13.3
Impact Factor
Theranostics 2022; 12(1):260-276. doi:10.7150/thno.63417 This issue Cite
Research Paper
Driver mutations of intrahepatic cholangiocarcinoma shape clinically relevant genomic clusters with distinct molecular features and therapeutic vulnerabilities
1. Department of General Surgery, Huashan Hospital, Fudan University, Shanghai, China
2. Cancer Metastasis Institute, Fudan University, Shanghai, China
3. Department of Laboratory Medicine, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China
4. Department of Infectious Diseases, Huashan Hospital, Fudan University, Shanghai, China
5. Biotech Research and Innovation Centre (BRIC), Department of Health and Medical Sciences, University of Copenhagen, Copenhagen N, Denmark
6. Molecular and Cell Biology Lab, Institutes of Biomedical Sciences, and the Shanghai Key Laboratory of Medical Epigenetics, and the Key Laboratory of Metabolism and Molecular, Ministry of Education, Fudan University, Shanghai, China
# These authors contribute equally to this work
Abstract
Purpose: To establish a clinically applicable genomic clustering system, we investigated the interactive landscape of driver mutations in intrahepatic cholangiocarcinoma (ICC).
Methods: The genomic data of 1481 ICCs from diverse populations was analyzed to investigate the pair-wise co-occurrences or mutual exclusivities among recurrent driver mutations. Clinicopathological features and outcomes were compared among different clusters. Gene expression and DNA methylation profiling datasets were analyzed to investigate the molecular distinctions among mutational clusters. ICC cell lines with different gene mutation backgrounds were used to evaluate the cluster specific biological behaviors and drug sensitivities.
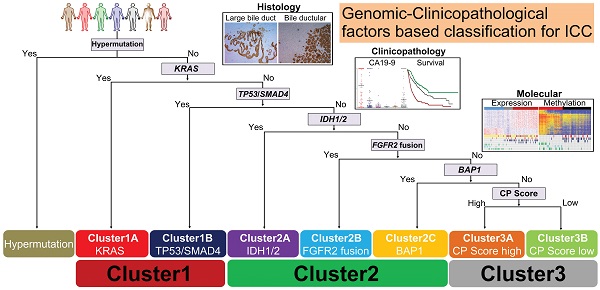
Results: Statistically significant mutation-pairs were identified across 21 combinations of genes. Seven most recurrent driver mutations (TP53, KRAS, SMAD4, IDH1/2, FGFR2-fus and BAP1) showed pair-wise co-occurrences or mutual exclusivities and could aggregate into three genetic clusters: Cluster1: represented by tripartite interaction of KRAS, TP53 and SMAD4 mutations, exhibited large bile duct histological phenotype with high CA19-9 level and dismal prognosis; Cluster2: co-association of IDH/BAP1 or FGFR2-fus/BAP1 mutation, was characterized by small bile duct phenotype, low CA19-9 level and optimal prognosis; Cluster3: mutation-free ICC cases with intermediate clinicopathological features. These clusters showed distinct molecular traits, biological behaviors and responses to therapeutic drugs. Finally, we identified S100P and KRT17 as “cluster-specific”, “lineage-dictating” and “prognosis-related” biomarkers, which in combination with CA19-9 could well stratify Cluster3 ICCs into two biologically and clinically distinct subtypes.
Conclusions: This clinically applicable clustering system can be instructive to ICC prognostic stratification, molecular classification, and therapeutic optimization.
Keywords: Genome sequencing, Driver mutation, ICC diversity.
 Global reach, higher impact
Global reach, higher impact