGF-1R | Enhance proliferation, migration, and invasion | [90] |
| Exosomal circRNAs and tumor metastasis | ||||||
| circ-PDE8A (hsa_circ_0036627) | Pancreatic ductal adenocarcinoma | Pancreatic ductal adenocarcinoma | Up | circ-PDE8A/miR-338/MACC1/MET | Lymphatic invasion and tumor progression, tumor infiltrating lymphocytes | [60]. |
| circPUM1 | Ovarian cancer | Ovarian cancer | Up | circPUM1/miR-615-5p/ miR-6753-5p/NF-κB/ MMP2 | Act on peritoneal mesothelial cells and promote metastasis | [92] |
| circNRIP1 | Gastric cancer | Gastric cancer | Up | circNRIP1/miR-149-5p/AKT1/mTOR | Promote EMT and metastasis | [93] |
| circPTGR1 | Hepatocellular carcinoma | Hepatocellular carcinoma | Up | circPTGR1/miR449a/MET | Promote metastasis and invasion | [95] |
| circWHSC1 | Ovarian cancer | Ovarian cancer | Up | circWHSC1/miR-145 /miR-1182/MUC1 /hTERT | Induce tumor metastasis through acting on peritoneal mesothelium | [96] |
| Exosomal circRNAs and tumor metabolism | ||||||
| circ-DB | Adipocytes | Hepatocellular carcinoma | Up | circ-DB/miR-34a/USP7/Cyclin A2 | Promote tumorigenesis and metastasis | [109] |
| ciRS-133 (circ-0010522) | Gastric cancer cells | Gastric cancer | Up | ciRS-133/miR-133/ PRDM16 | Promote white adipose browning, aggravate tumor cachexia and increase oxygen consumption | [110] |
| circ-MEMO1 | Non-small cell lung cancer | Non-small cell lung cancer | Up | circ-MEMO1/miR-101-3p/KRAS | Facilitate progression and glycolysis | [112] |
| circ-133 | Hypoxic colorectal cancer cells | Colorectal cancer | Up | circ-133 /miR-133a/GEF-H1/RhoA | Promote tumor metastasis and migration | [119] |
| circHIF1A | Hypoxic cancer-associated fibroblasts | Breast cancer | Up | circHIF1A/miR-580-5p/CD44 | Promote breast cancer cell proliferation and stemness | [121] |
| circSLC7A6 | Cancer-associated fibroblasts | Colorectal cancer | Up | circSLC7A6/ CXCR5 | Promote colorectal cancer cell proliferation and invasion | [122] |
Exosomal circRNAs regulate tumor immunity
As the richest cellular components of the TME, immune cells have been reported as promising targets for their anti-cancer cytotoxic capabilities [41]. Interactions among immune cells, cancer cells, and exosomes associated intercellular communication are important in tumor immunoregulation, which can create immunosuppressive environments fostering cancer development and progression [42, 43]. Hence, identifying key regulators within this interplay may provide candidates for therapeutic interventions. Recently, dramatic dysregulation of many exosomal circRNAs in the TME were observed in diverse cancer types [8]. Additionally, some exosomal circRNAs regulate immune cells including lymphocytes, macrophage, NK cells, and dendritic cells (DCs), thereby providing evidence for the functional roles of exosomal circRNAs in immune regulation.
As a major component of adaptive immunity, the majority of T lymphocytes (also called T cells) exhausted in the TME induce tumor immune escape [44]. Programmed death ligand 1 (PD-L1, CD274), known as an essential immune checkpoint protein, is a crucial constituent of tumor immunosuppression that binds to programmed death 1 (PD-1) on T-cells [45]. The ability of PD-L1/PD-1 to suppress T cell activation and to augment the immune tolerance of tumor cells is crucial to achieve tumor immune escape [46]. Interestingly, multiple observations have exhibited that exosomal circRNAs severed as crucial regulators of immune escape. A recent study detected plasma exosomal circRNA-002178 in lung adenocarcinoma (LUAD) patients and suggested that circRNA-002178 facilitates PD-L1 expression by acting as miR-34 sponge in LUAD, thus inducing T-cell exhaustion. Therefore, circRNA-002178 could be attractive as a promising noninvasive biomarker for the early detection of LUAD, and could enhance PD-L1 expression to mediate interactions between cancer cells and T cells (Figure 2A) [47]. Similarly, exosomal circ-0001068 is transferred into T cells, where it caused an upregulation of PD1 expression by sponging miR-28-5p in ovarian cancer (OC) [48]. Furthermore, Hong et al. demonstrated that non-small-cell lung cancer (NSCLC) cells release PD-L1-containing exosomes that boost cell stemness and upregulate the resistance of NSCLC cells to cisplatin [49]. NSCLC cells also induce the inactivation of CD8+ T cells in a delivered PD-L1-dependent manner. Furthermore, this finding indicates that circ-CPA4 positively mediates exosomal PD-L1, while NSCLC cells with circ-CPA4 deletion exhibit reactivation of CD8+ T cells in the co-culturing system. Therefore, deciphering exosomal circRNA-induced suppressive mechanisms of T cell activation will accelerate the discovery and development of novel immune checkpoint inhibitors for targeting complement-mediated immunoregulation.
Macrophages, the most prevalent immune cells in the TME, are an integral factor that hinders tumor progression and influences the antitumor immunity response [50]. Exosomes can act as a communicator connecting tumor cells and macrophages [51]. For instance, exosomal hsa-circ-0048117 has been reported to be upregulated in esophageal squamous cell carcinoma (ESCC). Exosomal hsa-circ-0048117 could be delivered to macrophages to induce M2 macrophage polarization, thereby remodeling the hypoxic microenvironment and regulating ESCC progression [52]. In the TME, tumors are able to recruit and induce macrophages to develop into tumor-associated macrophages (TAMs) that facilitate tumor progression. For example, circFARSA is elevated in NSCLC cells and associated with the elevation of EMT and metastasis. Moreover, circFARSA derived from NSCLC cells induces tumor TAMs polarization to M2 phenotype by promoting PTEN ubiquitination and subsequent degradation, thereby activating the PI3K/AKT signaling pathway [53].
NK cells are the first line of defense for host immune surveillance and, as such exert pivotal effects on antitumor immunotherapy. Due to the fact that NK cells have no major histocompatibility complex limitations on the recognition and damage of target cells, accumulating immunostimulants can be created to enhance cell killing [54]. Moreover, the density and activity of NK cells in the TME are correlated with prognoses of various cancers [55]. NK cell-derived exosomal circRNAs also serve as crucial players in the innate antitumor immune response. For instance, Zhang et al. recently showed that exosomal circUHRF1 secreted by hepatocellular carcinoma (HCC) cells induce NK cell exhaustion, thus mediating resistance to anti-PD1 therapy. Additionally, high expression of circUHRF1 in plasma exosomes correlates with a reduced proportion of NK cells as well as NK cell tumor infiltration. Moreover, HCC-derived exosomal circUHRF1 is transferred into NK cells, sponging miR-449c-5p to upregulate TIM-3 expression in turn inducing NK cell exhaustion, thereby favoring immunosuppression and resistance to anti-PD1 immunotherapy in HCC, offering a therapeutic target for patients with HCC (Figure 2A) [27].
Dysregulation of tumor infiltrating lymphocytes (TILs) and neutrophils in the TME is associated with overall survival [56, 57]. Pancreatic ductal adenocarcinoma (PDAC) is one of the most invasive and fatal forms of carcinomas with a low 5-year survival rate of 5%, due to of a high-risk of metastasis and recurrence [58, 59]. Nevertheless, breakthroughs have been made in exosome-mediated circRNAs crosstalk in PDAC. For instance, Li et al. reported that PDAC cell-derived exosomal circ-PDE8A is related to lymphatic invasion and tumor progression, thereby activating the MACC/MET/ERK or AKT pathways by sponging miR-338 [60]. Intriguingly, another recent study showed that high expression of cancer-derived exosomal circPACRGL promotes colorectal cancer (CRC) proliferation and metastasis. Functional assays revealed that circPACRGL sponges miR-142-3p/miR-506-3p to promote transforming growth factor-β1 (TGF-β1) expression, thereby contributing to CRC cell proliferation, migration and invasion, and differentiation of N1 to N2 neutrophils [61]. Currently, studies on the precise mechanisms of exosomal circRNAs in regulating immune cells are still in their infancy and more work is needed for further investigation to discover novel therapeutic targets.
Exosomal circRNAs mediate tumor angiogenesis
Angiogenesis is a complex process by which tumors not only obtain adequate nutritional support but also remove metabolic waste and carbon dioxide [11]. Overexpression of diverse angiogenic factors, as well as rapid growth of tumor cells in the TME, contribute to the development of vascular networks, thus leading to diverse structural and functional abnormalities [62, 63]. Vascular endothelial growth factor (VEGF), a potent angiogenic factor, plays an essential role in both physiological and pathological angiogenesis [64]. Meanwhile, cancer-associated endothelial cells are a significant component of the tumor stroma in the TME, that supports tumor neovasculature and blood vessel formation [70]. Furthermore, certain inhibitors (such as bevacizumab) can markedly decrease the expression of angiogenic factors (VEGF), thereby inhibiting angiogenesis [65]. Therefore, advancing our current understanding of the cellular and molecular mechanisms associated with tumor angiogenesis will enable the development of novel anti-angiogenic therapies [66].
Exosomal circRNAs can affect tumor angiogenesis by interacting with epigenetic regulators, thereby remodeling the TME to induce cancer initiation and progression. For instance, a recent study demonstrated that circ-CCAC1 expression is significantly upregulated in cancerous bile-resident EVs and tissues. Further analysis uncovered that cholangiocarcinoma (CCA)-derived EVs circ-CCAC1 is delivered to endothelial monolayer cells, thus destroying endothelial barrier integrity and promoting angiogenesis. Mechanistically, circ-CCAC1 upregulates cell leakiness via sequestering EZH2 in the cytoplasm, thereby increasing SH3GL2 expression to decrease intercellular junction proteins expression. Furthermore, in vivo studies revealed that elevated circ-CCAC1 levels in circulating EVs and cells facilitated both CCA tumorigenesis and metastasis (Figure 2B) [28].
Exosomal circRNAs also regulate angiogenesis by simultaneously acting as miRNA and RNA-binding protein (RBP) "sponges". For instance, current data provided by Xie et al. indicates that exosomes with elevated circSHKBP1 facilitate growth of cocultured gastric cancer (GC) cells. Mechanistically, circSHKBP1 acts as a competing endogenous RNA (ceRNA) for miR-582-3p to upregulate the expression of HUR, augmenting VEGF mRNA stability. Furthermore, circSHKBP1 directly interact with HSP90, thereby impeding the interaction between HSP90 and STUB1, subsequently suppressing ubiquitination of HSP90, and giving rise to the acceleration of GC development in vivo and in vitro. This study confirmed that exosomal circSHKBP1 modulated the miR-582-3p/HUR/VEGF axis to inhibit HSP90 degradation and facilitated the progression of GC (Figure 2B) [67]. Likely, Lu et al. revealed that circ-RanGAP1 is significantly upregulated in plasma exosomes of preoperative GC patients and mediates the miR-877-3p/VEGFA axis, thereby promoting the migration and invasion of GC [68]. Another study indicated that circFNDC3B-enriched exosomes can suppress CRC angiogenesis and liver metastasis via miR-97-5p/TIMP3 axis [69].
Exosome-mediated transfer of circRNAs can also modulate the permeability of endothelial cells, facilitating the dissemination and metastasis of cancer cells. Tumor metastasis is the major risk factor for cancer-associated death. Tumor endothelial cells obtain their specific characteristics in the TME, provoking the metastasis of tumor cells. Endothelial cells particularly serve as crucial regulators in the original stage of tumor metastasis [70]. Recent studies have elucidated the molecular mechanism by which exosomal circRNAs derived from tumor cells are involved in the regulation of the permeability of endothelial cells in the TME. For example, the pancreatic cancer (PC) cell-derived exosomal circRNA IARS can enter the human microvascular vein endothelial cells (HUVECs) via plasma exosomes and destroy the endothelial tight junctions, thus elevating endothelial cell permeability and facilitating the establishment of a microenvironment suitable for tumor invasion and metastasis. Further analysis revealed that the molecular axis of circRNA IARS/miR-122/RhoA/F-actin was responsible for these functions. Therefore, the behavior of exosomal circRNAs provides new insights into prospective endothelial monolayer permeability and contributes to preventing early tumor cell metastasis via suppressing endothelial cell permeability, suggesting potential targets for anti-angiogenesis strategy [71].
In addition to VEGF, other angiogenic factors are also regulated by exosome-delivered circRNAs, which either directly or indirectly affect tumor angiogenesis. A recent study reported that the upregulated exosomal circRNA-100338 in metastatic HCC cells, which are transported from HCC cells to HUVECs, influenced the permeability, angiogenesis, and proliferation of recipient HUVECs. Mechanistically, internalized circRNA can directly interact with NOVA2, a RBP, thereby mediating vascular development and achieving cell-to-cell communication [72].
Currently, certain clinical trials with antiangiogenic strategies failed to show clinical benefit in a subset of cancers, the overall outcome was not as promising as initially hoped for improved anticancer therapy. Current studies have demonstrated that exosomal circRNAs manipulate angiogenesis via different mechanisms to establish a favorable microenvironment, thereby providing dynamic information about the pathologic state of the exosome-producing cells. Targeting exosomal circRNAs from different cell types of the TME could be used as alternatives to the existing antiangiogenic therapies.
Exosomal circRNAs modulate drug resistance
Drug resistance is closely related to the TME. Exosomes, as key mediators of intercellular communication, are involved in drug resistance. Tumor and stromal cells in the TME can secrete drug-resistant exosomes containing distinct biomacromolecules including circRNAs. As a research hotspot in recent years, circRNAs secreted by exosomes exert multifunctional roles in the transfer of molecules related to drug resistance from primary cancer cells to recipient cells [6]. Accumulating evidence has shown that the emerging function and mechanistic axis of circRNAs are related to drug resistance. Intrinsic and extrinsic factors can exert an influence on cancer cell resistance to chemoradiation [73]. The extrinsic factors such as exosomal circRNAs in the TME can facilitate chemoradiation resistance and tumor recurrence, including the ECM, hypoxia, and the expression of angiogenic markers (VEGF and HIF1α) [74, 75]. As a result, the roles of exosomal circRNAs in the drug resistance could lead to the identification of novel therapeutic targets to develop more effective therapeutic agents for cancer treatment. Li. et al. observed that circRNA FLI1 exonic circular RNAs (FECRs) derived from serum exosomes was abnormally elevated in small cell lung cancer (SCLC) in comparison with control groups and was closely associated with the clinical response to chemotherapy and poor survival in SCLC patients. Mechanistically, FLI1 exonic circular RNAs (FECRs) act as a new oncogenic driver via the miR-584-ROCK1 pathway, thus determining the metastatic phenotype in SCLC [76]. This suggests a possible connection between exosomal circRNAs dysregulation and drug resistance. Moreover, exosomal circRNAs may function as intercellular signaling molecules to transfer drug-resistance properties from drug-resistant CRC cells to sensitive cells. A recent investigation identified ciRS-122 to be positively associated with chemoresistance in CRC. Exosomal ciRS-122 is delivered from chemoresistant cells to recipient cells. Further, ciRS-122 can function as a miR-122 sponge to elevate pyruvate kinase M2 (PKM2), thereby accelerating glycolysis and enhancing the resistance of sensitive CRC cells to oxaliplatin. Therefore, ciRS-122 might be a therapeutic target for the therapy of drug-resistant CRC (Figure 2C) [77]. Regarding chemoresistance in CRC, Hon et al. reported that serum exosomal hsa_circ_0000338 derived from fluorouracil/oxaliplatin/leucovorin (FOLFOX)-resistant CRC cells was elevated in comparison with levels in sensitive cell-derived exosomes. Further investigation showed that hsa_ circ_0000338 knockdown elevated the viability of FOLFOX-resistant CRC cells following exposure to 5-fluorouracil (5-FU) [78]. Similarly, another recent study on gliomas showed that exosomal circNFIX was highly expressed in the serum of temozolomide (TMZ)-resistant patients and was correlated with poor prognosis. Exosomal circNFIX is transferred from TMZ-resistant cells to targetable sensitive cells via the augmentation of cell migration and invasion as well as the suppression of cell apoptosis under TMZ exposure. Moreover, exosomal circNFIX promoted tumor growth, and its depletion elevated TMZ sensitivity in glioma cells by increasing miR-132 in vivo [29].
Moreover, exosomal circRNA-SORE is elevated in sorafenib-resistant HCC cells, while its depletion enhances the cell-killing ability of sorafenib. Mechanistically, circRNA-SORE interacts with YBX1, a master oncogenic protein that, inhibits the interplay between YBX1 and E3 ubiquitin ligase PRP19 thus impeding PRP19-induced YBX1 degradation. Furthermore, silencing of circRNA-SORE via siRNA can overcome sorafenib resistance in HCC mouse models [79]. Zhao et al. revealed that Cdr1as is decreased in serum exosomes of cisplatin-resistant patients and mediates the miR-1270/SCAI axis, thereby increasing the sensitivity of OC cells to cisplatin (Figure 2C) [80]. Research involving cisplatin resistance in NSCLC has recently revealed that hsa_circ_0014235 in exosomes derived from the serum is markedly upregulated in NSCLC and facilitates cisplatin chemoresistance, while modulating the miR-520a-5p/CDK4 pathway, thus increasing NSCLC development [81]. Regarding drug resistance in chronic lymphocytic leukemia (CLL), Wu et al. found that CLL-derived exosomal mc-COX2 (mitochondrial genome-derived circRNAs [mc]) is highly expressed in plasma and is positively correlated with leukemogenesis and the deteriorating survival of CLL patients. The endogenous decrease of mc-COX2 can influence mitochondrial functions, suppressing cell proliferation and inducing cell apoptosis [82].
The aformentioned studies show that cancer-associated cells are reprogrammed in the TME by utilizing various exosomal circRNAs to induce resistance of tumor cells to anticancer drugs or radiotherapy. Hence, these identified exosomal circRNAs may also be potential therapeutic targets in overcoming drug resistance, enhancing the clinical benefits in patients.
Exosomal circRNAs induce EMT and tumor cell migration
Accumulating evidence suggests that certain exosomal circRNAs play a diverse range of roles in EMT. Activation of EMT under pathological conditions plays crucial roles in tumor development and the initiation of metastasis and is involved in the procedure of transforming epithelial cells into mesenchymal cells, leading to tumor cell migration [83, 84]. Here are some examples regarding exosomal circRNAs-induced aberrant EMT activation, tumor cell motility and invasiveness. Chen. et al. revealed that serum and urine exosomal circPRMT5 was aberrantly upregulated in patients with urothelial carcinoma of the bladder (UCB). Further studies revealed that circPRMT5 could sponge miR-30c to promote UCB cell EMT and enhance the expression of its target genes E-cadherin and SNAIL1, thus enabling the cells to be more invasive (Figure 2D) [85]. Similarly, 97H-derived exosomal circ_MMP2 (hsa_circ_0039411) could be transmitted into L02 and HepG2 cells and its overexpression was correlated with a poor overall survival of HCC patients. Mechanistically, circ_MMP2 enrichment can abundantly sponge miR-136-5p to upregulate the expression of its host gene matrix metallopeptidase 2 (MMP2), thus effectively enhancing the EMT process and invasive abilities of HCC cells (Figure 2D) [86]. Feng et al. reported that circIFT80 expression was highly elevated in serum exosomes derived from CRC tissues and cells compared with that in exosomes derived from the normal counterparts. Functional assays revealed that circIFT80 sponges miR-1236-3p to facilitate HOXB7 expression, thus promoting CRC cell growth, proliferation, migration, and invasion via aberrant activation of EMT transcription factors [87].
Accumulating evidence also suggests that message transmission is crucial for tumor formation, and it is likely that exosomal circRNAs function as intercellular modulators in the process of carcinogenesis. For example, exosomal circRNA-100284 was transferred from malignant transformed cells into surrounding normal cells, leading to the malignant transformation of the non-transformed cells induced by arsenite. Functional assays revealed that exosomal circRNA_100284 sponges miR-217 to increase the expression of enhancer of zeste homolog 2 (EZH2) and cyclin D1, thus accelerating the cell cycle progression and promoting the proliferation of recipient cells [88]. A study reported that plasma exosomal circ-0000284 was upregulated in cholangiocarcinoma (CCA) patients compared to that in healthy controls. CCA cell-derived exosomal circ-0000284 was transported into surrounding cells, thus promoting the proliferation and migration of recipient cells and suppressing apoptosis via the miR-637/lymphocyte antigen 6 complex locus E (LY6E) axis [89]. Moreover, Tian et al. confirmed that serum exosome-derived circRASSF2 levels were abnormally elevated in laryngeal squamous cell carcinoma (LSCC) patients. circRASSF2 sponges miR-302b-3p to affect IGF-1R expression, promoting progression of LSCC [90].
Since exosomal circRNAs can be delivered into recipient cells and induce functional responses and phenotypic changes in cancers, it is important to design and conduct more experiments to confirm the functional role and underlying mechanisms of action of exosomal circRNAs in the TME to discover novel noninvasive prospective biomarkers and therapeutic targets for cancers.
Exosomal circRNAs mediate tumor metastasis
Tumor onset and metastasis are correlated with multiple oncogenes and involve multiple oncogenic signaling pathways [91]. Exosomal circRNAs play important roles in cancer cell-to-cell crosstalk and the surrounding stroma, contributing to metastasis. Li et al. first reported that exosomal circ-PDE8A levels were highly elevated in PDAC tissue and were significantly correlated with higher tumor-node-metastasis (TNM) stages, lymphatic invasion, and a poor survival rate. Furthermore, circ-PDE8A facilitated tumor cell growth by increasing the expression of MET (a tyrosine kinase receptor), encoded by one of the vital oncogenes for a subclass of epithelial tumors, containing PDA. Importantly, tumor-derived exosomal circ-PDE8A could be delivered into blood circulation, thereby mediating MACC1 and tumor metastasis by sponging miR-338 by the MACC/MET/ERK or AKT axis (Figure 2E) [60]. Guan et al. reported that circPUM1 could sponge miR-615-5p and miR-6753-5p and elevate the expression of NF-κB and MMP2, thus acting on peritoneal mesothelial cells and promoting OC metastasis (Figure 2E) [92]. Similarly, another study confirmed that exosomal circNRIP1 could be transported between GC cells, thus promoting cell proliferation, migration, and invasion by sponging miR-149-5p to activate the AKT1/mTOR signaling pathway [93]. These findings were also confirmed in breast cancer [94].
Exosomal circRNAs are known to exert critical effects on the development and metastasis of HCC. Wang et al. showed that the upregulation of serum exosomal circPTGR1 from patients with HCC was significantly linked to the clinical stage and prognosis. Further analysis revealed that exosomal circPTGR1 knockdown significantly inhibited tumor migration and invasion of non- and low-metastatic cell lines. Mechanistically, exosomal circPTGR1 can promote the development of HCC by sponging miR-449a [95]. Another study indicated that the elevated level of circWHSC1 in OC facilitated tumorigenesis by serving as miR-145 and miR-1182 sponge and the presence of these miRNAs in exosomes promoted tumor metastasis by acting on the peritoneal mesothelium. Functional assays revealed that exosomal circWHSC1 can be transmitted to peritoneal mesothelial cells to facilitate peritoneal dissemination by upregulating the expression of mucin 1 (MUC1) in recipient cells, thus promoting OC progression [96]. Taken together, the above-mentioned findings have corroborated the extensive and pivotal role of exosomal circRNAs in tumor metastasis by regulating TME. Therefore, exosomal circRNAs may act as potential therapeutic targets for management of tumor metastasis and provide new directions for cancer therapy. Further research on exosomal circRNAs is warranted to deepen our understanding of their detailed mechanisms of action in vivo as well as their potential clinical applications.
Exosomal circRNAs affect tumor metabolism
Since reprogramming energy metabolism is regarded as a new hallmark of cancer, tumor metabolism has attracted increasing attention in cancer research. To date, several metabolic interventions, including those associated with fatty acid oxidation and the respiratory complex I inhibitor, metformin [97, 98], have been successfully combined with immunotherapeutic agents to modulate anticancer effects [99]. Growing knowledge regarding the metabolism of tumor cells in the TME has facilitated the design of novel therapies that are safe and effective for cancer patients [100]. In fact, multiple therapeutic agents have been shown to affect tumor cell metabolism, glutaminolysis aerobic glycolysis, the adipose tissue microenvironment, as well as other metabolic targets [101-104]. New therapeutic strategies are also being developed to target metabolism of stromal and immune cells [105, 106].
As a key example of the metabolic interplay, adipocytes can offer nutrients such as alanine and lipids to the TME, which support malignant cells [107, 108]. For instance, Zhang et al. reported that the adipocyte-derived exocirc-deubiquitination (exo-circ-DB) can sponge miR-34a to activate the USP7/CyclinA2 signaling pathway, thus boosting HCC tumorigenesis and metastasis while reducing DNA damage (Figure 2F) [109]. Zhang et al. revealed that plasma-derived exosomal ciRS-133 (circ-0010522) levels are elevated in GC patients and are closely related to the browning of white adipose tissue and cancer-related cachexia. Further studies reported that exosomal ciRS-133 is delivered into preadipocytes, where it inhibits miR-133 and activates PR domain containing protein 16 (PRDM16), thus facilitating white adipose browning, aggravating tumor cachexia, and increasing oxygen consumption in GC patients, thereby highlighting the crucial role of exosomal circRNAs in the pathologic process (Figure 2F) [110].
Aerobic glycolysis is a hallmark of tumor cells, which allows them to meet their high demand for energy required for tumor growth and maintenance [111]. Meanwhile, exosomal circ-MEMO1 promotes the progression and glycolytic metabolism of NSCLC by modulating the miR-101-3p/KRAS axis [112]. Exosomal circRNAs also regulate tumor metabolism by inducing cell death, such as autophagy. Zhang et al. recently identified that exosomal circNRIP1 can be transferred between GC cells and promote EMT and tumor metastasis in vivo. Moreover, the exosomal circNRIP1/AKT1/mTOR axis can promote GC metastasis by altering metabolism and autophagy [93].
Tumor cells and stromal cells in the TME generally restrict the entry of nutrients and oxygen, inducing a hypoxic environment [113]. Accumulating evidence suggests that the hypoxic TME is closely involved in multiple "hallmarks of cancer" [114, 115]. Accordingly, studies are underway to assess hypoxia-regulated cancer biology, including exosome-mediated crosstalk in the TME. The TDEs, which are abundant in the TME, transport tumoral signaling to both stromal cells and tumor cells and exert an essential influence on numerous pathological scenarios [116]. As transcriptional regulators, HIFs acts as a pivotal regulator in mediating the proliferation and metastasis of tumor cells and the responses of the TME via activating the transcription of downstream oncogenes, including hypoxia-responsive elements, and mediating diverse signaling pathways [117, 118]. The exosomal circRNA-mediated cell interplay within hypoxic TME has been widely studied. Recently, Yang et al. reported that hypoxic cell-derived exosomal circ-133 is transferred into normoxic cells and regulates the E-cadherin membrane distribution through activation of the miR-133a/GEF-H1/RhoA signaling pathway, thus promoting CRC metastasis [119].
Carcinoma-associated fibroblasts (CAFs) plays a pivotal role in the TME at depositing and remodeling ECM [120]. Hypoxia is one of the key components in the TME that greatly affects fibroblast activation. For instance, a recent study demonstrated that circHIF1A (circ_0032138) expression is significantly elevated in the exosomes from hypoxic CAFs. Further analysis uncovered that exosomal circHIF1A from hypoxic CAFs could promote breast cancer stemness through miR-580-5p/CD44 axis [121]. Another study found that exosomal circSLC7A6 from CAFs could promote CRC progression. Additionally, matrine suppressed CRC tumorigenesis by modulating CXCR5 and blocking the secretion of exosomal circSLC7A6 [122]. These findings indicated that the specific role of CAFs in tumor progression could be a promising therapeutic strategy for the development of anti-tumor therapy.
Together, exosomal circRNAs execute a broad repertoire of functions to influence hypoxia and adipocyte-related regulators in the TME. As the TME contains different cell types with flexible metabolic programming, well-designed experiment will be required to unveil the metabolic demands, adaptations, and interactions among various cancer cells. Given that metabolic changes related to the TME contribute to cancer development and progression, targeting metabolic dysfunction through the use of different strategies may reshape the TME to improve efficacy of oncology drugs.
Potential clinical applications of exosomal circRNAs in cancer therapy
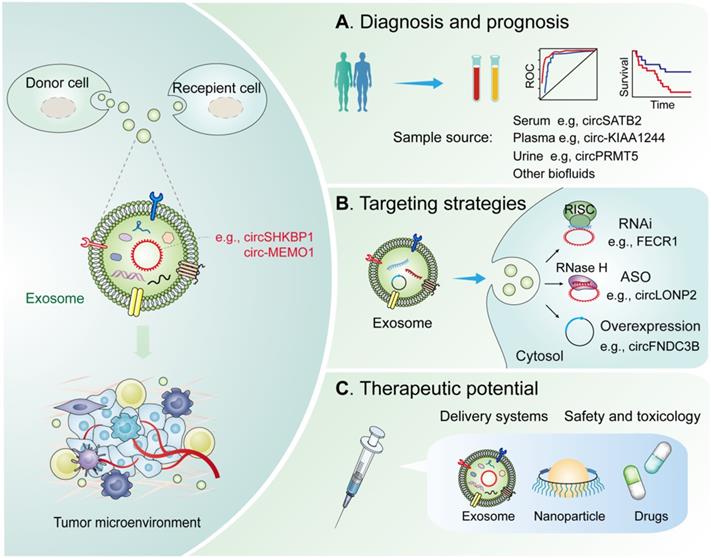
Characterizing the underlying mechanism associated with exosomal circRNAs in suitable models could provide further evidence for their potential clinical application in cancer therapy. Current research has established that exosomal circRNAs exhibit great potential as potential biomarkers and therapeutic targets in cancer treatment. The application of biomarkers in cancer therapy will allow oncologists to better optimize therapeutic strategies for cancer patients and practice precision medicine in clinical settings. In the following sections, we discuss how exosomal circRNAs could be utilized as potential biomarkers for diagnosis, prognosis, and treatment monitoring for different cancer types (Figure 3).
Potential clinical applications of exosomal circRNAs in the TME. Diagnostic and therapeutic potential of exosomal circRNAs in the TME. (A) Diagnosis and prognosis. These exosomal circRNAs can be measured from liquid biopsy samples and are novel noninvasive diagnostic and prognostic biomarkers. (B) Targeting strategies. Knockdown or overexpression technologies can be used to target circRNAs in the cytoplasm and nucleus. (C) Therapeutics. Exosomal circRNAs can be delivered via nanoparticles or antibody/receptor conjugates in vivo. Abbreviation: RNAi, RNA interference; ASO, antisense oligonucleotides; RISC, RNA-induced silencing complex; RNase H, Ribonuclease H.
Diagnostic potential of exosomal circRNAs
Accumulating evidence indicates that multiple circRNAs involved in the regulation of the TME are dysregulated in diverse cancers [123, 124]. Exosomal circRNAs are critical regulators of healthy and diseased states and may represent valuable biomarkers for diagnoses and prognoses of multiple cancers (Figure 3A).
The use of exosomal circRNAs as potential biomarkers in lung cancer has been proposed. For instance, Li et al. reported that serum Friend leukemia virus integration 1 (FLI1) is aberrantly upregulated in SCLC and is inextricably linked to poor survival and the clinical response to chemotherapy [76]. Similarly, Xian et al. reported that circ_0007761, circ_0056285, and circ_0047921 can be used for diagnosis of NSCLC. In differentiating between NSCLC cases and healthy controls, a panel of the above-mentioned circRNAs possessed an AUC of 0.919 (95% CI, 0.877-0.962) in the validation set and an AUC of 0.926 (95% CI, 0.895-0.956) in the training set. Moreover, circ_0047921 can effectively discriminate between NSCLC patients from chronic obstructive pulmonary disease (COPD) controls (AUC, 0.890; 95% CI, 0.831-0.948), whereas the combination of circ_0056285 and circ_0007761 could differentiate between NSCLC patients and tuberculosis controls (AUC, 0.820; 95% CI, 0.739-0.902). Similar findings were found for these circRNAs in distinguishing early-stage NSCLC patients from COPD controls, tuberculosis controls, or healthy controls for early diagnosis of NSCLC. Further, circ_0056285 expression was linked to lymph node metastasis and clinical stage [125]. Zhang et al. showed that serum exosomal circSATB2 was highly elevated in lung cancer patients with high specificity and sensitivity for clinical detection. Interestingly, exosomal, circSATB2 is correlated well with lung cancer metastasis, suggesting it as a prognostic biomarker for NSCLC [126].
Exosomal circRNAs as valuable biomarkers for the diagnosis and prognosis of GC have been reported. For example, Shao et al. reported that the plasma exosome-derived hsa_circ_0065149 expression levels are reduced in early-stage GC patients. Moreover, exosomal hsa_circ_0065149 expression has higher sensitivity and specificity in early-stage GC screening compared to the traditional clinical biomarkers [127]. More importantly, exosomal circ-KIAA1244 may be stable in plasma due to the protection of exosomes from degradation. Hence, exosomal circ-KIAA1244 derived from GC might be an exploitable circulating biomarker for GC screening [128].
CircRNAs associated with TDEs might represent promising biomarkers in CRC as well. For instance, the expression of serum exosomal hsa_circ_0004771 is markedly elevated in CRC patients, compared with that in healthy individuals and benign intestinal disease (BID) patients, and is closely associated with cancer metastasis and TNM stage in CRC patients. Moreover, the area under the ROC curves (AUCs) of circulating exosomal hsa-circ-0004771 can effectively differentiate benign intestinal diseases (BIDs), stage I/II CRC patients, and CRC patients from healthy controls. Notably, the AUC value that differentiate stage I/II CRC patients from patients with BIDs was 0.816 (95% CI, 0.728-0.9) [129]. Additionally, serum exosomal circFMN2 levels have been shown to be significantly upregulated in CRC patients, contributing to the progression of CRC via the miR-1182/hTERT signaling pathway [130]. Other studies have indicated that numerous circRNAs are markedly downregulated in KRAS mutant cells and can be transmitted by exosomes secreted by tumor cells [131]. In CRC cell lines, many circRNAs were also detected in secreted EVs including exosomes, indicating that EV or exosomal circRNAs may act as potential cancer biomarkers [131].
Recently, studies have reported that exosomal circRNAs are also potential diagnostic and prognostic biomarkers for HCC patients. For example, plasma exosomal hsa_circ_0051443 is expressed at lower levels in HCC patients compared to healthy controls. Normal cell-derived exosomal hsa_circ_0051443 is transmitted into HCC cells and suppresses malignant characteristics, thereby modulating intercellular crosstalk during HCC carcinogenesis [132]. Analogously, in another report, Zhong et al. identified eight extracellular vesicle long RNAs (exLRs), including mRNA, circRNA, and lncRNA, that are potential biomarkers with high diagnostic performance in the testing cohort (AUC, 0.9627; 95% CI, 0.9263-0.9991), the training cohort [AUC, 0.9527; 95% CI, 0.9170-0.9883], and the validation cohort (AUC, 0.9825; 95% CI, 0.9606-1).Thus, numerous exLRs in human plasma are capable of serving as potential biomarkers for diagnosis of cancers [133].
Currently, certain exosomal circRNAs have been confirmed as biomarkers in papillary thyroid carcinoma (PTC). For instance, Wu et al. reported that serum exosomal levels of circRASSF2 and circ_0006156 (circFNDC3B) are elevated in PTC patients and mediate PTC progression via the miR-1178/TLR4 pathway, thereby highlighting exosomal circRNAs as biomarkers for diagnosis, prognosis and treatment of PTC [134, 135].
To date, multiple diagnostic clinical trials are underway to validate the utility of emerging exosome-based biomarkers for diagnosis and prognosis as well as treatment prediction in various types of cancer. For example, RNA profiling from circulating exosomes can be used as a biomarker for lung metastases of primary high-grade osteosarcoma (NCT03108677). For updated and complete information on exosomal circRNA clinical trials, please refer to ClinicaTrials.gov database.
Together, exosomal circRNAs as valuable biomarkers for diagnosis, prognosis and treatment of diverse cancers provide insights into the normal or pathological state of circRNA-producing cells. Due to their high stability, it is important to validate exosomal circRNAs in serum/plasma or other biofluids as biomarkers for diagnosis, prognosis, and treatment prediction of cancers. However, the main limitation of exosomal circRNAs as cancer biomarkers is that the utility of these potential biomarkers has not yet been validated with large sample size in multi-centre trails. Given the heterogeneity of tumor-associated circRNAs in different human populations, it is difficult to compare results between study groups from populations in different geographical locations. Therefore, well designed clinical trials should be conducted to validate diagnostic and prognostic value of these exosomal circRNAs in a wide range of cancers.
Strategies to target exosomal circRNAs
Considering the crucial biological functions of exosomal circRNAs in tumor progression and metastasis, as the exosomal circRNAs may prove to be promising diagnostic and therapeutic options in cancer therapy. Hence, many ongoing studies have been designed to either regulate exosome production or block exosome uptake pathways. An increasing amount of research has emphasized that circRNAs may represent oncogenic drivers or tumor-suppressive mediators in a variety of cancers [136, 137]. Currently, studies have focused on the clinical utility of circRNAs as therapeutic targets of cancer [136, 138]. Effective techniques for gene overexpression or knockdown may provide clues for the therapeutic targeting of circRNAs.
RNA-based therapeutic strategies primarily include RNA interference (RNAi) and antisense oligonucleotides (ASOs), which can be designed to target a large and heterogeneous class of transcripts (Figure 3B) [139]. For oncogenic circRNAs, specific shRNAs or siRNAs targeting the back-splice junction, which can be interfered with by homologous linear mRNA expression, were utilized to reach circRNA-specific knockdown [140]. In terms of intron circularized circRNAs, researchers have carefully designed complementary paired siRNAs to target intron region sequences to destroy RNA formation, inducing circRNA knockdown [141]. Serum exosomal FECR1 is a novel oncogenic circRNA in small cell lung cancer, which targeting it with shRNA can inhibit tumor metastasis in nude mice and improve overall survival [76]. ASOs have shown promise in targeting of ncRNAs in patients with cancers. For example, circLONP2 is increased in metastasis-initiating CRC subgroups, acting as a key metastasis driver of CRC. Meanwhile, targeting circLONP2 by ASOs restricts metastasis to foreign organs, thereby functioning as a promising anti-metastatic therapeutic approach [142]. For tumor-suppressive circRNAs, overexpression vectors fostering back-splicing consist of flanking introns with reverse complementary sequences and circRNA-forming exons [143]. Recent preclinical studies have highlighted protein-coding genes derived functional circRNAs might provide a new repertoire of candidates for RNA-based therapeutics such as circRNAs. LoVo cell-derived exosomes with circFNDC3B overexpression have been used to treat CRC in animal model [69]. Engineered exosome-based therapy has been applied in a phase I clinical trial for the treatment of pancreatic cancer patients with KrasG12D mutation (NCT03608631). Together, more well-designed circRNA-specific therapies are expected in the near future.
Engineered exosomes are designed for clinical applications by artificially optimizing a combination of specific cargo, including RNA-based therapy and tumor drugs. As they deliver specific bioactive molecules identified in the membrane of their progenitor cells, exosomes can be improved through the application of specific factors capable of targeting their transport to the TME. For instance, immature DC-derived exosomes modified with tumor targeting ligands such as the αv integrin-specific iRGD peptide can be employed for the transportation of DOX to tumors. Therefore, this method represents a promising strategy for targeted tumor therapy [144]. A previous report has confirmed that the transmission of miRNA or siRNA payloads through exosomes is a prospective clinical tool in exosome-mediated therapies for various cancer treatments [5]. Together, exosomal circRNAs (e.g., circ-MEMO1) might be a promising therapeutic target, while targeting circRNAs might be a novel therapeutic approach for cancer therapy. Development of more advanced technology that allows for the clinical applications of RNA-based therapy for cancer treatment remains warranted.
Therapeutic potential of exosomal circRNAs
Current studies have identified exosomes as delivery tools for circRNA-targeting agents and circRNA expression vectors (Figure 3C) [145, 146]. Using exosomes, one study has delivered a siRNA targeting ciRS-122, ultimately inhibiting CRC glycolysis and reverse resistance to oxaliplatin in mice [77]. Another example used exosomes as delivery tools for circRNA expression vector showed that delivery of engineered rabies virus glycoprotein-circSCMH1-EVs promoted functional recovery in both and nonhuman primate models [147]. Therefore, exosome engineering is a prospective tool in circRNA-based therapeutic strategy with clinical application; however, certain limitations such as the limited efficacy of the exosome route of injection or the quantity of exosomes administered, cell line employed, or the site of infusion remain to be addressed [148]. Therefore, new strategies are necessary to advance the clinical potential of exosome-based therapeutics.
Exosomes are more biocompatible than man-made nanoparticles for drug and RNA delivery, which have been well demonstrated in cancer therapy [149]. Although exosomes may be more biocompatible than synthetic nanoparticles, this system faces multiple challenges as a delivery system due to difficulty in large-scale production and loading of RNA payloads. Nevertheless, human red blood cells (RBC) are being considered an ideal source due to safety and abundance. Additionally, RBC-derived EVs or exosomes lack both nuclear and mitochondrial DNA, while on the other hand, RBCs have been used safely and routinely for blood transfusions. Therefore, RBC-derived EVs or exosomes could be used as a natural delivery system of therapeutic RNAs for cancer therapy [150]. Hence, RBC derived EVs or exosomes act as natural carriers of anticancer agents (e.g. circRNAs) and may represent an ideal delivery system because of their negligible antigenicity, minimal cytotoxicity. The development of standardized protocols and proper analytical approaches are crucial to improve the reproducibility in exosome research, thereby ensuring the applicable translation of exosomes in anticancer therapeutics [151]. Development of specific and effective strategies to target circRNAs in vivo will accelerate research progress of circRNA-based therapeutics.
Concluding remarks and future perspectives
It is a hot research areas to investigate the roles of circRNAs in the TME, including in immune escape, angiogenesis, drug resistance, EMT, tumor metastasis, and tumor metabolism. Accumulating evidence suggested that the roles of exosomal circRNAs in the TME are involved in diverse molecular mechanisms such as serving as miRNA sponges, binding to proteins or DNAs, etc. Potential clinical applications of exosomal circRNAs in cancer diagnosis and prognosis are also being explored.
Recently, advances in the functions and mechanisms of exosomal circRNAs hold great promises for diagnostic and therapeutic applications for cancers. However, there are a series of key questions that remain to be answered: (i) Could exosomal circRNAs in body fluids be a novel source of candidate biomarkers for cancer diagnosis? Currently, it is too early to determine that since the research on exosomal circRNAs is still in its infancy and much more research is warranted to explore their potential application in cancer diagnosis. Nevertheless, circRNAs have been demonstrated to have better stability, promoting their storage, isolation and detection. (ii) If targeting circRNAs could be used as a novel therapeutic strategy in cancer therapy? Given that the current circRNAs studies have only been conducted in two-dimensional cell culture models, which lack the TME, the available data should be further verified using three-dimensional tumor organoid models. (iii) How to develop appropriate preclinical models to assess the safety, pharmacokinetic, and pharmacodynamic characteristics for clinical dose predictions? Various in vitro experimental strategies should be adopted to achieve this including the stability test of exosomal circRNAs under light and different temperature, the cytotoxicity test of exosomal circRNAs, and pharmacokinetic investigation using in vitro and in vivo models [152].
Exosomes have emerged as new organelles of interest in cancer diagnostics and treatment [153]. Although some research progress has been made in the roles of exosomal circRNAs in the TME, there is a gap between basic findings and clinic application for diagnostic and prognostic biomarkers, and therapeutic target as well. Further large-scale, multicentered prospective cohort clinical studies are warranted to confirm the potential utility of exosomal circRNA in development of new therapy to improve cancer patient outcomes.
Abbreviations
ASOs: antisense oligonucleotides; AUCs: area under the ROC curves; CCA: cholangiocarcinoma; ceRNA: competing endogenous rna; CircRNA: circular RNA; ciRNAs: intronic circRNAs; CLL: chronic lymphocytic leukemia; CRC: colorectal cancer; DCs: dendritic cells; GC: gastric cancer; ecircRNAs: exonic circRNAs; EIciRNAs: exonic-intronic circRNAs; EVs : extracellular vesicles; exLRs: extracellular vesicle long RNAs; EZH2: enhancer of zeste homolog 2; HCC: hepatocellular carcinoma; HUVECs: human microvascular vein endothelial cells; LSCC: laryngeal squamous cell carcinoma; LUAD: lung adenocarcinoma; LY6E: lymphocyte antigen 6 complex locus E; MMP2: matrix metallopeptidase 2; MUC1: mucin 1; MVB: multivesicular body; NcRNAs: noncoding RNAs; NK: natural killer; NSCLC: non-small-cell lung cancer; PC: pancreatic cancer; PD-L1: programmed death ligand 1; PDAC: pancreatic ductal adenocarcinoma; OC: ovarian cancer; PKM2: pyruvate kinase M2; PRDM16: pr domain containing protein 16; PTC: papillary thyroid carcinoma; RBP: RNA-binding protein; RIG-I: retinoic acid-inducible gene I; SCLC: small cell lung cancer; SRY: sex-determining region Y; TDEs: tumor-derived exosomes; TILs: tumor infiltrating lymphocytes; TGF-β1: transforming growth factor-β1; TME: tumor microenvironment; TMZ: temozolomide; TNM: tumor-node-metastasis; tricRNAs: circRNAs produced from tRNAs; Tregs: regulatory t cells; UCB: urothelial carcinoma of the bladder; VEGF: vascular Endothelial growth factor.
Acknowledgements
This work was supported by the following research grants from the Natural Science Foundation of Hubei Province (2019CFB591 to Z.M.); the National Medical Research Council, Singapore [NMRC/CSASI/0006/2016 and NMRC/CG/M005/2017_NCIS] (Goh BC), and the Joint NCIS and NUS Cancer Program Seed Funding Grants [NUHSRO/2020/122/MSC/07/Cancer] (B.G. & L.W.) and the Singapore Ministry of Health's National Medical Research Council [NMRC/CNIG/1146/2016, L. W.]
Author Contributions
J.L. and G.Z. collected the related paper and drafted the manuscript. G.Z. and C.G.L. prepared the figures. X.X., M.L., and G.S. helped to revise the manuscript. Z.M. conceived the review article. Z.M., B.G. and L.W. designed the review and provided supervision. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mathieu M, Martin-Jaular L, Lavieu G, Thery C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21:9-17
2. Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: small particle, big player. J Hematol Oncol. 2015;8:83
3. Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255-89
4. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020 367
5. Nam GH, Choi Y, Kim GB, Kim S, Kim SA, Kim IS. Emerging prospects of exosomes for cancer treatment: from conventional therapy to immunotherapy. Adv Mater. 2020;32:e2002440
6. Li I, Nabet BY. Exosomes in the tumor microenvironment as mediators of cancer therapy resistance. Mol Cancer. 2019;18:32
7. Shi X, Wang B, Feng X, Xu Y, Lu K, Sun M. circRNAs and exosomes: A mysterious frontier for human cancer. Mol Ther Nucleic Acids. 2020;19:384-92
8. Seimiya T, Otsuka M, Iwata T, Shibata C, Tanaka E, Suzuki T. et al. Emerging roles of exosomal circular RNAs in cancer. Front Cell Dev Biol. 2020;8:568366
9. Hinshaw DC, Shevde LA. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019;79:4557-66
10. Bejarano L, Jordao MJC, Joyce JA. Therapeutic targeting of the tumor microenvironment. Cancer Discov. 2021;11:933-59
11. Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346-54
12. Xiao Y, Yu D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol Ther. 2021;221:107753
13. Wang M, Yu F, Li P, Wang K. Emerging function and clinical significance of exosomal circRNAs in cancer. Mol Ther Nucleic Acids. 2020;21:367-83
14. Bai H, Lei K, Huang F, Jiang Z, Zhou X. Exo-circRNAs: a new paradigm for anticancer therapy. Mol Cancer. 2019;18:56
15. Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W. et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18:116
16. van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213-28
17. Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569-79
18. Choi DS, Kim DK, Kim YK, Gho YS. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics. 2013;13:1554-71
19. Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475-90
20. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852-6
21. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675-91
22. Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309-22
23. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423-37
24. Maia J, Caja S, Strano Moraes MC, Couto N, Costa-Silva B. Exosome-based cell-cell communication in the tumor microenvironment. Front Cell Dev Biol. 2018;6:18
25. Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New technologies for analysis of extracellular vesicles. Chem Rev. 2018;118:1917-50
26. Hu W, Liu C, Bi ZY, Zhou Q, Zhang H, Li LL. et al. Comprehensive landscape of extracellular vesicle-derived RNAs in cancer initiation, progression, metastasis and cancer immunology. Mol Cancer. 2020;19:102
27. Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM. et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19:110
28. Xu Y, Leng K, Yao Y, Kang P, Liao G, Han Y. et al. A novel circular RNA, circ-CCAC1, contributes to CCA progression, induces angiogenesis, and disrupts vascular endothelial barriers. Hepatology. 2020
29. Ding C, Yi X, Wu X, Bu X, Wang D, Wu Z. et al. Exosome-mediated transfer of circRNA CircNFIX enhances temozolomide resistance in glioma. Cancer Lett. 2020;479:1-12
30. Zomer A, Steenbeek SC, Maynard C, van Rheenen J. Studying extracellular vesicle transfer by a Cre-loxP method. Nat Protoc. 2016;11:87-101
31. Pucci F, Garris C, Lai CP, Newton A, Pfirschke C, Engblom C. et al. SCS macrophages suppress melanoma by restricting tumor-derived vesicle-B cell interactions. Science. 2016;352:242-6
32. Verweij FJ, Bebelman MP, Jimenez CR, Garcia-Vallejo JJ, Janssen H, Neefjes J. et al. Quantifying exosome secretion from single cells reveals a modulatory role for GPCR signaling. J Cell Biol. 2018;217:1129-42
33. Men Y, Yelick J, Jin S, Tian Y, Chiang MSR, Higashimori H. et al. Exosome reporter mice reveal the involvement of exosomes in mediating neuron to astroglia communication in the CNS. Nat Commun. 2019;10:4136
34. Zhu L, Sun HT, Wang S, Huang SL, Zheng Y, Wang CQ. et al. Isolation and characterization of exosomes for cancer research. J Hematol Oncol. 2020;13:152
35. Coumans FAW, Brisson AR, Buzas EI, Dignat-George F, Drees EEE, El-Andaloussi S. et al. Methodological Guidelines to Study Extracellular Vesicles. Circ Res. 2017;120:1632-48
36. Syn NL, Wang L, Chow EK, Lim CT, Goh BC. Exosomes in cancer nanomedicine and immunotherapy: Prospects and challenges. Trends Biotechnol. 2017;35:665-76
37. Lu M, Huang B, Hanash SM, Onuchic JN, Ben-Jacob E. Modeling putative therapeutic implications of exosome exchange between tumor and immune cells. Proc Natl Acad Sci U S A. 2014;111:E4165-74
38. Xu Z, Zeng S, Gong Z, Yan Y. Exosome-based immunotherapy: a promising approach for cancer treatment. Mol Cancer. 2020;19:160
39. Zhou Z, Sun B, Huang S, Zhao L. Roles of circular RNAs in immune regulation and autoimmune diseases. Cell Death Dis. 2019;10:503
40. Chen X, Yang T, Wang W, Xi W, Zhang T, Li Q. et al. Circular RNAs in immune responses and immune diseases. Theranostics. 2019;9:588-607
41. Lei X, Lei Y, Li JK, Du WX, Li RG, Yang J. et al. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126-33
42. Roma-Rodrigues C, Mendes R, Baptista PV, Fernandes AR. Targeting tumor microenvironment for cancer therapy. Int J Mol Sci. 2019 20
43. Wellenstein MD, de Visser KE. Cancer-cell-intrinsic mechanisms shaping the tumor immune landscape. Immunity. 2018;48:399-416
44. Madden MZ, Rathmell JC. The complex integration of T-cell metabolism and immunotherapy. Cancer Discov. 2021;11:1636-43
45. Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677-704
46. Jiang X, Wang J, Deng X, Xiong F, Ge J, Xiang B. et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer. 2019;18:10
47. Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan Y. et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11:32
48. Wang X, Yao Y, Jin M. Circ-0001068 is a novel biomarker for ovarian cancer and inducer of PD1 expression in T cells. Aging (Albany NY). 2020;12:19095-106
49. Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA circ-CPA4/ let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC). J Exp Clin Cancer Res. 2020;39:149
50. Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14:399-416
51. Guo W, Li Y, Pang W, Shen H. Exosomes: A potential therapeutic tool targeting communications between tumor cells and macrophages. Mol Ther. 2020;28:1953-64
52. Lu Q, Wang X, Zhu J, Fei X, Chen H, Li C. Hypoxic tumor-derived exosomal circ0048117 facilitates M2 macrophage polarization acting as miR-140 sponge in esophageal squamous cell carcinoma. Onco Targets Ther. 2020;13:11883-97
53. Chen T, Liu Y, Li C, Xu C, Ding C, Chen J. et al. Tumor-derived exosomal circFARSA mediates M2 macrophage polarization via the PTEN/PI3K/AKT pathway to promote non-small cell lung cancer metastasis. Cancer Treat Res Commun. 2021;28:100412
54. Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991-8
55. Wang WT, Zhu HY, Wu YJ, Xia Y, Wu JZ, Wu W. et al. Elevated absolute NK cell counts in peripheral blood predict good prognosis in chronic lymphocytic leukemia. J Cancer Res Clin Oncol. 2018;144:449-57
56. Hu X, Li YQ, Li QG, Ma YL, Peng JJ, Cai SJ. ITGAE defines CD8+ tumor-infiltrating lymphocytes predicting a better prognostic survival in colorectal cancer. EBioMedicine. 2018;35:178-88
57. Jaillon S, Ponzetta A, Di Mitri D, Santoni A, Bonecchi R, Mantovani A. Neutrophil diversity and plasticity in tumour progression and therapy. Nat Rev Cancer. 2020;20:485-503
58. Herbst B, Zheng L. Precision medicine in pancreatic cancer: treating every patient as an exception. Lancet Gastroenterol Hepatol. 2019;4:805-10
59. Paulson AS, Tran Cao HS, Tempero MA, Lowy AM. Therapeutic advances in pancreatic cancer. Gastroenterology. 2013;144:1316-26
60. Li Z, Yanfang W, Li J, Jiang P, Peng T, Chen K. et al. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237-50
61. Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B. et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-beta1 axis. Mol Cancer. 2020;19:117
62. Gacche RN, Meshram RJ. Targeting tumor micro-environment for design and development of novel anti-angiogenic agents arresting tumor growth. Prog Biophys Mol Biol. 2013;113:333-54
63. Li D, Xie K, Zhang L, Yao X, Li H, Xu Q. et al. Dual blockade of vascular endothelial growth factor (VEGF) and basic fibroblast growth factor (FGF-2) exhibits potent anti-angiogenic effects. Cancer Lett. 2016;377:164-73
64. Weis SM, Cheresh DA. Tumor angiogenesis: molecular pathways and therapeutic targets. Nat Med. 2011;17:1359-70
65. Jayson GC, Hicklin DJ, Ellis LM. Antiangiogenic therapy-evolving view based on clinical trial results. Nat Rev Clin Oncol. 2012;9:297-303
66. Liu CG, Li J, Xu Y, Li W, Fang SX, Zhang Q. et al. Long non-coding RNAs and circular RNAs in tumor angiogenesis: From mechanisms to clinical significance. Mol Ther Oncolytics. 2021;22:336-54
67. Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F. et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer. 2020;19:112
68. Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie JW. et al. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38-48
69. Zeng W, Liu Y, Li WT, Li Y, Zhu JF. CircFNDC3B sequestrates miR-937-5p to derepress TIMP3 and inhibit colorectal cancer progression. Mol Oncol. 2020
70. Chouaib S, Kieda C, Benlalam H, Noman MZ, Mami-Chouaib F, Ruegg C. Endothelial cells as key determinants of the tumor microenvironment: interaction with tumor cells, extracellular matrix and immune killer cells. Crit Rev Immunol. 2010;30:529-45
71. Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K. et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37:177
72. Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH. et al. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res. 2020;39:20
73. Kelderman S, Schumacher TN, Haanen JB. Acquired and intrinsic resistance in cancer immunotherapy. Mol Oncol. 2014;8:1132-9
74. McGee HM, Jiang D, Soto-Pantoja DR, Nevler A, Giaccia AJ, Woodward WA. Targeting the Tumor Microenvironment in Radiation Oncology: Proceedings from the 2018 ASTRO-AACR Research Workshop. Clin Cancer Res. 2019;25:2969-74
75. Moeller BJ, Dreher MR, Rabbani ZN, Schroeder T, Cao Y, Li CY. et al. Pleiotropic effects of HIF-1 blockade on tumor radiosensitivity. Cancer Cell. 2005;8:99-110
76. Li L, Li W, Chen N, Zhao H, Xu G, Zhao Y. et al. FLI1 Exonic Circular RNAs as a novel oncogenic driver to promote tumor metastasis in Small Cell Lung Cancer. Clin Cancer Res. 2019;25:1302-17
77. Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T. et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14:539-55
78. Hon KW, Ab-Mutalib NS, Abdullah NMA, Jamal R, Abu N. Extracellular Vesicle-derived circular RNAs confers chemoresistance in Colorectal cancer. Sci Rep. 2019;9:16497
79. Xu J, Ji L, Liang Y, Wan Z, Zheng W, Song X. et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct Target Ther. 2020;5:298
80. Zhao Z, Ji M, Wang Q, He N, Li Y. Circular RNA Cdr1as upregulates SCAI to suppress cisplatin resistance in ovarian cancer via miR-1270 suppression. Mol Ther Nucleic Acids. 2019;18:24-33
81. Xu X, Tao R, Sun L, Ji X. Exosome-transferred hsa_circ_0014235 promotes DDP chemoresistance and deteriorates the development of non-small cell lung cancer by mediating the miR-520a-5p/CDK4 pathway. Cancer Cell Int. 2020;20:552
82. Wu Z, Sun H, Wang C, Liu W, Liu M, Zhu Y. et al. Mitochondrial genome-derived circRNA mc-COX2 functions as an oncogene in chronic lymphocytic leukemia. Mol Ther Nucleic Acids. 2020;20:801-11
83. Taki M, Abiko K, Ukita M, Murakami R, Yamanoi K, Yamaguchi K. et al. Tumor immune microenvironment during epithelial-mesenchymal transition. Clin Cancer Res. 2021
84. Cheng JT, Wang L, Wang H, Tang FR, Cai WQ, Sethi G. et al. Insights into biological role of lncRNAs in epithelial-mesenchymal transition. Cells. 2019 8
85. Chen X, Chen RX, Wei WS, Li YH, Feng ZH, Tan L. et al. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin Cancer Res. 2018;24:6319-30
86. Liu D, Kang H, Gao M, Jin L, Zhang F, Chen D. et al. Exosome-transmitted circ_MMP2 promotes hepatocellular carcinoma metastasis by upregulating MMP2. Mol Oncol. 2020;14:1365-80
87. Feng W, Gong H, Wang Y, Zhu G, Xue T, Wang Y. et al. circIFT80 Functions as a ceRNA of miR-1236-3p to Promote Colorectal Cancer Progression. Mol Ther Nucleic Acids. 2019;18:375-87
88. Dai X, Chen C, Yang Q, Xue J, Chen X, Sun B. et al. Exosomal circRNA_100284 from arsenite-transformed cells, via microRNA-217 regulation of EZH2, is involved in the malignant transformation of human hepatic cells by accelerating the cell cycle and promoting cell proliferation. Cell Death Dis. 2018;9:454
89. Wang S, Hu Y, Lv X, Li B, Gu D, Li Y. et al. Circ-0000284 arouses malignant phenotype of cholangiocarcinoma cells and regulates the biological functions of peripheral cells through cellular communication. Clin Sci (Lond). 2019;133:1935-53
90. Tian L, Cao J, Jiao H, Zhang J, Ren X, Liu X. et al. CircRASSF2 promotes laryngeal squamous cell carcinoma progression by regulating the miR-302b-3p/IGF-1R axis. Clin Sci (Lond). 2019;133:1053-66
91. Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat Rev Cancer. 2003;3:453-8
92. Guan X, Zong ZH, Liu Y, Chen S, Wang LL, Zhao Y. circPUM1 promotes tumorigenesis and progression of ovarian cancer by sponging miR-615-5p and miR-6753-5p. Mol Ther Nucleic Acids. 2019;18:882-92
93. Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z. et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18:20
94. Wang J, Zhang Q, Zhou S, Xu H, Wang D, Feng J. et al. Circular RNA expression in exosomes derived from breast cancer cells and patients. Epigenomics. 2019;11:411-21
95. Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J. et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine. 2019;40:432-45
96. Zong ZH, Du YP, Guan X, Chen S, Zhao Y. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. J Exp Clin Cancer Res. 2019;38:437
97. Zhang Q, Wang H, Mao C, Sun M, Dominah G, Chen L. et al. Fatty acid oxidation contributes to IL-1beta secretion in M2 macrophages and promotes macrophage-mediated tumor cell migration. Mol Immunol. 2018;94:27-35
98. Scharping NE, Menk AV, Whetstone RD, Zeng X, Delgoffe GM. Efficacy of PD-1 blockade is potentiated by Metformin-induced reduction of tumor hypoxia. Cancer Immunol Res. 2017;5:9-16
99. Ocana MC, Martinez-Poveda B, Quesada AR, Medina MA. Metabolism within the tumor microenvironment and its implication on cancer progression: An ongoing therapeutic target. Med Res Rev. 2019;39:70-113
100. Ringel AE, Drijvers JM, Baker GJ, Catozzi A, Garcia-Canaveras JC, Gassaway BM. et al. Obesity Shapes Metabolism in the Tumor Microenvironment to Suppress Anti-Tumor Immunity. Cell. 2020
101. Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491:364-73
102. Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10:671-84
103. Katt WP, Cerione RA. Glutaminase regulation in cancer cells: a druggable chain of events. Drug Discov Today. 2014;19:450-7
104. Quail DF, Dannenberg AJ. The obese adipose tissue microenvironment in cancer development and progression. Nat Rev Endocrinol. 2019;15:139-54
105. Kaymak I, Williams KS, Cantor JR, Jones RG. Immunometabolic Interplay in the Tumor Microenvironment. Cancer Cell. 2021;39:28-37
106. Yang E, Wang X, Gong Z, Yu M, Wu H, Zhang D. Exosome-mediated metabolic reprogramming: the emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduct Target Ther. 2020;5:242
107. Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR. et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17:1498-503
108. Zhang M, Di Martino JS, Bowman RL, Campbell NR, Baksh SC, Simon-Vermot T. et al. Adipocyte-derived lipids mediate melanoma progression via FATP proteins. Cancer Discov. 2018;8:1006-25
109. Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K. et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38:2844-59
110. Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng T. et al. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int J Cancer. 2019;144:2501-15
111. Talekar M, Boreddy SR, Singh A, Amiji M. Tumor aerobic glycolysis: new insights into therapeutic strategies with targeted delivery. Expert Opin Biol Ther. 2014;14:1145-59
112. Ding C, Xi G, Wang G, Cui D, Zhang B, Wang H. et al. Exosomal Circ-MEMO1 promotes the progression and aerobic glycolysis of Non-small Cell Lung Cancer through targeting miR-101-3p/KRAS Axis. Front Genet. 2020;11:962
113. Wei X, Chen Y, Jiang X, Peng M, Liu Y, Mo Y. et al. Mechanisms of vasculogenic mimicry in hypoxic tumor microenvironments. Mol Cancer. 2021;20:7
114. Ma Z, Xiang X, Li S, Xie P, Gong Q, Goh BC. et al. Targeting hypoxia-inducible factor-1, for cancer treatment: Recent advances in developing small-molecule inhibitors from natural compounds. Semin Cancer Biol. 2020 Doi: 10.1016/j.semcancer.2020.09.011
115. Ma Z, Wang LZ, Cheng JT, Lam WST, Ma X, Xiang X. et al. Targeting hypoxia-inducible factor-1-mediated metastasis for cancer therapy. Antioxid Redox Signal. 2021;34:1484-97
116. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74
117. Patel A, Sant S. Hypoxic tumor microenvironment: Opportunities to develop targeted therapies. Biotechnol Adv. 2016;34:803-12
118. You L, Wu W, Wang X, Fang L, Adam V, Nepovimova E. et al. The role of hypoxia-inducible factor 1 in tumor immune evasion. Med Res Rev. 2021;41:1622-43
119. Yang H, Zhang H, Yang Y, Wang X, Deng T, Liu R. et al. Hypoxia induced exosomal circRNA promotes metastasis of Colorectal Cancer via targeting GEF-H1/RhoA axis. Theranostics. 2020;10:8211-26
120. Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM. et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020;20:174-86
121. Zhan Y, Du J, Min Z, Ma L, Zhang W, Zhu W. et al. Carcinoma-associated fibroblasts derived exosomes modulate breast cancer cell stemness through exonic circHIF1A by miR-580-5p in hypoxic stress. Cell Death Discov. 2021;7:141
122. Gu C, Lu H, Qian Z. Matrine reduces the secretion of exosomal circSLC7A6 from cancer-associated fibroblast to inhibit tumorigenesis of colorectal cancer by regulating CXCR5. Biochem Biophys Res Commun. 2020;527:638-45
123. Zhang Q, Wang W, Zhou Q, Chen C, Yuan W, Liu J. et al. Roles of circRNAs in the tumour microenvironment. Mol Cancer. 2020;19:14
124. Ma Z, Shuai Y, Gao X, Wen X, Ji J. Circular RNAs in the tumour microenvironment. Mol Cancer. 2020;19:8
125. Xian J, Su W, Liu L, Rao B, Lin M, Feng Y. et al. Identification of three circular RNA cargoes in serum exosomes as diagnostic biomarkers of Non-Small-Cell Lung Cancer in the Chinese population. J Mol Diagn. 2020;22:1096-108
126. Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu M. et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer. 2020;19:101
127. Shao Y, Tao X, Lu R, Zhang H, Ge J, Xiao B. et al. Hsa_circ_0065149 is an indicator for early gastric cancer screening and prognosis prediction. Pathol Oncol Res. 2020;26:1475-82
128. Tang W, Fu K, Sun H, Rong D, Wang H, Cao H. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. 2018;17:137
129. Pan B, Qin J, Liu X, He B, Wang X, Pan Y. et al. Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet. 2019;10:1096
130. Li Y, Li C, Xu R, Wang Y, Li D, Zhang B. A novel circFMN2 promotes tumor proliferation in CRC by regulating the miR-1182/hTERT signaling pathways. Clin Sci (Lond). 2019;133:2463-79
131. Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM. et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016;6:37982
132. Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L. et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020;475:119-28
133. Li Y, Zhao J, Yu S, Wang Z, He X, Su Y. et al. Extracellular Vesicles Long RNA Sequencing Reveals Abundant mRNA, circRNA, and lncRNA in Human Blood as Potential Biomarkers for Cancer Diagnosis. Clin Chem. 2019;65:798-808
134. Wu G, Zhou W, Lin X, Sun Y, Li J, Xu H. et al. circRASSF2 acts as ceRNA and promotes papillary thyroid carcinoma progression through miR-1178/TLR4 signaling pathway. Mol Ther Nucleic Acids. 2020;19:1153-63
135. Wu G, Zhou W, Pan X, Sun Z, Sun Y, Xu H. et al. Circular RNA profiling reveals exosomal circ_0006156 as a novel biomarker in papillary thyroid cancer. Mol Ther Nucleic Acids. 2020;19:1134-44
136. Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in cancer: biogenesis, function, and cinical significance. Trends Cancer. 2020;6:319-36
137. Su M, Xiao Y, Ma J, Tang Y, Tian B, Zhang Y. et al. Circular RNAs in Cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer. 2019;18:90
138. Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19:30
139. Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov. 2021;20:629-51
140. Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nat Rev Genet. 2008;9:554-66
141. Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233-47
142. Han K, Wang FW, Cao CH, Ling H, Chen JW, Chen RX. et al. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA-17. Mol Cancer. 2020;19:60
143. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428-42
144. Tian Y, Li S, Song J, Ji T, Zhu M, Anderson GJ. et al. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35:2383-90
145. Herrmann IK, Wood MJA, Fuhrmann G. Extracellular vesicles as a next-generation drug delivery platform. Nat Nanotechnol. 2021;16:748-59
146. He AT, Liu J, Li F, Yang BB. Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal Transduct Target Ther. 2021;6:185
147. Yang L, Han B, Zhang Z, Wang S, Bai Y, Zhang Y. et al. Extracellular vesicle-mediated delivery of circular RNA SCMH1 promotes functional recovery in rodent and nonhuman primate ischemic stroke models. Circulation. 2020;142:556-74
148. Wiklander OP, Nordin JZ, O'Loughlin A, Gustafsson Y, Corso G, Mager I. et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J Extracell Vesicles. 2015;4:26316
149. Fu P, Zhang J, Li H, Mak M, Xu W, Tao Z. Extracellular vesicles as delivery systems at nano-/micro-scale. Adv Drug Deliv Rev. 2021: 113910.
150. Usman WM, Pham TC, Kwok YY, Vu LT, Ma V, Peng B. et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat Commun. 2018;9:2359
151. Pirisinu M, Pham TC, Zhang DX, Hong TN, Nguyen LT, Le MT. Extracellular vesicles as natural therapeutic agents and innate drug delivery systems for cancer treatment: Recent advances, current obstacles, and challenges for clinical translation. Semin Cancer Biol. 2020
152. Li S, Yu Y, Bian X, Yao L, Li M, Lou YR. et al. Prediction of oral hepatotoxic dose of natural products derived from traditional Chinese medicines based on SVM classifier and PBPK modeling. Arch Toxicol. 2021;95:1683-701
153. Shehzad A, Islam SU, Shahzad R, Khan S, Lee YS. Extracellular vesicles in cancer diagnostics and therapeutics. Pharmacol Ther. 2021;223:107806
Author contact
![]() Corresponding authors: Z. Ma, E-mail: zhaowu823com, Tel:+86 15972188216; B.C. Goh, E-mail: phcgbcedu.sg, Tel: +65 67724641; L. Wang, E-mail: csiwledu.sg, Tel: +65 65168925.
Corresponding authors: Z. Ma, E-mail: zhaowu823com, Tel:+86 15972188216; B.C. Goh, E-mail: phcgbcedu.sg, Tel: +65 67724641; L. Wang, E-mail: csiwledu.sg, Tel: +65 65168925.
 Global reach, higher impact
Global reach, higher impact