13.3
Impact Factor
Theranostics 2022; 12(1):459-473. doi:10.7150/thno.65739 This issue Cite
Research Paper
Comprehensive omics analyses profile genesets related with tumor heterogeneity of multifocal glioblastomas and reveal LIF/CCL2 as biomarkers for mesenchymal subtype
1. Department of Neurosurgery, Xinqiao Hospital, Third Military Medical University (Army Medical University), Chongqing, China.
2. Institute of Pathology and Southwest Cancer Center, Southwest Hospital, Third Military Medical University (Army Medical University), Chongqing, China.
3. Biobank of Institute of Pathology, Southwest Hospital, Third Military Medical University (Army Medical University), Chongqing, China.
4. K2 Oncology Co., Ltd, Beijing, China.
* These authors contributed equally to this work.
Abstract
Rationale: Around 10%-20% patients with glioblastoma (GBM) are diagnosed with more than one tumor lesions or multifocal GBM (mGBM). However, the understanding on genetic, DNA methylomic, and transcriptomic characteristics of mGBM is still limited.
Methods: In this study, we collected nine tumor foci from three mGBM patients followed by whole genome sequencing, whole genome bisulfite sequencing, RNA sequencing, and immunohistochemistry. The data were further examined using public GBM databases and GBM cell line.
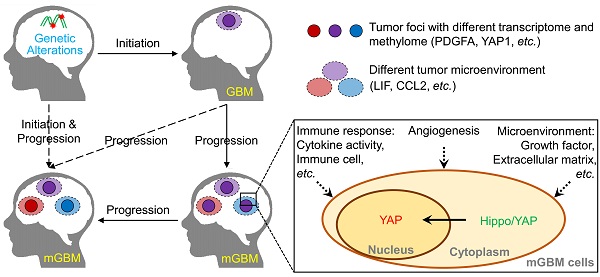
Results: Analysis on genetic data confirmed common features of GBM, including gain of chr.7 and loss of chr.10, loss of critical tumor suppressors, high frequency of PDGFA and EGFR amplification. Through profiling DNA methylome of individual tumor foci, we found that promoter methylation status of genes involved in detection of chemical stimulus, immune response, and Hippo/YAP1 pathway was significantly changed in mGBM. Although both CNV and promoter methylation alteration were involved in heterogeneity of different tumor foci from same patients, more CNV events than promoter hypomethylation events were shared by different tumor foci, implying CNV were relatively earlier than promoter methylation alteration during evolution of different tumor foci from same mGBM. Moreover, different tumor foci from same mGBM assumed different molecular subtypes and mesenchymal subtype was prevalent in mGBM, which might explain the worse prognosis of mGBM than single GBM. Interestingly, we noticed that LIF and CCL2 was tightly correlated with mesenchymal subtype tumor focus in mGBM and predicted poor survival of GBM patients. Treatment with LIF and CCL2 produced mesenchymal-like transcriptome in GBM cells.
Conclusions: Together, our work herein comprehensively profiled multi-omics features of mGBM and emphasized that components of extracellular microenvironment, such as LIF and CCL2, contributed to the evolution and prognosis of tumor foci in mGBM patients.
Keywords: multifocal GBM, molecular subtype, extracellular matrix, immune response, LIF, CCL2
 Global reach, higher impact
Global reach, higher impact