13.3
Impact Factor
Theranostics 2022; 12(5):2406-2426. doi:10.7150/thno.69189 This issue Cite
Review
Molecular imprinting of glycoproteins: From preparation to cancer theranostics
1. Center for Supramolecular Chemical Biology, State Key Laboratory of Supramolecular Structure and Materials, School of Life Sciences, Jilin University, Changchun 130023, China.
2. State Key Laboratory of Bioelectronics, National Demonstration Center for Experimental Biomedical Engineering Education, Southeast University, Nanjing 210096, China.
3. Center for Climate Research and Development COMSATS University Islamabad, Park Road, Tarlai Kalan, 45550, Islamabad, Pakistan.
4. HEJ Research Institute of Chemistry, International Center for Chemical and Biological Sciences, University of Karachi, Karachi-75270, Pakistan.
5. College of Chemistry, Beijing Key Laboratory of Environmentally Harmful Chemical Analysis, Beijing University of Chemical Technology, Beijing, 100029, China.
Received 2021-11-17; Accepted 2022-1-30; Published 2022-2-28
Abstract
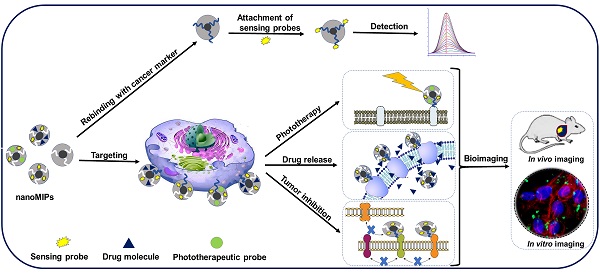
Glycoprotein imprinted polymers have rapidly grown as excellent receptors for cancer targeting, diagnostics, inhibition, and nanomedicines as they specifically target glycans and glycosites overexpressed in various tumors. Compared to natural antibodies, they are easy to synthesize, stable, and cost-efficient. Currently, no study specifically discusses glycoproteins imprinting strategies for cancer theranostics. In this review, firstly we explored various factors involved in designing and synthesis of glycoprotein imprinted materials, including, the characteristics and choice of monomers for imprinting, types of templates and their interactions involved, and the imprinting methods. Secondly, the integration of these MIPs with different probes that have been applied for in vitro and in vivo targeting for cancer diagnostics including biosensing and bioimaging, and image-guided therapeutic applications as nanomedicines. These Glycoprotein imprinted polymers have been found to specifically target the glycoprotein biomarkers and glycosylated cell receptors overexpressed in different cancers and have been reported as excellent diagnostic tools. As nanomedicines, they have been potentially employed in various modes of cancer therapy such as targeted drug delivery, photodynamic therapy, photothermal therapy, and nanoMIPs themselves as therapeutics for locally killing tumor cells. Although the research is still in its early stages and no real-world clinical trials on humans have been conducted, nanoMIPs have a promising future in this field. We believe these findings will pave the way for MIPs in advanced diagnostics, antibody treatment, and immunotherapy as future nanomedicine for real-world cancer theranostics.
Keywords: Molecularly imprinting, Glycoproteins, Biosensing, Cancer theranostics, Proteomics
Introduction
Glycoproteins perform vital roles in several physiological systems, such as molecular recognition, inter-and intra-cell signaling, immune response, sperm-egg interaction, and developmental control. The role of multiple glycoproteins, such as mucins, surface receptors, adhesive proteins, proteoglycans, their transcription factors, and binding ligands is influenced by irregular transcription of sugar moieties, leading to an increase in pancreatic cancer severity and a cancer-friendly microenvironment [1-3]. Glycoprotein identification, targeting, and quantification are crucial in life sciences and medical research, clinical diagnostics, medical instruments, and aging [4-7]. The progression of malignancies is linked to aberrant structural alterations and improper expressions. As a result, several glycoproteins, glycans, and monosaccharides are frequently utilized as biomarkers for disease diagnosis and treatment. Nevertheless, substantial challenges limit their detection and targeting, especially selective identification of glycoprotein and glycans structural properties [8-11]. In addition, glycoproteins have a low concentration in complex biological samples and co-exist with cellular substances, non-glycoproteins, peptides, and small molecules. Consequently, glycoprotein targeting, isolation, purification, and quantification are substantially hindered by the complex matrix. Therefore, it is indeed critical to selectively and specifically recognize the glycosylated molecules for accurate diagnostic and therapeutic applications.
Glycoprotein-induced antibodies are unique in recognizing protein units regardless of specific glycan structures [12]. However, they are constrained in their capacity to identify a complete glycan structure. Besides, lectins serve as affinity tools for glycans but cannot deliver structural details about the glycoprotein's fragments [13]. Furthermore, the generation of highly efficient antibodies is expensive, tedious, and challenging. Often, antibodies' stability and reproducibility are troublesome [14, 15]. Recognizing these shortcomings in biological affinity tools, there is a considerable opportunity and potential for the development of synthetic affinity tools, skilled in exceptionally accurate molecular targeting for specific challenging biological macromolecules [16].
A great challenge in molecular imprinting includes a method of template-induced creation of molecular cavities with identification sites in a substance that arises as a very interesting idea [17]. Molecularly imprinted polymers (MIPs) are developed employing several interactions such as covalent bonding, hydrogen bonding, van der Waals, hydrophobic and electrostatic interactions in different monomers and crosslinkers. These monomers are engineered to build recognition sites in imprinted nanoscale cavities complementary template molecules, compatible with shape, dimensions, and functionalities, indicating a vital role in supporting target affinity and selectivity [18, 19]. They are simple to synthesize, cost-effective, durable, and stable in a broad pH range, a variety of solvents, and thermal conditions [20]. Moreover, in a broad range of analytical tools including chromatographic columns or microextraction systems, MIPs can be conveniently integrated. MIPs have been commonly used as affinity materials for sample pretreatment. Recently, employing MIPs before mass spectrometry, electrochemical techniques [21], surface plasmon resonance [22], surface-enhanced resonance Raman scattering [23, 24], colorimetry [25], and fluorescence [26], have gained increasing interest. In addition, these glycoprotein imprinted nanoparticles have also been potentially applied in cancer diagnostics and treatment [27]. Based on their capability to capture covalently and reversibly with cis-diols of glycosylated molecules, boronic acid has appeared as a front-runner to serve as engineered receptors in imprinted cavities for targeting glycoprotein and glycosites that are overexpressed in most malignancies. This unique mechanism enables boronic acids as interesting ligands for a lot of applications targeting specific cancer sites, drug delivery, biosensing, isolation, and self-assembly [16, 28, 29].
In recent years, several reviews have been reported on the application of molecularly imprinted polymers in separation [30], sensing [31], microbe-analysis [32], and cell detection [32]. Despite a large number of studies, no review specifically discusses the molecular imprinting strategies for glycoproteins and their applications in sensing glycoprotein cancer markers, tumor imaging, and treatment. In this review, we discussed rational designing regarding the selection of the polymer matrix, templates, immobilizing and capturing mechanisms and some novel synthetic approaches for glycoprotein imprinting. Particularly, integration of these MIPs with sensing probes and tumor drugs explored for in vitro and in vivo targeting, cancer diagnostics, bioimaging, and therapeutic applications. The existing challenges and prospects of glycoprotein imprinted technology for targeted drug delivery, bioimaging, and clinical cancer biosensing are also discussed.
2. Designing and synthesis of MIPs
2.1 Brief comparison of different molecularly imprinted materials for glycoprotein
Different molecularly imprinted materials applied for glycoproteins have specific characteristics, their properties are tunable and there are several monomers available. Proper combination and orientation of monomers may lead to enhanced affinity and high selectivity for the target glycoproteins. Some commonly available monomers and crosslinkers are summarized in Table 1.
Comparing these molecularly imprinted materials for glycoproteins, several observations can be made; 1) Facile synthesis is an important parameter to choose the materials for imprinting and in the reported materials, silica [47], polydopamine [47, 48] and polyether sulphone [49] are more facile to prepare as no harsh conditions are required; 2) Easy removal of template molecule can be achieved by time-controlled thickness or by smart imprinted materials. Time-controlled thickness is especially important for surface imprinting because the template is usually buried inside the polymer layer in case of the uncontrolled thickness of the imprinting layer. The thickness of silica [50-52] and polydopamine [47] imprinting layer can be tuned by controlling the reaction time. Smart polymer materials such as poly (N-isopropylacrylamide), whose imprinting cavity channels can be opened by switching the temperature or pH of the solution, allowing the easy removal of the template molecule; 3) Availability of the multifunctional monomers is also very important for high specificity and strong binding, and numerous monomers of silica and acrylate with special functionality are commercially available as described in Table 1; 4) So far, compatibility with boronate affinity, silica, and polydopamine imprinting layer is developed at pH (8.5) suitable for the binding of boronate affinity [45, 48, 53]. Considering the various properties of these materials, the preference of which polymer should be used is determined by the particular use. Some of the important features of these materials are compared in Table 2.
Types and functions of functional monomers in glycoprotein imprinting.
| Types of monomers | Name | Structure | Functions | References |
|---|---|---|---|---|
| Acrylate Monomers | Acrylic acid | 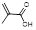 | Chelator | [33] |
| Vinylbenzyl iminodiacetic acid | 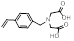 | Strong chelator | [34] | |
| Methacryl amide | 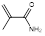 | Impart hydrophilicity | [16] | |
| Aminoethyl methacrylate | 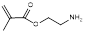 | Impart hydrophilicity | [35] | |
| N-isopropylacrylamide | 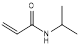 | Impart thermo-responsive properties | [36] | |
| 2-(Dimethylamino) ethyl methacrylate |  | Impart pH-responsive properties | [37] | |
| (3-acrylamidopropyl) trimethyl ammonium chloride | 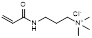 | Anion exchanger | [35] | |
| N-phenylacrylamide | 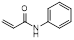 | π-π interaction facilitator | [34] | |
| N, N'-methylene bis(acrylamide) | 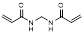 | Hydrophilic crosslinker | [38] | |
| 4-Vinylphenylboronic acid |  | Capture cis-diols | [39] | |
| 2,4-Difluoro-3-formylphenylboronic acid | 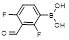 | Capture cis-diols at reduced binding pH | [40] | |
| 6-Vinylbenzoxaborole |  | Capture cis-diols at reduced binding pH (improved Wulff affinity) | [41] | |
| Silica Monomers | Tetraethyl orthosilicate |  | Initial silica precursor | [42] |
| 3-Aminopropyltriethoxysilane | 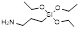 | Facilitate amine-group for functionalization | [43] | |
| N-octyltrimethoxysilane | Facilitate hydrophobicity | [43] | ||
| 3-Ureidopropyl-triethoxysilane |  | Two-fold cyclic hydrogen bond donor | [44] | |
| 3-Methacryloyloxypropyl trimethoxysilane |  | Coupling silica with an acrylate polymer | [38] | |
| Polydopamine monomers | Dopamine | 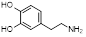 | Facilitate coating on any hydrophilic or hydrophobic surface | [45] |
| 4-aminophenyl boronic acid |  | Capture cis-diols | [40] | |
| Polyaniline monomers | Aniline | Basic hydrophobic polyaniline precursor | [46] |
Summary of important features of aforementioned molecularly imprinted materials for glycoproteins
| Imprinting materials | Facile synthesis | Time controlled thickness | Availability of functional monomers | Compatibility with boronate affinity | Flexibility or easy template removal |
|---|---|---|---|---|---|
| Silica | +++ | +++ | ++ | +++ | ++ |
| Polydopamine | +++ | +++ | + | +++ | ++ |
| Polyacrylate | + | + | +++ | ++ | + |
| Smart Polyacrylate | + | + | +++ | ++ | +++ |
| Polyaniline | + | + | + | ++ | + |
| Polyether Sulphone | +++ | + | + | + | + |
The accessibility is measured by the quantity of “+” symbols.
2.2 Types of Templates and their characteristics to develop MIPs
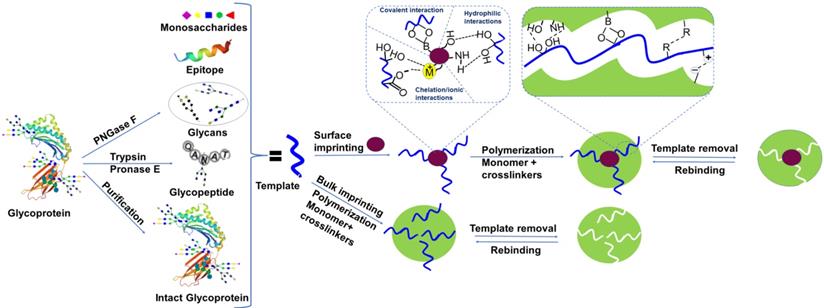
The application of protein imprinted materials as artificial antibodies has increased significantly in the last decade due to their captivating features such as tolerance to severe conditions, storage durability, and reusability [54]. Template molecules are also crucial for glycoprotein imprinted polymer materials. Conventionally, native glycoproteins were used as templates and this simple methodology did not require any special procedure to prepare the template and usually results in appropriate binding capacity and high specificity. However, this approach has apparent drawbacks if native proteins are used as templates [17]. First, while ensuring the native conformation of template proteins, the available polymerization methods are limited. Second, acquiring a large amount of high-purity template protein, especially for low-abundance proteins, is challenging. Third, several smaller non-targets bind comparatively large imprinted sites, leading to lower selectivity. To address these issues, some modern technologies are introduced where synthetic receptors of the target protein [55, 56] such as glycan, a monosaccharide, an oligosaccharide [57], or an epitope [58] are used as templates. These strategies offer a more versatile template selection, allowing the imprinting of uncommon, inaccessible, and unstable target glycoproteins. In addition, these segments as templates are small in size and easily removed from the reaction system. However, binding affinity becomes a key challenge in this approach, and larger template coverage is required. Bie et al. [59] used purified glycans for glycan-oriented surface imprinting, obtained by the digestion of target proteins using glycosidase enzyme. This methodology is used as an efficient tool for the precise enrichment of target glycoproteins and their characteristic fragments. These types of MIPs are highly consistent with MS-analysis and structural characterization because they easily release the captured target analyte [60]. However, this strategy is not appropriate for the imprinting of long-chain glycopeptides and even cannot be applied to the O-glycans.
Another approach called in situ dual enzymatic digestion is also recommended to fabricate the desired length of glycopeptides. In this methodology, the native glycoprotein carrying the required glycans is immobilized via boronate affinity of the substrate, followed by the in situ enzymatic digestion to get glycopeptides of the desired length. Then, a precise imprinting layer is developed and the template is removed by acidic solution. This strategy eliminates tedious fabrication steps template purification and is useful for imprinting both N-glycans and O-glycans [61]. The same group also used glycoproteins as a semi template in the organic phase, allowing the imprinting of glycans to control the hydrolysis of silica and consequently thickness of the imprinting layer [62]. However, these approaches are not ideal for imprinting glycoproteins, glycans, and monosaccharides without free cis-diols, such as glycosaminoglycans. In such cases, a specific recognizable part of the protein, called epitope is used as a template for the specific glycoprotein. Epitope templates are readily synthesized via artificial fabrication, including rare glycoproteins. In comparison, since they mimic the antibody's recognition domain, epitope-imprinted cavities are more analogous to natural recognition sites, contributing to higher specificity against the target [58]. Inspired by the recognition of the 3-dimensional shape of epitopes by antibodies, instead of a linear shape, some studies also used the conformational peptide as a template to get more specificity [63]. However, there are also some disadvantages of epitope imprinting. In certain techniques, epitopes are fixed on substrates before imprinting through a functional group such as -SH [64] and -NH2 [65] present on the terminal sites. Nevertheless, the coexistence of the same functional groups with other amino acids also contributes to an unfavorable epitope arrangement on the substrate. Epitopes are not immobilized on a substrate in some methods, and the imprinting direction is random, resulting in poor imprinting performance because a major proportion of templates are concealed inside the polymer. Certain approaches are only applicable to restricted situations, and the imprinting of multiple epitopes typically involves tedious conditional selection. Monosaccharides and disaccharides as template molecules have also been reported [66, 67]. Monosaccharide templates are stable, easily available, and monosaccharide imprinted materials revealed a high binding capacity, however, they offer poor selectivity. The template preparation approaches have been depicted in Figure 1.
2.3 Interaction of template molecules with molecularly imprinted materials
There are two main types of interactions in the imprinting of glycoproteins, covalent and noncovalent, between the template and the specific monomers employed for the production of molecularly imprinted materials (Figure 1). For covalent interactions, boronic acid ligands bind covalently in a reversible manner to form cyclic esters of 1,2, or 1,3-cis-diols in an alkaline environment (typically the pKa < solution pH). This covalent linkage is broken when pH is switched to an acidic environment, thus the selectivity of the recognition sites is typically very high. Furthermore, several novel boronate affinity ligands have been reported, with lower pKa values which can bind in physiological as well as in a slightly acidic environment with strong binding affinity [48]. This strong binding affinity enables boronate affinity materials to selectively enrich trace glycosylated biomolecules. Such chemistry brings some important advantages to boronic acid-functionalized materials, including comparatively stronger affinity (as opposed to non-specific bindings), broad-spectrum selectivity (a ligand can bind a wide range of molecules), pH-dependent capture and release, and rapid adsorption/desorption kinetics. Since slightly acidic conditions may release the target compounds from the specimen well, separation based on boronate affinity and sample pretreatment is very consistent with x-omics analysis based on mass spectrometry (MS), where an acidic medium is used for ionization. However, three obvious limitations are associated with traditional boronate affinity materials; (i) non-biocompatible binding pH, (ii) weak affinity, and (iii) comparatively low selectivity, hampering their real-world applications. The issue of non-biocompatible binding pH can be handled in four ways, i.e., the introduction of electron-withdrawing phenylboronic acids, such as carbonyl, sulfonyl, and fluoro on the phenyl ring [68], boronic acids comprising intramolecular tetracoordinate bonds B-N or B-O (Wulff-type), boronic acids with intramolecular coordinated bonds B-O (improved Wulff-type), and heterocyclic bonds [69]. Since reversible condensation reactions are limited, the covalent interaction technique is less versatile. Besides, thermodynamic equilibrium is very difficult to achieve because strong covalent associations result in sluggish capturing and dissociation [70].
Non-covalent approaches use hydrophilic, hydrophobic [71], electrostatic, chelation interactions [72], and van der Waals forces to capture the template glycoproteins in the imprinting cavity. As MIPs are the “plastic” antibodies or antibody mimics, thus inspired by the natural antibodies' capturing mechanism, non-covalent interactions are more reliable for targeting glycoproteins. These approaches are more general and offer standard interactions for molecularly imprinted materials because of their utility, fast binding kinetics, and ease of template removal. However, minor disturbance in the interactions between the template and the monomer is sensitive. Since non-covalent interactions are typically weaker than covalent interactions, the selectivity of these materials with non-covalent interactions is generally not high. More precisely, boronate affinity-based covalent interactions are more suitable to immobilize the template glycosylated molecule on the surface of a substrate and non-covalent interactions are preferably developed in the following polymerization step with suitable functional monomers.
2.4 Imprinting approaches to develop MIPs
Usually, MIPs are fabricated by free radical polymerization in desired formats such as monoliths (bulk imprinting approach), microparticles, nanoparticles, thin films (surface imprinting), nanocomposites, etc. However, condensation polymerization is more suitable for surface imprinting due to controlled thickness and mild reaction conditions [73]. The commonly used molecularly imprinted materials for glycoprotein and their characteristics have already been discussed in the previous section. Here, we will discuss the types of imprinting methods adopted to develop the glycoprotein imprinted nanoparticles for theranostic application.
2.4.1 Bulk Imprinting
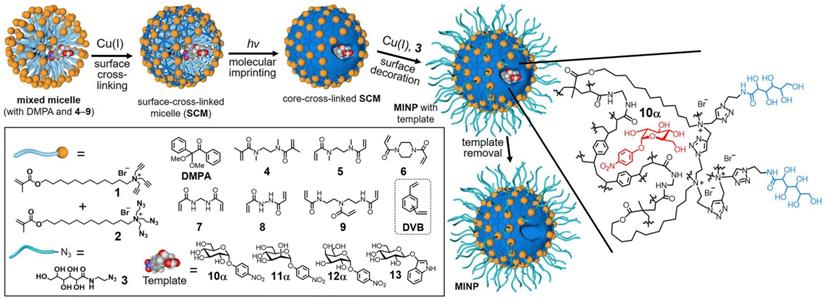
In bulk imprinting, the functional monomers, crosslinkers, initiators, and template molecules are mixed in the desired porogenic solvent, and polymerization is executed followed by crushing, grinding, and sieving to get the suitably sized particles. Then, the template is removed and obtained product is ready to use. This approach has clear advantages since it offers fast and facile synthesis, no need for complicated or expensive apparatus, and the imprinted materials are extremely pure. Liu and coworkers [39] reported a general and simple method to develop glycoprotein imprinted material via photolithographic approach using VPBA, which performed key roles, not only UV initiated polymerization to produce a thin layer imprinting array but also facilitated strong covalent affinity to capture the target glycoprotein. The facile synthesis via UV initiation takes less than 3 h for whole processing, has a high tolerance to interference, and is used in a wide range of pH. Lectins are a natural class of proteins, which capture carbohydrates with high specificity in physiological conditions as affinity tools for glycans but cannot deliver structural details about the glycoprotein's fragments [13]. Considering the chemistry of lectins and the complexity of glycosylation, Zangiabadi et al. [57] developed a molecularly imprinted micelle via a convenient one-pot method, to specifically capture the target glycosides, glycans, and even the glycoproteins (Figure 2). In water, physiologically viable micromolar binding affinities are revealed, and small structural variations amongst glycans are identified. Among acrylate polymers, smart polymers using NIPAM as a monomer, have gained increasing attention since they change their characteristics like volume and hydrophilicity by modulating temperature, pH, or salt concentration, also have the potential to solve the existing issues in large-sized protein imprinting such as low mass transfer, low binding capacity and especially easy template removal as the pores are open by varying their environmental conditions. Xie et al. [74] proposed a quick and common molecular imprinting approach for specific capture of glycoprotein via dual pH-responsive polymer imprinted magnetic spheres using DMA as a pH-sensitive monomer along with VBA as an affinity monomer and MBA as a crosslinker containing OVA as template molecules.
Schematic overview of templates used in glycoproteomics, molecularly imprinted polymer synthesis, and types of interactions involved.
Preparation of glycan-binding MINP via molecular imprinting, with a schematic representation of the crosslinked structure using a mixture of DVB and 7 as the free radical cross-linkers. Adapted with permission from [57], copyright 2020 Nano Letters.
Usually, the conditions needed for the imprinting of native proteins as a template may disturb the protein structure and cause difficulty in removing the protein template. To tackle this issue, Yang et al. [49] synthesized epitope-imprinted polyethersulfone (PES) polymer via PES phase inversion in a nonsolvent using epitope of TRF comprising of N-terminal sequence MRLAVGALL in dimethylacetamide solution. The phase inversion occurred when the mixture was transferred to the water by quick exchange of dimethylacetamide and water, the formation of TRF epitope imprinted solidified PES-particles appeared. Furthermore, due to an appropriate polarity of dimethylacetamide, it dissolved three different types of epitopes regarding their physical and chemical characteristics, so, the same strategy was used for the imprinting of three different epitopes, with a different GRAVY for the recognition of HAS, IgG, and TRF [75]. It should be remembered, however, that there are also some significant disadvantages of bulk imprinting: 1) A lot of the recognition sites are positioned within the particles, leading to the consumption of a long time for saturation; 2) Captured targets are difficult to elute, causing poor recovery; 3) The efficiency of the template is limited, decreasing the binding potential and recognition capability of captured targets. The imprinting process can be updated in the ways to overcome these problems as follows: 1) By expanding the channel pores on the surface to allow a high mass transfer or using the smart polymers; 2) By reducing the size of polymer particles, including the nano-size, to enhance surface recognition sites. Some attempts have recently been made to fabricate nanoparticles via an updated phase inversion process [76], to benefit the future solution of the above-mentioned problems.
2.4.2 Surface Imprinting
Surface imprinting is more advantageous, as the template may be perfectly oriented on solid support followed by the polymerization, which may give rise to a certainly aligned cavity, easily accessible to the rebinding of template molecules[77]. Compared to bulk imprinting, surface imprinting facilitates complete removal of the template and offers significant availability to target moieties, particularly for large peptides, proteins, and cells. The imprinted cavities in bulk-imprinted materials are usually incapable of binding these large molecules[78]. However, the controlled and appropriate thickness of the imprinting layer is very important in surface imprinting because the small thickness does not provide the desired specificity, and too large thickness buries the template inside the imprinting layer. A lot of surface imprinting strategies have been reported in the past few years, which described excellent efficiency for glycosylated molecules. Here, we have summarized some of the important and recent strategies. Pan and co-workers [46] developed a strategy based on enhanced binding affinity to specifically capture the glycoproteins via multiple boronic acid sites introducing polyethylene polyamine as a scaffold applying surface imprinting. Boronate affinity-based magnetic MIPs have low binding capacity because of less surface area due to the smoothness of silica-like polymers or magnetite surfaces and narrow working pH ranges offered by boronic acid ligands. Sun et al. [79] proposed a magnetic molecular imprinting approach by functionalizing porous TiO2, coated Fe3O4 with boronate affinity, followed by controlled polymerization of dopamine and named as Fe3O4@pTiO2@MIP. Compared to silica, flowerlike TiO2 coated on the Fe3O4 exhibits a high surface area and offers more binding sites and high binding capacity. Due to the strong electron-withdrawing effect of Ti (IV), Fe3O4@pTiO2@MIP selectively captured glycoproteins in a wide range of pH (6.0 to 9.0). Tian and co-workers [80] also explored a facile method using chelation interaction action for molecular imprinting of glycoproteins, where OVA was immobilized on the surface of MCM-48 via Cu2+-affinity chelation interactions, followed by the formation of an imprinting layer using PDA in a controlled manner. The prepared MIP exhibited fast binding kinetics and completely captured OVA within 30 min and showed the binding capacity of 395.30 mg g-1.
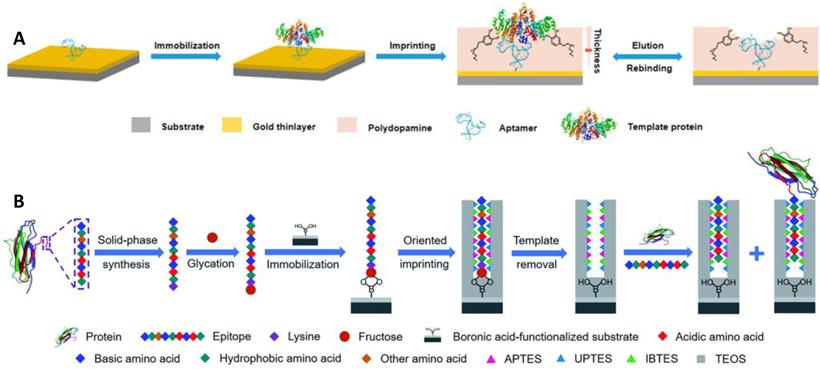
Aptamers have a lot of special advantages, including in vitro synthesis, facile modification, bio-compatibility, and a wide variety of possible targets [81]. However, they usually stuck to poor affinity and low specificity due to their dynamic structures. The integration of MIPs and aptamers may produce ideal hybrid alternatives with desired characteristics. Li et al. [71] reported a novel approach based on the incorporation of aptamer and MIPs in the controlled and facile way for specific and high-affinity recognition of glycoproteins (Figure 3A). Thiol-functionalized aptamer, with less affinity and low specificity for a protein target (alkaline phosphatase, ALP), was fixed on the gold surface via S-Au bond and ALP was immobilized, followed by the controlled development of a thin imprinting layer via self-polymerization of DA. The resulting aptamer-MIP hybrid exhibited enhanced affinity and specificity towards ALP, cross-reactivity of 3.2-5.6%, and a dissociation constant of 1.5 nM. In the aforementioned strategies, the glycoproteins were used as a template, however, the template glycoproteins are unstable, adopted for the polymerization of the monomers for imprinting, and consequently, the imprinting cavity could specifically recognize the template glycoproteins, and importantly its characteristic fragments too. Bie et al. [59] developed lectin-like MIPs based on boronate-affinity glycan-oriented surface imprinting to tackle these issues. Glycans (multiple or single) were obtained by digesting target protein using glycosidase followed by purification via ultrafiltration and bound to the surface of the nano-magnetic cores using boronate affinity, imprinting layer was developed and glycan templates were removed. The resulting MIPs were capable of specifically capturing glycoproteins, their glycopeptides, and glycans as well. Moreover, MIPs exhibited an excellent IF value of 8.4 for RNase B and 21.8 for TRF with IE values of 44.8% and 43.5%, respectively. Another solution to the aforementioned problem is epitope imprinting [64, 65]. However, epitope imprinting is stuck to epitope immobilization-related issues as mentioned above. Xing et al. [44] reported an epitope imprinting approach, where C-terminus epitope was glycated and attached to a boronic acid-modified core as a template, accompanied through controllable oriented surface imprinting, by reacting multiple silylating reagents and functionalities interacted with the epitope, such as APTES, UPTES, IBTES, and TEOS as depicted in Figure 3B. C-terminal nonapeptides for human β2-Microglobulin (B2M) and Mb were KIVKWDRDM and NYKELGFQG, respectively, selected from protein structure databases such as UniProt. The prepared epitope imprinting material exhibited Kd and Qmax value of (2.08 ±0.11) ×10-7 M and (169.03 ±5.50) nmol g-1, respectively, for β2-Microglobulin. Qin et al. [36] also developed a novel thermo-responsive dual template epitope imprinting approach, where C-terminal peptides of transferrin and human serum albumin were employed as templates and zinc acrylate as affinity monomer to produce a six-membered ring with epitope via metal chelation, NIPAAm as a thermos-responsive monomer, and EGDMA as a crosslinker for the polymerization which was executed in 30 min to generate the imprinting cavity.
(A) Schematic illustration of the principle and procedure for preparing aptamer-MIP hybrid. (B) Schematic of the principle and procedure of controllable oriented surface imprinting of boronate affinity-anchored epitopes. Adapted with permission from [71], copyright, 2019 Analytical chemistry and [44], copyright 2019 Chemical science.
3. Cancer diagnostics and imaging potential of glycoprotein imprinted nanoMIPs
Cancer must be diagnosed early to minimize death rates. Despite significant advancements in the area of improved diagnostic and prognostic tools, only a tiny percentage of tumor victims are identified in early stages. The identification of circulating tumor cells is an efficient method for predicting cancer prognosis at early stages [82]. Unfortunately, due to the limited quantity of circulating tumor cells in circulating blood, effective techniques for their selective identification and separation are highly desired. Traditionally, antibodies have been in practice using immunoassays by specific recognition of the target, but the methodology suffers from massive cost, long time of fabrication, and instability [83]. As artificial antibodies, MIPs are the effective alternative to natural antibodies, regarding their cost and stability, and have been effectively employed in the field of cancer diagnostics because of their captivating characteristics such as cost-effectiveness, high specificity, facile synthesis, high stability, and reusability. Additionally, MIPs show good compatibility with commonly used detection tools in sensor development. In addition, bioimaging is also one of the key foundations in biomedical study, since it enables early identification, surgical guiding, therapy impact monitoring, and clinical prognosis [84, 85]. Conventionally, imaging modes include magnetic resonance imaging (MRI), X-ray, ultrasound, positron emission tomography (PET), and computed tomography (CT) [86], however nanoprobe-based fluorescence imaging is considered as one of the developing techniques, and it is growing rapidly in combination with nanotechnology for cancer research.
3.1. MIP-based biosensing technology for cancer diagnostics
NanoMIPs outperform natural antibodies in terms of thermal, physical, and chemical stability, giving biosensors a considerable advantage in identification even when exposed to environmental stresses [87]. These nanoMIPs can be coupled with different types of detection systems to produce efficient biosensors for glycoproteins, however, glycoprotein imprinted MIPs-based sensors for tumor monitoring are discussed here. Ouyang et al. [88] reported a glycoprotein biomarker imprinted biofuel cell incorporating boronic acid anchored biliroxidase-carbon nanotube as a biocatalyst and signaling probe, and exhibited high sensitivity and specificity for AFP in human serum sample with a LOD value of 1 ng mL-1. Cuschieri and co-workers [89] used a nine-amino acid epitope of VEGF as a template followed by imprinting and resulting nanoMIPs were used to target human-VEGF. These nanoMIPs were integrated with quantum dots to make them fluorescent by embedding and surface attachment. The resulting material exhibited high affinity (Kd = 1.6 nM) for target human-VEGF and low affinity for non-target molecules. Although no definite imprinting parameters on the protein level were described, QD-MIPs directly targeted VEGF-overexpressed human melanoma cell xenografted in zebrafish embryos.
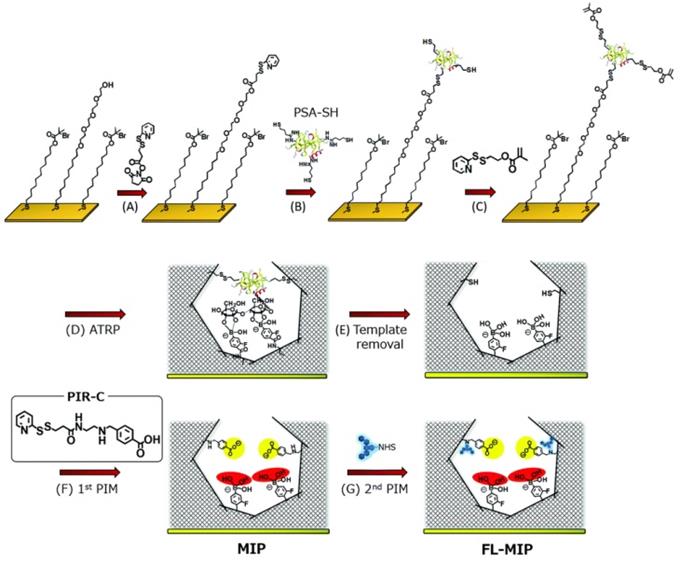
For electrochemical sensing, the conductivity of the exposed surface is highly important to depict the strong response. Inspired by the electrical conductivity of PANI, Moreira, and Sales [90] reported an electrochemical sensor for CEA (a colorectal cancer biomarker). The integrated device consisted of a MIP-based sensor unit and a dye-sensitized solar cell as a voltaic cell. The imprinting layer was developed using CEA on a highly conducting PANI-counter electrode by electropolymerization and the template was removed using proteinase K action. The resulting sensor was assessed by electrochemical impedance spectroscopy and square-wave voltammetry as detection modes. The self-powered detection of CEA was similar to common biosensors with excellent sensitivity due to the incorporation of MIPs on the counter electrode. Irradiating the light on the surface of the photoanode of the dye-sensitized solar cell, a current was generated to run the device. Xing et al. [91] reported a dual MIP-based plasmonic immunosandwich assay for the sensitive sensing of the cancer glycoprotein marker. The technology was made up of two imprinted parts: Ag@SiO2 nanoparticles with an N-terminal epitope and a gold nanoparticle monolayer-coated glass slide with a C-terminal epitope. The strategy integrated high specificity of dual-MIPs and excellent sensitivity of SERS detection and evaluated for model glycoproteins and neuron-specific enolase (NSE) in human serum. Furthermore, relative to traditional ELISA tests, the MIP-PISA showed significant features, such as convenience, faster response, reduced sample amount, and a wider linear range. Zhou et al. also reported a similar dual MIP-based plasmonic immunosandwich assay comprising two imprinted parts where one of the parts is N-terminal or C-terminal (similar to the aforementioned study) of the polypeptide and the other part will be the glycosite, and other criterions are same [56]. Saeki et al. [54] prepared PSA fluorescent sensor coupling molecular imprinting and following multistep post-imprinting modifications with a novel designed multifunctional reagent, fluorescent-signaling molecularly-imprinted nanocavities with orthogonal dual recognition sites for the sensing of prostate cancer biomarker glycoprotein, as shown in Figure 4. Zhang et al. [92] also presented a molecularly imprinted strategy coupled with a highly sensitive electrochemical detection system for the detection of PSA. The sensing substrate was made of MoS2 and gold nanoparticles, the surface imprinted cavities were made of PSA template and 4-mercaptophenylboronic acid, and the tracing tag was made of gold polymerized methylene blue composites labeled with 4-mercaptophenylboronic acid.
Tawfik et al. [93] developed a cost-effective, simple-to-use, and enzyme-free, cancer biomarker detection assay based on a new method involving fluorescent molecular imprinting conjugated polythiophenes (FMICPs). With a PLQY of up to 55 percent, the attractive conjugated polythiophenes structure offered a simple and inexpensive technique for free-enzyme signal production. Interestingly, the paper-printed probe was integrated with a low-cost smartphone and accessible prototype testing equipment for point-of-care cancer detection. In addition, the FMICP-nanofibers exhibited 80 times higher sensitivity than that of native FMICPs. For real sample applications, the proposed sensors were effectively used for the rapid detection of AFP in patients with liver cancer, with outcomes that were consistent with clinical ELISA results. Among these sensors, MIPs coupled electrochemical and fluorescent sensors are easy to miniaturize to develop a point-of-care device for rapid and on-site cancer testing just like the glucose sensors. Therefore, there is a need to develop these types of low-cost, easy-to-operate, highly sensitive POC devices, urgently. Some of the recent studies on biosensors for sensing of different types of glycoprotein cancer biomarkers and features of these materials are compared in Table 3.
Synthesis procedure of molecularly-imprinted polymer (MIP) thin layer possessing orthogonal dual interaction sites and fluorescent reporting groups via molecular imprinting and sequential post-imprinting modifications (PIMs). Adapted with permission from [54], copyright, 2019 Journal of Materials Chemistry B.
Summary of MIPs-coupled biosensors for detection of different types of glycoprotein cancer biomarkers.
| Sensor type | Monomers | Imprinting type | Target protein | LOD | Cancer type | Linear dynamic range | Sample | Reference |
|---|---|---|---|---|---|---|---|---|
| Electrochemical | ANI | Bulk imprinting | CEA | 0.10 pg mL-1 | Colorectal cancer | 0.025 ng mL-1 | Human urine | [94] |
| APBA/o-PD | Surface imprinting | PSA | 0.03 pg mL-1 | Prostate cancer | 1.0×10-4 to 1.0×104 ng·mL-1 | [92] | ||
| Phenol | CA 125 | 0.5 U mL-1 | Ovarian cancer | 0.5-400 U mL-1 | [95] | |||
| Fluorescence | FMB | PSA | 2.35 ng mL-1 | Prostate cancer | 5 to 150 ng mL-1 | [54] | ||
| PYM, MPC/MBAA | AFP | 0.27 ng mL-1 | HCC | - | [96] | |||
| SERS | APTES, UPTES, IBTES, TEOS | CEA | 5.6 × 10-14 M | Colon cancer | 1 ng mL-1 -10 μg mL-1 | Human serum | [56] | |
| NSE | 10 pg mL-1 | small cell lung cancer | 100 pg mL-1 - 10 μg mL-1 | [91] |
3.2. MIPs-based imaging technology for cancer diagnostics
The capacity to track biomolecules and related compounds with excellent chronic and spatial resolution is crucial for understanding biological systems at the molecular level [93]. Initially, it was started with the discovery of green fluorescent protein (GFP) and its derivatives contributed towards the progression of bioimaging [97] and evolved with the development of organic dyes [98] followed by the quantum dots [99] for both in vitro and in vivo studies. However, GFP and organic dyes-based imaging technologies are limited in sensitivity and long-term stability. In this regard, QDs are gaining increasing interest due to their splendid optical characteristics, dimension-dependent tunable emission wavelength, and consecutive excitation of QDs using a single light and wider spectral windows [100]. Antibodies coupled with QDs have just been used to effectively monitor a broad variety of biological phenomena among the cells and animals, offering the site and morphology of cells [101]. It revealed the process of tumorigenesis, growth, and metastasis by shedding light on the functioning of physiological processes.
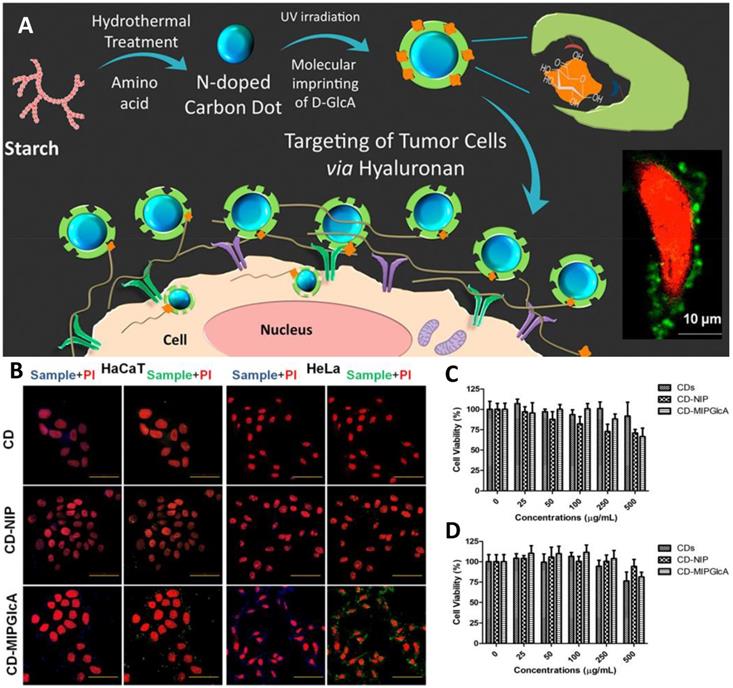
Initially, small molecules such as nitrobenzoxadiazoles were coupled with nanoMIPs for developing fluorescent molecular imprinting probes for cell imaging [67]. The surroundings of tumor tissue and the intracellular microenvironment both are complex, thus, enzymes there and ambient pH may result in quick fluorescence quenching [102]. On the other hand, quantum dot-coupled imprinted particles provided better imaging performance. The imaging quality of QD-integrated MIPs is an alternative to immunofluorescent probes. Using QDs emissions, stimulated by UV light, Haupt's group suggested a unique local photopolymerization technique for producing MIP-based QDs (QDs@MIPs) for the identification of glucuronic acid and N-acetylneuraminic acid functionalized on the cells [103]. Wang et al. [104] created several QD-based MIPs for cell pattern identification via multiple imaging by attaching to three different types of monosaccharides. Various fluorescence signals were detected, indicating the properties of these three monosaccharides on different cells, exposing their similarities and differences, and enabling the identification of particular cancer cells. Haupt's group then used MIPs with carbon nanodots (CD) to track hyaluronan on cancer cells. CDs, as opposed to QDs, exhibit more steady luminosity without blinking [105] as depicted in Figure 5. Furthermore, unlike QDs, they do not comprise heavy metals and are more biocompatible. Medina Rangel et al. [106] described a novel and effective approach based on CD-based MIPs, efficient for probing and staining hyaluronic acid existing on human epidermal cells, aided by a solid-phase method. Therefore, similar to polyclonal antibodies in probing and staining mechanism. Monosaccharide imprinted MIPs is not so specific to targeted sites, however, the epitope importing and glycoprotein imprinting technology is a more specific and direct approach and may lead to effective targeted imaging.
Epidermal growth factor receptors (EGFR), also commonly recognized as HER family, are expressed by the EGFR gene, and belong to tyrosine kinase receptors. Their overexpression may lead to the abnormal growth of the cell. Zhang et al. developed an imaging probe using carbon-dots (CDs) as a fluorescent-core followed by imprinting using the epitope imprinting technique [107]. MIPs were prepared by reverse emulsion polymerization in a mixture of water/hexane combination in the presence of dioctyl sulfosuccinate sodium salt and Brij-30 as surfactants using an extracellular N-terminal nonapeptide epitope (EEKKVCQGT) modified with palmitic acid as a template. The function of palmitic acid was to keep the conformation of the epitope at the oil-water interface. The resulting imaging revealed that MIP-target HeLa cells (high-expression EGFR) glow more brightly than MIP-target MCF-7 cells (low-expression EGFR), indicating that the C-MIP targets tumor cells that overexpress EGFR. Due to its low tissue penetration, the single fluorescent imaging mode is not favorable for correct diagnostics. Considering the issue, Ren et al. [108] proposed glycan-oriented MIPs, using the BACOSI technique for high-resolution fluorescent and magnetic resonance imaging (MRI). First Gd-comprised fluorescent silica nanoparticles were fabricated and used as a core, followed by the functionalization of boronic acid to immobilize the epitope (Gal-NAc) of Tn antigen, and then surface imprinting was performed. Gd enhanced the fluorescent and MRI-imaging resolution as it is a well-known fact that doping of heteroatom (B, P) or metal ions in SiNPs can significantly boost the intrinsic properties [109, 110]. Resulting NPs exhibited a size of 31.8 nm and revealed stable fluorescence capability with an adsorption capacity of 1.15 μM g-1, IF of 7.5, and low cytotoxicity. It has great potential for targeted cancer cell imaging with high resolution in real samples and MIPs integrated with imaging probes for targeted tumor imaging has been summarized in Table 4.
(A) Schematic diagram for tracking hyaluronan on cancer cell targeting and imaging by molecularly imprinted carbon dots; (B) Confocal microscope images of fixed HaCaT and HeLa cells treated with CDs, CD-NIP, and CD-MIPGlcA; Cell viability (MTT) assay of (C) HaCaT and (D) HeLa cells in the presence of CDs, CD-MIPGlcA, and CD-NIP for 24 h. Data are means from five different experiments. Adapted with permission from [105], copyright 2018 ACS Applied Materials & Interfaces.
Summary of MIPs-coupled imaging technology for cancer diagnostics.
| Imaging type | Template | Monomers | Tumor model | Imaging probe | Cancer type | Reference |
|---|---|---|---|---|---|---|
| Fluorescence | Epitopes of EGFR | Aam/MBAA | HeLa cells (over-expression EGFR), HeLa-tumor-bearing mice | CDs | Cervical cancer | [107] |
| Glucuronic acid | AB, MAM/EGDMA | HA on HaCaT cells | N-doped CDs | [105] | ||
| Hyaluronan | NIPAM/EbAm | Rh B | [111] | |||
| Fluorescence & MRI | Gal-NAc | TEOS | MCF-7 cells | Gd-doped SiNPs | Breast cancer | [108] |
| SERS | Sialic acid | HepG-2 | Raman-active AgNPs | HCC | [112] |
4. Image-guided cancer treatment potential of glycoprotein imprinted nanoMIPs
Cancer is the world's second-highest cause of death and a serious public health issue [113]. Mainly, chemotherapy has been in practice for cancer treatment, however, it has some obvious drawbacks, such as being non-specific and harmful to healthy normal tissues and producing significant adverse effects. Unaddressed medical need is the development of an innovative way to address the aforementioned problem. Furthermore, tumor progression is usually occurred along with aberrant glycosylation, which gives rise to the possible markers for cancer cells. Alterations are often found during cancer, including abnormal branching of N-glycans, shortened O-linked chains, fucosylation, sialylation, and glycosphingolipid expression [114, 115]. Several glycoproteins are tumor biomarkers linked to tumor cells. Therefore, glycoprotein targeted drug delivery is highly important, however, there are few ligands [12] that can identify or target important glycan biomarkers, limiting the establishment of glycan-targeting nanomedicine for cancer treatment.
4.1 NanoMIPs as a carrier for drug delivery
Among the advanced nanomaterials, molecularly imprinted polymers have numerous benefits of simple fabrication procedures, low cost, excellent stability, and facile incorporation with nanomaterials. Recently, their potential medical applications are found in the field of tumor targeting [116] and targeted drug delivery [117, 118]. They are produced in novel formats and are a promising tool for cancer nanomedicines with a targeted drug release system. Premature drug release during blood circulation of chronically given cancer nanomedicines might limit the quantity of medication reaching tumors while also causing adverse effects [119, 120]. Once the nanomedicines reach the target site, they intend to release the medicine in the required effective therapeutic amount. In addition, drug release kinetics are also very important and the antitumor drug should have a continuous stimuli-responsive. These problems may be tackled using nanoMIPs. Initially, non-covalent interactions were employed to capture the target monosaccharide of the glycoside but with the evolution of boronate affinity, nanoMIPs were capable to capture the template moieties with higher affinity, specificity, and pH-dependent detachment. For drug loading, non-covalent interactions are usually used to capture the drug molecules, tuned by pH or temperature according to the need, and hydrogen bonding is most common in practice. Because the terminal of glycoproteins and lipids on the cell membrane of cancer cells generally overexpress SA. Therefore, SA-imprinted mesoporous nanocarriers (SIMNs) exhibit cancer selectivity and targeted drug delivery. Considering the phenomenon, Yin et al. [121] fabricated a sialic acid-imprinted hydrogel layer on mesoporous silica-NPs to specifically bind the sialic acid moieties present on the glycoproteins and lipids attached to the cell membrane of cancer cells. The common pharmaceutical medication doxorubicin (DOX) was loaded in the mesopores of the silica-NPs and human hepatocellular carcinoma cells (HepG-2) were targeted for the drug release using sialic acid imprinted silica-NPs. These NPs targeted SA-overexpressed cancer cells and increased cancer cellular reception, ultimately resulting in better suppression of cancer cell growth.
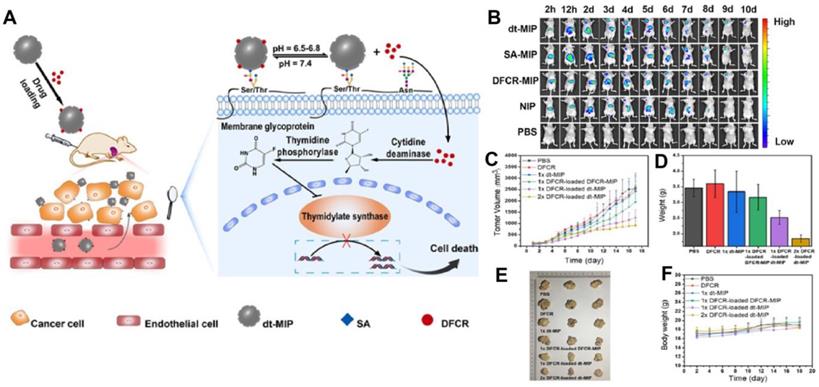
Prodrugs are inert progenitors of effective therapeutic agents that are biologically converted or triggered in the body after delivery to enhance the parent drug's pharmacokinetic characteristics [122, 123], and more effective way as compared to direct chemotherapy. Liu and co-workers [116] integrated nanoMIPs with prodrug to produce a smart prodrug delivery probe, which exhibited longer retention, tumor microenvironment-triggered releasing, and specific targeting. 5'-deoxy-5-fluorocytidine (DFCR) was used as a prodrug, SA as a tumor marker, and this combination was used as a co-template, and the imprinting was done using a silica layer and resulted in a smart nano-MIP carrier. Prodrug-loaded smart carrier collected precisely and effectively at the tumor site before progressively releasing the drug (Figure 6). This MIP-based prodrug delivery is liver-independent instead of tumor-dependent, unlike other prodrug designs that frequently need in-liver bioconversion.
Using monosaccharide imprinted MIPs is not so specific to the targeted sites, however, the glycoprotein imprinted materials are more specific and direct and lead to the development of effective targeting nanomedicines. Considering this, Han et al. [124] fabricated CA125-imprinted graphene oxide, using DA as imprinting monomer revealing an IF of 6.4. While DOX was loaded on the CA125-imprinted GO, it exhibited enhanced cytotoxicity to the tumor cells and high specificity to the overexpressed CA125 sites. To prevent the conformational changes in the larger structure of glycoprotein, aqueous and compatible reactions conditions are necessary. Furthermore, the removal of proteins templates is difficult due to their large size and numerous interactions with different ligands which give rise to high cross-reactivity [125]. As a result, the use of intact protein-imprinted MIPs for targeted delivery in cancer nanomedicine has been constrained.
Epitope-imprinting is an attractive way for the recognition of receptor glycoproteins on the cancer cells, where a short peptide is used as a template [126]. The epitope-imprinting shows considerable benefits over whole protein-templated MIPs, including an increased affinity to target glycoproteins, decreased nonspecific bindings, simple template production, and broad synthesis conditions [127, 128]. Based on these fascinating features, epitope imprinted NPs are extensively implemented for preparing nanoMIPs in tumor treatment. Hamid et al. [129] assessed HER2 receptors and produced the most effective epitope. The fabricated epitope and DOX were used as templates and PDA-based imprinting was executed on silica-NPs. The nanoMIP was directly used for ovarian cancer, indicated by increased DOX concentrations in tumor tissues, improved antitumor effectiveness, and longer mouse life.
(A) Schematic of the drug transport and action mechanism in the dual-templated MIP-based smart prodrug delivery system, (B) In vivo fluorescence imaging of HepG-2 tumor (left upper chest) and liver site (upper abdomen) after intravenous injection ofNIR797-doped dt-MIP, SA-MIP, DFCR-MIP, NIP and PBS for different times, (C) Mean tumor volume, (D) Mean tumor weights after excision at 18 days, (E) Representative photographs of mice in different groups after treatment for 18 days, (F) Body weight of the mice in different groups at different time intervals. Adapted with permission from [116], copyright 2021 Angewandte Chemie.
Recently, metal-organic frameworks have gained increasing attention due to their well-ordered arrangement and unique porous structure with tunable porosity, especially when they are reduced in size to the nanoscale, they have become efficient drug carriers [130] and imaging probes [131] for delivering drugs, chemotherapy, and phototherapy [132]. It is worth mentioning that zeolitic imidazolate framework-8 (ZIF-8) is biodegraded completely in an acidic environment. Considering this, Qin et al. [133] fabricated fluorescent zeolitic imidazolate framework-8 incubated it with DOX, as a core, and developed MIP using epitope of CD59 glycoprotein as a template, to specifically target MCF-7 cancer cells, termed as FZIF-8/DOX-MIP. This biodegradable FZIF-8/DOX-MIP can specifically target the CD59 cell membrane glycoprotein present on MCF-7 cancer cells. MIPs-layer was capable to be disrupted in the tumor acidic environment, allowing FZIF-8/DOX-core to further degradation in an acidic environment to release the DOX and demonstrated fluorescent imaging and active drug release. Relative to other cells, MCF-7 cells were more capable of phagocytosing FZIF-8/DOX-MIPs, and FZIF-8/DOX-MIPs were more deadly to MCF-7 cells.
4.2 NanoMIPs for photodynamic and photothermal therapy
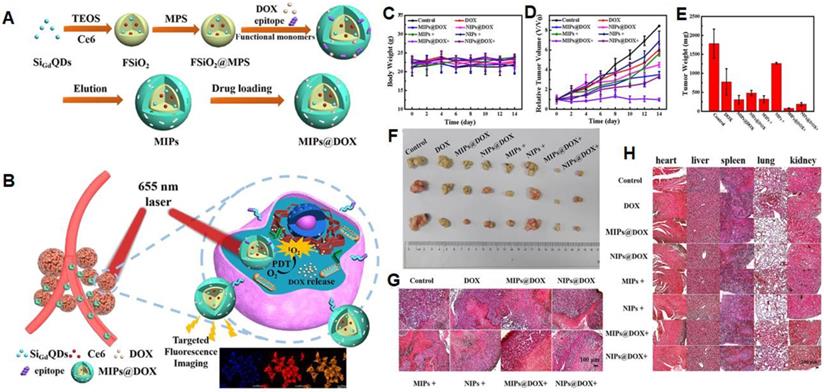
NanoMIPs have potential applications in photodynamic and photothermal therapies for cancer treatment. In photodynamic therapy, a photosensitizer is introduced which produces the cytotoxic reactive oxygen species by the irradiation of light. These produced reactive oxygen species kill targeted cells at a particular irradiated site. Liu et al. [134] described an inducible epitope imprinting approach where an imprinting template, a folate receptor-a (FR-a) epitope (QTRIAWARTELLNVAMNAKH) was folded to an a-helix shape and used as a template to produce nanoMIPs to capture the target peptide alter it from disordered-to-ordered conformational change, consequently inducing binding sites in the target receptors. To make the MIPs more specific to the target, strong epitopes are created by obtaining the sequence and 3-dimensional arrangement from data banks and identifying epitopes using in silico techniques. In comparison to NIP, MIPs show enhanced binding affinity to FITC-modified, peptide, and recombinant FR in fluorescence polarization assays. For fluorescence imaging and flow cytometry, polymers tagged with the fluorescent dye Nile red demonstrated stronger binding with human cervical cancer HeLa cells (FR- positive) than with lung cancer A549 (low FR) cells. Compared to NIP-NPs and NPs imprinted with the twisted peptide, MIP-NPs encapsulate the near-infrared dye IR-783 accumulated at tumor sites of animals transplanted with HeLa cells. Methylene blue (MB) was incorporated in the pre-polymerization mixture as a photosensitizer to develop nanoMIPs with a size of 78 nm for the generation of reactive oxygen species and these nanoMIPs demonstrated a strong inhibition effect on HeLa cancer xenografts in the following photodynamic treatment. In another study, Zhang et al. [63] used an N-terminal α-helix of the p32, an overexpressed glycoprotein on the tumor cell membrane that was grafted on apamin substrate, as a template. As a result, apamin sites were substituted with topologically comparable residues from p32's N-terminal -helix, however, the residues with the highest -helix affinity were preserved. The peptide was linked with palmitic acid at the free amino group of the side - chain to guarantee that it is well-orientated at the contact of the water and oil regions throughout polymerization. Microemulsion polymerization was carried out at room temperature to produce sub-40 nm-sized nanoparticles that were specifically bound to P32, P32-up regulated cancer cells, and effectively performed the targeted photodynamic treatment in vivo. Recently, Peng et al. [135] demonstrated dual template nanoMIPs revealing two different therapies, simultaneously as shown in Figure 7. The silica nanoparticles were used as a core, comprising fluorescent gadolinium-silicon quantum dots for fluorescence imaging and MR-imaging, and photosensitizers (Ce6) for liberating toxic O2 by 655 nm laser radiations to kill the tumor cells. The polymerization was done in the presence of two templates, the epitope of CD59 (YNCPNPTADCK) and DOX, and after the removal of the template, the resulting nanoMIPs were capable to specifically load the drug and target the CD59-overexpressed tumor sites. In vitro and in vivo experimentations revealed, after specific targeting the CD59-overexpressed tumor sites, nanoMIPs effectively released DOX in acidic microenvironment and irradiating 655 nm laser radiations, photosensitizers (Ce6) killed the tumor cells and allowed clear fluorescent and MR-imaging.
Photothermal therapy is frequently used in cancer care since it promotes the apoptotic or necrotic behavior of cancer cells while also suppressing tumor development by creating a localized thermal effect [136]. Yin et al. [137] reported sialic acid imprinted nanoMIPs for targeted photothermal tumor therapy. The nanoMIPs were fabricated by immobilizing sialic acid template on boronic acid-functionalized AuNRs followed by controlled imprinting. HepG-2 as overexpressing SA cells and L-02 as normal cells were treated with FITC-doped MIP nanorods to differentiate cancer cells from normal cells, and confocal microscopy and flow cytometry demonstrated that these nanoMIPs efficiently target the HepG-2 cells. In vitro investigations revealed that after 6 minutes of illumination with a 750 nm laser beam, MIP nanorods caused apoptosis in HepG-2 cells as measured by the MTT assay, however, the survival of L- 02 cells maintained greater than 85%. For in vivo studies, nanoMIPs demonstrated a substantial decrease in tumor volume in 2 weeks after laser irradiation and a strong photothermal impact on the ablation of HepG-2 tumors in mice. AuNR-MIPs might be used as selective medicine for tumor ablation employing photothermal treatment without harming normal tissue.
(A) Schematic Illustration for the Preparation of MIPs@DOX; (B) Schematic Illustration of MIPs@DOX for Targeted Chemo-Photodynamic Synergistic Treatment of Tumor in vivo. Body weight changes (C) and relative tumor volume (D) in 14 days after various treatments. The average tumor weight (E) and corresponding tumor tissues (F) after various treatments for 14 days. H&E staining of tumor sections (G) and main organs (heart, liver, spleen, lung, and kidney; H) were collected from various treatments groups on the 14th day. Adapted with permission from [135], copyright 2020 ACS Applied Materials & Interfaces.
4.3 NanoMIPs themselves as therapeutics
Human epidermal growth factor receptor-2 (HER2) belongs to the epidermal growth factor receptor (EGFR) class and is upregulated in 20-30% of breast cancers [138, 139]. The most powerful EGFR signaling dimer among the heterodimers is HER2/HER3 dimer [140]. Prevention of HER2 dimerization with other EGFRs, particularly HER3, offers an appropriate therapy for HER2-positive breast cancer. Dong et al. [141] presented a novel methodology incorporating molecular imprinting technique for the inhibition of HER2+ breast cancer growth. The glycan-imprinted nanoparticles were prepared using HER2 N-glycans as a template prepared through the digestion of HER2 protein by PNGase F, immobilized on the fluorescent silica nanoparticles via boronate affinity followed by the imprinting employing a cutting-edge imprinting technique known as boronate-affinity-controllable oriented surface imprinting. The fabricated nanoMIPs may capture nearly all HER2 glycans and prevent HER2 dimerization to other HER2 receptors, limiting downstream signaling pathways and reducing the development of HER2+ breast cancer. The in-vitro studies revealed that nanoMIPs attached to HER2+ cells specifically, thereby inhibiting cell proliferation up to 30%. In vivo experimentation demonstrated that nanoMIP-treated group's average tumor volume was only approximately half that of non-treated groups. The study offers a novel way to treat HER2-positive breast cancer and adds to the growing body of data that nanoMIPs may be used to treat cancer. Among the techniques, the pristine protein imprinted polymers are difficult to manufacture, although the procedures are basic and straightforward for producing nanoMIP. This is especially true when the proteins are unstable or not readily available in their pure condition. Although epitope-imprinting is an effective way to tackle the issue, but still needs further improvement in the conformational preservation of epitope peptide throughout the imprinting process.
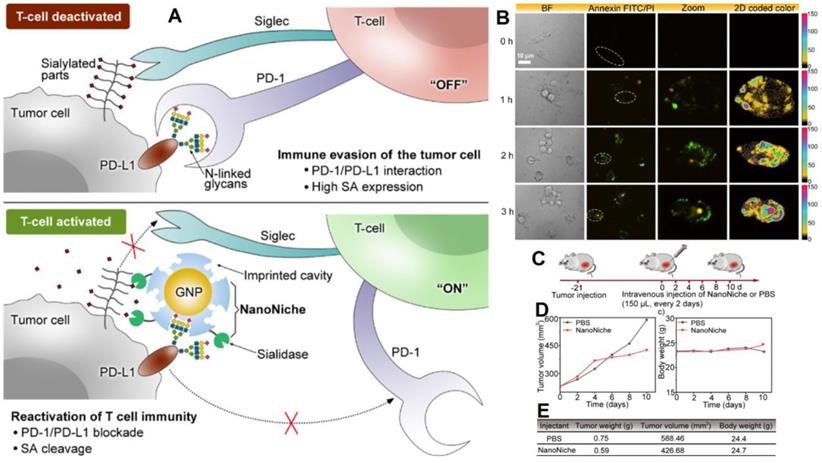
Discovering of programmed death 1 (PD-1) and programmed death-ligand 1 (PD-L1) signaling pathways have altered clinical cancer therapy since their inhibition plays a key role in the regulation of immunological T-cells [142-144]. Inhibiting PD-1 or PD-L1 to reactivate T-cell response has proven an effective cancer therapy technique, and antibodies targeting PD-1 or PD-L1 have been extensively applied for the treatment of many malignancies [145, 146]. However, N-linked glycosylation of PD-L1 can make its antigenic epitopes unidentifiable by traditional PD-L1 antibodies. Zhou et al. presented “NanoNiche”, a synthetic PD-L1 antibody integrated with desialylation functioning to boost up immune cell activation [147], as shown in Figure 8. NanoNiche was prepared by immobilizing the glycans of PD-L1 on gold nanoparticles followed by the formation of a silica imprinting layer. Interestingly, sialidase was covalently attached to the nanoMIPs for desialylation on the surface of the cell. NanoNiche has been shown to target the PD-L1 on the surface of tumor cells, causing desialylation and inhibiting the proliferation of MDA-MB-231 cells in vitro. In vivo studies in tumor cells reveal improved treatment effectiveness. As a result, the NanoNiche-sialidase conjugate is a viable immune-checkpoint blocking treatment option. Some of the recent studies depicting the current cancer treatment, targeting specific cells with different mechanisms of treatment and features of these materials are compared in Table 5.
NanoMIPs executes multitasks for cancer therapy targeting specific sites.
| Type of therapy | Template | Monomers | Tumor model | Drug | Targeted cancer | Reference |
|---|---|---|---|---|---|---|
| Drug delivery | CA125 | DA | SMMC-7721 | DOX | Liver cancer | [124] |
| SA | NAPMAAm, NIPAAm, Aam/MBAA | SA over expressed HepG-2 cells | [121] | |||
| N-terminal epitope of CD59 glycoprotein | TFMA, DMAEMA, NIPAAm, TBAm/ BAC | MCF-7 cancer cells, MCF-7-tumor-bearing mice | Lung cancer, breast cancer | [133] | ||
| Linear peptide of HER2 | Aam, ZnA/ EGDMA | SK-BR-3 cells | HER2- breast cancer | [148] | ||
| SA | TEOS | HepG-2, MCF-7 cells, HepG-2 tumor-bearing mice | DFCR (prodrug) | Hepatic cancer & breast cancer | [116] | |
| Photodynamic therapy | N-terminal peptide of FR-α | TFMA, Aam/MBAA | HeLa cells (FR-α over-expressed cells), HeLa-tumor-bearing mice | MB (photosensitizer) | Cervical cancer | [134] |
| Disulfide-linked α-helix-containing peptide-apamin | Aam/ MBAA | 4T1 murine cells and BxpC-3 human cells, 4T1tumor-bearing mice | Breast cancer | [63] | ||
| Photodynamic therapy + Drug delivery | Linear peptides of CD59 DOX | NIPAAm, TBAm, Aam/ MBAA | MCF-7 cancer cells | DOX + photosensitizers Ce6 liberating 1O2 | [135] | |
| Photothermal therapy | SA | TEOS | HepG-2, HepG-2 tumor-bearing mice | AuNRs | Liver cancer | [137] |
| Inhibition of HER2 breast cancer | N-glycans of HER2 glycoprotein | SKBR-3, SKBR-3 tumor-bearing mice | NanoMIPs themselves as therapeutic | HER2- breast cancer | [141] | |
| Immune checkpoint blockade therapy | N-glycans of PD-L1 glycoprotein | MDA-MB-231, MDA-MB-231 tumor-bearing mice | NanoNiche itself as therapeutic | Human breast cancer | [147] |
(A) The principle of tumor cell immune evasion from T-cells and the reactivation of T-cell immunity by blocking PD-L1 through N-glycan-imprinted NanoNiche with a desialylation function schematic illustration; (B) Microscopy images showing the T-cell-mediated tumor cell-killing effect in MDA-MB-231 cells incubated with NanoNiche (200 μL) after treatment for 0, 1, 2, and 3 h; Schedule of various treatments in tumor-bearing mice; (C) Average tumor volumes after various treatments at different intervals of time; (D) Body weights of mice in various groups after treatments at various time intervals; (E) In vivo anticancer activity outcomes of mice in various groups after a 10-day therapy. Adapted with permission from [147], copyright 2021 ACS Applied Bio Materials.
4. Conclusion and Future Perspectives
Recognizing both diverse functions of glycoproteins in different physiological processes and natural mechanisms, a lot of glycoproteins imprinted approaches have been engineered or updated to meet the standards of targeted proteomics analysis. MIPs are widely used for generating antibody-like substitutes that have allowed complex samples to specifically enrich target glycoproteins. Because of their outstanding targeting capability, biocompatibility, high specificity, excellent anti-interference performance, and pH-controlled binding/dissociation, they have shown promising applications in proteome analysis, sensing, disease diagnostics, tissue engineering, and drug delivery. To facilitate its intensified use, much advancement is needed in glycoprotein imprinting. Regarding the prospects for glycoprotein imprinting, there are certain substantial aspects such as dimensions of imprinting material, complexity, the fragility of the structure of glycoproteins, difference in solubility of monomers, and template molecule, purity, and availability of a template, and uncontrolled thickness of imprinting layer. The size of imprinting material and overall particle should be reduced and have a flexible network for facile removal of template molecules and in-vivo applications. In addition, novel smart temperature-responsive or polymer-materials should be considered for facile template removal and fast extraction/desorption kinetics. Due to the complex structure of glycoprotein, the selection of monomers and the mechanism of interaction should be well studied, not only on an experimental basis but also by theoretical simulations. Moreover, for the specific extraction of diverse glycoproteins, novel ligands such as aptamers could be valuable options. Furthermore, glycoproteins are water-soluble but their monomers are limited, which need to be developed. On the other hand, using different types of molecular structural modeling programs such as the Gaussian program can facilitate the Gauss View for molecular imprinting of glycoproteins.
Another barrier is the uncontrolled thickness of the imprinting layer and at present, a limited number of biocompatible and self-polymerizable ligands are available for imprinting of glycoproteins. Among these polymer materials, time-dependent controllability of self-polymerization has been revealed by dopamine/APBA and TEOS. This challenge can be addressed by finding more effective functional monomers or changing the current monomers with unique functionalities or structures. Moreover, ATRP and RAFT polymerization methods are also useful for controlling the thickness of the imprinting layer. PolyNIPAM is approved by FDA due to its biocompatibility and other polyacrylamides are less toxic revealed by MTT assay in most of the living cells. For practical applications, MIPs should have monoclonal antibody-like features such as high affinity, fast recognition kinetics, and high specificity. An effective nanoMIP-imaging probe for clinical usage is biodegradable or quickly eliminated, with minimal toxicity and a strong imaging response. The imprinted particles should have a diameter less than 8 nm to be excreted by kidneys otherwise larger-sized particles are cleared by the liver. As the renal clearance is ideal due to rapid response, but size-controlled development at such a small scale is challenging. Owing to their advantages, nanoMIPs also have potent cancer therapeutic characteristics and revealed excellent performance in precise targeting, a systemic circulation, and controlled drug delivery systems. NanoMIPs can also combine several functionalities into a single system, making them a potent and adaptable substrate for complementary medications to cure cancers. For real-world cancer therapeutic applications and future clinical translation, multifunctional MIP-based nanomedicines are urgently required, considering their biocompatibility, specific targeting, controlled drug release kinetics, excellent clear imaging, and relevant animal models for in vivo studies, which must be evaluated and explored for future clinical translation. Regarding blood compatibility, their effect on blood coagulation and inflammatory reaction must also be evaluated.
Abbreviations
Am: acrylamide; AFP: α-fetoprotein; APBA: 3-acrylamidophenyl boronic acid; APTES: 3-aminopropyltriethoxysilane; APTMS: 3-aminopropyl trimethoxysilane; BAC: N, N'-diacrylylcystamine; BACOSI: boronate affinity controllable oriented surface imprinting; CD: circular dichroism; CEA: carcinoembryonic antigen; DMA: 2-(dimethylamino) ethyl methacrylate; DMAEMA: dimethylaminoethyl methacrylate; EGDMA: ethylene glycol dimethacrylate; FITC: fluorescein isothiocyanate isomer I; FM: 4-[2-(N-methacrylamido) ethylaminomethyl] benzoic Acid; FMB: 4-(2-methacrylamidoethylcarbamoyl)-3-fluorophenylboronic; GRAVY: grand average of hydropathicity; HEMA: 2-hydroxyethylmethacrylate; HSA: human serum albumin; IBTES: isobutyltriethoxysilane; IE: imprinting efficiency; IF: imprinting factor; MAA: methacrylic acid; Mb: myoglobin; MB: methylene blue; MBA: N, N′-methylenebisacrylamide; MIPs: molecularly imprinted polymers; MNPs: magnetic nanoparticles; MPC: 2-methacryloyloxyethyl phosphorylcholine; MPTMS: 3-methacryloyloxypropyl trimethoxysilane; NAPMAAm: N-(3-aminopropyl) methacrylamide hydrochloride; NIPAAm: N-isopropylacrylamide; NPs: nanoparticles; o-PD: o-phenylene- diamine; OVA: ovalbumin; PISA: plasmonic immunosandwich assay; PSA: prostate-specific antigen; RNase B: ribonuclease B; SELEX: systematic evolution of ligands by exponential enrichment; SERS: surface-enhanced Raman scattering; TBAm: N-tert-butyl acrylamide; TFMA: 2-(trifluoromethyl) acrylic acid; TRF: transferrin; UPTES: 3-ureidopropyl-triethoxysilane; VEGF: vascular endothelial growth factor; VPBA: 4-vinylphenylboronic acid; ZnA: zinc acrylate.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (21676061, 21874006) and the State Key Laboratory of Analytical Chemistry for Life Science (Nanjing University, SKLACLS2002).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bonduelle C, Lecommandoux Sb. Synthetic glycopolypeptides as biomimetic analogues of natural glycoproteins. Biomacromolecules. 2013;14:2973-83
2. Hayashi T, Sun Y, Tamura T, Kuwata K, Song Z, Takaoka Y. et al. Semisynthetic lectin-4-dimethylaminopyridine conjugates for labeling and profiling glycoproteins on live cell surfaces. J Am Chem Soc. 2013;135:12252-8
3. Burnham-Marusich AR, Berninsone PM. Multiple proteins with essential mitochondrial functions have glycosylated isoforms. Mitochondrion. 2012;12:423-7
4. Gilgunn S, Conroy PJ, Saldova R, Rudd PM, O'kennedy RJ. Aberrant PSA glycosylation—a sweet predictor of prostate cancer. Nat Rev Urol. 2013;10:99
5. Hudak JE, Bertozzi CR. Glycotherapy: new advances inspire a reemergence of glycans in medicine. Chem Biol. 2014;21:16-37
6. PM L. Gage FH. Varki AP. The glycans of stem cells. Curr Opin Chem Biol. 2007;11:373-80
7. Vasconcelos-dos-Santos A, Oliveira IA, Lucena MC, Mantuano NR, Whelan SA, Dias WB. et al. Biosynthetic machinery involved in aberrant glycosylation: promising targets for developing of drugs against cancer. Front Oncol. 2015;5:138
8. Sun S, Shah P, Eshghi ST, Yang W, Trikannad N, Yang S. et al. Comprehensive analysis of protein glycosylation by solid-phase extraction of N-linked glycans and glycosite-containing peptides. Nat Biotechnol. 2016;34:84-8
9. Xiao H, Suttapitugsakul S, Sun F, Wu R. Mass spectrometry-based chemical and enzymatic methods for global analysis of protein glycosylation. Acc Chem Res. 2018;51:1796-806
10. Ali MM, Zhu Z, Wang M, Hussain D, Gao X, Wang J. et al. Melamine foam assisted in-tip packed amine-functionalized titanium metal-organic framework for the selective enrichment of endogenous glycopeptides. J Chromatogr A. 2021;1636:461711
11. Ali MM, Hussain D, Tang Y, Sun X, Shen Z, Zhang F. et al. Boronoisophthalic acid as a novel affinity ligand for the selective capture and release of glycoproteins near physiological pH. Talanta. 2021;225:121896
12. Sterner E, Flanagan N, Gildersleeve JC. Perspectives on anti-glycan antibodies gleaned from development of a community resource database. ACS Chem Biol. 2016;11:1773-83
13. Zhang L, Luo S, Zhang B. The use of lectin microarray for assessing glycosylation of therapeutic proteins. MAbs: Taylor & Francis. 2016 p. 524-35
14. Baker M. Reproducibility crisis: Blame it on the antibodies. Nat News. 2015;521:274
15. Baker M. Antibody anarchy: a call to order. Nature. 2015;527:545-51
16. Ali MM, Hussain D, Xu B, Sun T, Du Z. Diethylenetriamine assisted functionalization of boronic acid on poly GMA-MAA-DVB for selective enrichment of glycoproteins and glycopeptides. Talanta. 2020;219:121178
17. Chen L, Xu S, Li J. Recent advances in molecular imprinting technology: current status, challenges and highlighted applications. Chem Soc Rev. 2011;40:2922-42
18. Mourão ClA, Bokeloh F, Xu J, Prost E, Duma L, Merlier F. et al. Dual-oriented solid-phase molecular imprinting: toward selective artificial receptors for recognition of nucleotides in water. Macromolecules. 2017;50:7484-90
19. Kitayama Y, Yoshikawa K, Takeuchi T. Post-cross-linked molecular imprinting with functional polymers as a universal building block for artificial polymeric receptors. Macromolecules. 2017;50:7526-34
20. Zhang G, Jiang L, Zhou J, Hu L, Feng S. Epitope-imprinted mesoporous silica nanoparticles for specific recognition of tyrosine phosphorylation. Chem Commun. 2019;55:9927-30
21. Akiba U, Anzai J-i. Recent progress in electrochemical biosensors for glycoproteins. Sensors. 2016;16:2045
22. Saeki T, Sunayama H, Kitayama Y, Takeuchi T. Orientationally fabricated zwitterionic molecularly imprinted nanocavities for highly sensitive glycoprotein recognition. Langmuir. 2018;35:1320-6
23. Muhammad P, Tu X, Liu J, Wang Y, Liu Z. Molecularly imprinted plasmonic substrates for specific and ultrasensitive immunoassay of trace glycoproteins in biological samples. ACS Appl Mater Interfaces. 2017;9:12082-91
24. Ye J, Chen Y, Liu Z. A boronate affinity sandwich assay: an appealing alternative to immunoassays for the determination of glycoproteins. Angew Chem Int Ed. 2014;53:10386-9
25. Wang H, Wang J, Wang Y, Liu Y, Liu R, Wang X. et al. Oriented boronate affinity-imprinted inverse opal hydrogel for glycoprotein assay via colorimetry. Microchim Acta. 2020;187:1-9
26. Jiang L, Lu R, Ye L. Towards detection of glycoproteins using molecularly imprinted nanoparticles and boronic acid-modified fluorescent probe. Polymers. 2019;11:173
27. Xu S, Wang L, Liu Z. Molecularly imprinted polymer nanoparticles: an emerging versatile platform for cancer therapy. Angew Chem Int Ed. 2021;60:3858-69
28. Wang H-C, Zhou H, Chen B, Mendes PM, Fossey JS, James TD. et al. A bis-boronic acid modified electrode for the sensitive and selective determination of glucose concentrations. Analyst. 2013;138:7146-51
29. Ali MM, Hussain D, Tang Y, Sun X, Shen Z, Fengxia Z. et al. Boronoisophthalic Acid as a Novel Affinity Ligand for the Selective Capture and Release of Glycoproteins near physiological pH. Talanta. 2020: 121896.
30. Rutkowska M, Płotka-Wasylka J, Morrison C, Wieczorek PP, Namieśnik J, Marć M. Application of molecularly imprinted polymers in analytical chiral separations and analysis. TrAC, Trends Anal Chem. 2018;102:91-102
31. Yang Q, Li J, Wang X, Peng H, Xiong H, Chen L. Strategies of molecular imprinting-based fluorescence sensors for chemical and biological analysis. Biosens Bioelectron. 2018;112:54-71
32. Jia M, Zhang Z, Li J, Ma X, Chen L, Yang X. Molecular imprinting technology for microorganism analysis. TrAC, Trends Anal Chem. 2018;106:190-201
33. You M, Yang S, Tang W, Zhang F, He P-G. Ultrasensitive electrochemical detection of glycoprotein based on boronate affinity sandwich assay and signal amplification with functionalized SiO2@ Au nanocomposites. ACS Appl Mater Interfaces. 2017;9:13855-64
34. Haupt K, Medina Rangel PX, Bui BTS. Molecularly imprinted polymers: Antibody mimics for bioimaging and therapy. Chem Rev. 2020;120:9554-82
35. Xu J, Miao H, Wang J, Pan G. Molecularly imprinted synthetic antibodies: from chemical design to biomedical applications. Small. 2020;16:1906644
36. Qin Y-P, Jia C, He X-W, Li W-Y, Zhang Y-K. Thermosensitive metal chelation dual-template epitope imprinting polymer using distillation-precipitation polymerization for simultaneous recognition of human serum albumin and transferrin. ACS Appl Mater Interfaces. 2018;10:9060-8
37. Qin Y, Xie J, Li S, Cai C, Chen X, Zhong G. et al. A boronate affinity MIP-based resonance light scattering sensor for sensitive detection of glycoproteins. Anal Methods. 2018;10:5112-7
38. Saylan Y, Üzek R, Uzun L, Denizli A. Surface imprinting approach for preparing specific adsorbent for IgG separation. J Biomater Sci Polym Ed. 2014;25:881-94
39. Li L, Lu Y, Bie Z, Chen HY, Liu Z. Photolithographic boronate affinity molecular imprinting: a general and facile approach for glycoprotein imprinting. Angew Chem Int Ed. 2013;52:7451-4
40. Chen W, Fu M, Zhu X, Liu Q. A close-packed imprinted colloidal array for naked-eye detection of glycoproteins under physiological pH. Biosens Bioelectron. 2019;142:111499
41. Mitchell P, Tommasone S, Angioletti-Uberti S, Bowen J, Mendes PM. Precise generation of selective surface-confined glycoprotein recognition sites. ACS Appl Bio Mater. 2019;2:2617-23
42. Luo J, Huang J, Cong J, Wei W, Liu X. Double recognition and selective extraction of glycoprotein based on the molecular imprinted graphene oxide and boronate affinity. ACS Appl Mater Interfaces. 2017;9:7735-44
43. Lin Z, Sun L, Liu W, Xia Z, Yang H, Chen G. Synthesis of boronic acid-functionalized molecularly imprinted silica nanoparticles for glycoprotein recognition and enrichment. J Mater Chem B. 2014;2:637-43
44. Xing R, Ma Y, Wang Y, Wen Y, Liu Z. Specific recognition of proteins and peptides via controllable oriented surface imprinting of boronate affinity-anchored epitopes. Chem Sci. 2019;10:1831-5
45. Mrówczyński R, Markiewicz R, Liebscher J. Chemistry of polydopamine analogues. Polym Int. 2016;65:1288-99
46. He P, Zhu H, Ma Y, Liu N, Niu X, Wei M. et al. Rational design and fabrication of surface molecularly imprinted polymers based on multi-boronic acid sites for selective capture glycoproteins. Chem Eng J. 2019;367:55-63
47. Lee H, Dellatore SM, Miller WM, Messersmith PB. Mussel-inspired surface chemistry for multifunctional coatings. Science. 2007;318:426-30
48. Chen Y, Huang A, Zhang Y, Bie Z. Recent advances of boronate affinity materials in sample preparation. Anal Chim Acta. 2019;1076:1-17
49. Yang K, Liu J, Li S, Li Q, Wu Q, Zhou Y. et al. Epitope imprinted polyethersulfone beads by self-assembly for target protein capture from the plasma proteome. Chem Commun. 2014;50:9521-4
50. Shi Y, Wan Y, Zhao D. Ordered mesoporous non-oxide materials. Chem Soc Rev. 2011;40:3854-78
51. Asefa T, Tao Z. Mesoporous silica and organosilica materials—Review of their synthesis and organic functionalization. Can J Chem. 2012;90:1015-31
52. Brady R, Woonton B, Gee ML, O'Connor AJ. Hierarchical mesoporous silica materials for separation of functional food ingredients—A review. Innov Food Sci Emerg Technol. 2008;9:243-8
53. Di Renzo F, Testa F, Chen J, Cambon H, Galarneau A, Plee D. et al. Textural control of micelle-templated mesoporous silicates: the effects of co-surfactants and alkalinity. Microporous Mesoporous Mater. 1999;28:437-46
54. Saeki T, Takano E, Sunayama H, Kamon Y, Horikawa R, Kitayama Y. et al. Signalling molecular recognition nanocavities with multiple functional groups prepared by molecular imprinting and sequential post-imprinting modifications for prostate cancer biomarker glycoprotein detection. J Mater Chem B. 2020;8:7987-93
55. Duan L, Zangiabadi M, Zhao Y. Synthetic lectins for selective binding of glycoproteins in water. Chem Commun. 2020;56:10199-202
56. Zhou L, Wang Y, Xing R, Chen J, Liu J, Li W. et al. Orthogonal dual molecularly imprinted polymer-based plasmonic immunosandwich assay: A double characteristic recognition strategy for specific detection of glycoproteins. Biosens Bioelectron. 2019;145:111729
57. Zangiabadi M, Zhao Y. Selective Binding of Complex Glycans and Glycoproteins in Water by Molecularly Imprinted Nanoparticles. Nano Lett. 2020;20:5106-10
58. Yang K, Li S, Liu L, Chen Y, Zhou W, Pei J. et al. Epitope imprinting technology: progress, applications, and perspectives toward artificial antibodies. Adv Mater. 2019;31:1902048
59. Bie Z, Chen Y, Ye J, Wang S, Liu Z. Boronate-affinity glycan-oriented surface imprinting: a new strategy to mimic lectins for the recognition of an intact glycoprotein and its characteristic fragments. Angew Chem Int Ed. 2015;54:10211-5
60. Xing R, Wang S, Bie Z, He H, Liu Z. Preparation of molecularly imprinted polymers specific to glycoproteins, glycans and monosaccharides via boronate affinity controllable-oriented surface imprinting. Nat Protoc. 2017;12:964-87
61. Bie Z, Xing R, He X, Ma Y, Chen Y, Liu Z. Precision imprinting of glycopeptides for facile preparation of glycan-specific artificial antibodies. Anal Chem. 2018;90:9845-52
62. Guo Z, Xing R, He H, Liu Z. Boronate affinity controllable oriented surface imprinting of glycans in organic phase using intact glycoproteins as semi-templates. Chin Sci Bull. 2018;64:1418-26
63. Zhang Y, Deng C, Liu S, Wu J, Chen Z, Li C. et al. Active targeting of tumors through conformational epitope imprinting. Angew Chem. 2015;127:5246-9
64. Dechtrirat D, Jetzschmann KJ, Stöcklein WF, Scheller FW, Gajovic-Eichelmann N. Protein rebinding to a surface-confined imprint. Adv Funct Mater. 2012;22:5231-7
65. Tang A-n, Duan L, Liu M, Dong X. An epitope imprinted polymer with affinity for kininogen fragments prepared by metal coordination interaction for cancer biomarker analysis. J Mater Chem B. 2016;4:7464-71
66. Poma A, Guerreiro A, Whitcombe MJ, Piletska EV, Turner AP, Piletsky SA. Solid-phase synthesis of molecularly imprinted polymer nanoparticles with a reusable template-“plastic antibodies”. Adv Funct Mater. 2013;23:2821-7
67. Shinde S, El-Schich Z, Malakpour A, Wan W, Dizeyi N, Mohammadi R. et al. Sialic acid-imprinted fluorescent core-shell particles for selective labeling of cell surface glycans. J Am Chem Soc. 2015;137:13908-12
68. Li Q, Lü C, Li H, Liu Y, Wang H, Wang X. et al. Preparation of organic-silica hybrid boronate affinity monolithic column for the specific capture and separation of cis-diol containing compounds. J Chromatogr A. 2012;1256:114-20
69. Liu Z, He H. Synthesis and applications of boronate affinity materials: from class selectivity to biomimetic specificity. Acc Chem Res. 2017;50:2185-93
70. Wu X, Wang X, Lu W, Wang X, Li J, You H. et al. Water-compatible temperature and magnetic dual-responsive molecularly imprinted polymers for recognition and extraction of bisphenol A. J Chromatogr A. 2016;1435:30-8
71. Li W, Zhang Q, Wang Y, Ma Y, Guo Z, Liu Z. Controllably prepared aptamer-molecularly imprinted polymer hybrid for high-specificity and high-affinity recognition of target proteins. Anal Chem. 2019;91:4831-7
72. Wang P, Zhu H, Liu J, Ma Y, Yao J, Dai X. et al. Double affinity integrated MIPs nanoparticles for specific separation of glycoproteins: A combination of synergistic multiple bindings and imprinting effect. Chem Eng J. 2019;358:143-52
73. O'Sullivan M, Zhang Z, Vincent B. Silica-Shell/Oil-Core Microcapsules with Controlled Shell Thickness and Their Breakage Stress. Langmuir. 2009;25:7962-6
74. Xie J, Zhong G, Cai C, Chen C, Chen X. Rapid and efficient separation of glycoprotein using pH double-responsive imprinted magnetic microsphere. Talanta. 2017;169:98-103
75. Yang K, Li S, Liu J, Liu L, Zhang L, Zhang Y. Multiepitope templates imprinted particles for the simultaneous capture of various target proteins. Anal Chem. 2016;88:5621-5
76. Liu L, Yang K, Li S, Zhang L, Zhang Y. Poly (ether sulfone) nanoparticles and controllably modified nanoparticles obtained through temperature-dependent cryogelation. J Appl Polym Sci. 2019;136:47485
77. Dong C, Shi H, Han Y, Yang Y, Wang R, Men J. Molecularly imprinted polymers by the surface imprinting technique. Eur Polym J. 2021;145:110231
78. Ding X, Heiden PA. Recent developments in molecularly imprinted nanoparticles by surface imprinting techniques. Macromolecular Materials and Engineering. 2014;299:268-82
79. Sun X-Y, Ma R-T, Chen J, Shi Y-P. Magnetic boronate modified molecularly imprinted polymers on magnetite microspheres modified with porous TiO 2 (Fe 3 O 4@ pTiO 2@ MIP) with enhanced adsorption capacity for glycoproteins and with wide operational pH range. Microchim Acta. 2018;185:1-10
80. Guo B, Bi S, Zhang B, Tong Y, Chen X, Tian M. Synthesis of nanoparticles with a combination of metal chelation and molecular imprinting for efficient and selective extraction of glycoprotein. Microchem J. 2021;167:106262
81. Li J, Dai Y, Wang S, Han C, Xu K. Aptamer-tagged green-and yellow-emitting fluorescent silver nanoclusters for specific tumor cell imaging. Sens Actuators B Chem. 2016;232:1-8
82. Bidard F-C, Fehm T, Ignatiadis M, Smerage JB, Alix-Panabières C, Janni W. et al. Clinical application of circulating tumor cells in breast cancer: overview of the current interventional trials. Cancer Metastasis Rev. 2013;32:179-88
83. Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L. et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235-9
84. Fass L. Imaging and cancer: a review. Mol Oncol. 2008;2:115-52
85. Schmidt MA, Payne GS. Radiotherapy planning using MRI. Phys Med Biol. 2015;60:R323
86. Elsabahy M, Heo GS, Lim S-M, Sun G, Wooley KL. Polymeric nanostructures for imaging and therapy. Chem Rev. 2015;115:10967-1011
87. Luan J, Xu T, Cashin J, Morrissey JJ, Kharasch ED, Singamaneni S. Environmental stability of plasmonic biosensors based on natural versus artificial antibody. Anal Chem. 2018;90:7880-7
88. Ouyang J, Liu Z, Han Y, Zeng K, Sheng J, Deng L. et al. Fabrication of surface protein-imprinted biofuel cell for sensitive self-powered glycoprotein detection. ACS Appl Mater Interfaces. 2016;8:35004-11
89. Cecchini A, Raffa V, Canfarotta F, Signore G, Piletsky S, MacDonald MP. et al. In vivo recognition of human vascular endothelial growth factor by molecularly imprinted polymers. Nano Lett. 2017;17:2307-12
90. Moreira FT, Sales MGF. Autonomous biosensing device merged with photovoltaic technology for cancer biomarker detection. J Electroanal Chem. 2019;855:113611
91. Xing R, Wen Y, Dong Y, Wang Y, Zhang Q, Liu Z. Dual Molecularly Imprinted Polymer-Based Plasmonic Immunosandwich Assay for the Specific and Sensitive Detection of Protein Biomarkers. Anal Chem. 2019;91:9993-10000
92. Zhang G, Yu Y, Zhang L, Lin B, Wang Y, Guo M. et al. Precise detection of prostate specific antigen in serum: a surface molecular imprinted sensor based on novel cooperated signal amplification strategy. Sens Actuators B Chem. 2020;302:126998
93. Tawfik SM, Elmasry MR, Sharipov M, Azizov S, Lee CH, Lee Y-I. Dual emission nonionic molecular imprinting conjugated polythiophenes-based paper devices and their nanofibers for point-of-care biomarkers detection. Biosens Bioelectron. 2020;160:112211
94. Moreira FTC, Sales MGF. Autonomous biosensing device merged with photovoltaic technology for cancer biomarker detection. J Electroanal Chem. 2019;855:113611
95. Viswanathan S, Rani C, Ribeiro S, Delerue-Matos C. Molecular imprinted nanoelectrodes for ultra sensitive detection of ovarian cancer marker. Biosens Bioelectron. 2012;33:179-83
96. Morishige T, Takano E, Sunayama H, Kitayama Y, Takeuchi T. Post-Imprinting-Modified Molecularly Imprinted Nanocavities with Two Synergetic, Orthogonal, Glycoprotein-Binding Sites to Transduce Binding Events into Fluorescence Changes. ChemNanoMat. 2019;5:224-9
97. Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509-44
98. Zhang Y, Fang F, Li L, Zhang J. Self-Assembled Organic Nanomaterials for Drug Delivery, Bioimaging, and Cancer Therapy. ACS Biomater Sci Eng. 2020;6:4816-33
99. Hola K, Zhang Y, Wang Y, Giannelis EP, Zboril R, Rogach AL. Carbon dots—Emerging light emitters for bioimaging, cancer therapy and optoelectronics. Nano Today. 2014;9:590-603
100. AndrewáWang Y. Cadmium-free quantum dots as time-gated bioimaging probes in highly-autofluorescent human breast cancer cells. Chem Commun. 2013;49:624-6
101. Wu Y, Chen Z, Zhang P, Zhou L, Jiang T, Chen H. et al. Recombinant-fully-human-antibody decorated highly-stable far-red AIEdots for in vivo HER-2 receptor-targeted imaging. Chem Commun. 2018;54:7314-7
102. Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M. et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541-50
103. Panagiotopoulou M, Salinas Y, Beyazit S, Kunath S, Duma L, Prost E. et al. Molecularly imprinted polymer coated quantum dots for multiplexed cell targeting and imaging. Angew Chem. 2016;128:8384-8
104. Wang S, Wen Y, Wang Y, Ma Y, Liu Z. Pattern Recognition of Cells via Multiplexed Imaging with Monosaccharide-Imprinted Quantum Dots. Anal Chem. 2017;89:5646-52
105. Demir B, Lemberger MM, Panagiotopoulou M, Medina Rangel PX, Timur S, Hirsch T. et al. Tracking Hyaluronan: Molecularly Imprinted Polymer Coated Carbon Dots for Cancer Cell Targeting and Imaging. ACS Appl Mater Interfaces. 2018;10:3305-13
106. Rangel PXM, Laclef S, Xu J, Panagiotopoulou M, Kovensky J, Bui BTS. et al. Solid-phase synthesis of molecularly imprinted polymer nanolabels: affinity tools for cellular bioimaging of glycans. Sci Rep. 2019;9:1-9
107. Zhang Y, Li S, Ma X-T, He X-W, Li W-Y, Zhang Y-K. Carbon dots-embedded epitope imprinted polymer for targeted fluorescence imaging of cervical cancer via recognition of epidermal growth factor receptor. Microchim Acta. 2020;187:228
108. Ren X-H, Wang H-Y, Li S, He X-W, Li W-Y, Zhang Y-K. Preparation of glycan-oriented imprinted polymer coating Gd-doped silicon nanoparticles for targeting cancer Tn antigens and dual-modal cell imaging via boronate-affinity surface imprinting. Talanta. 2021;223:121706
109. Shen S, Huang B, Guo X, Wang H. A dual-responsive fluorescent sensor for Hg 2+ and thiols based on N-doped silicon quantum dots and its application in cell imaging. J Mater Chem B. 2019;7:7033-41
110. Sugimoto H, Yamamura M, Sakiyama M, Fujii M. Visualizing a core-shell structure of heavily doped silicon quantum dots by electron microscopy using an atomically thin support film. Nanoscale. 2018;10:7357-62
111. Medina Rangel PX, Laclef S, Xu J, Panagiotopoulou M, Kovensky J, Tse Sum Bui B. et al. Solid-phase synthesis of molecularly imprinted polymer nanolabels: Affinity tools for cellular bioimaging of glycans. Sci Rep. 2019;9:3923
112. Yin D, Wang S, He Y, Liu J, Zhou M, Ouyang J. et al. Surface-enhanced Raman scattering imaging of cancer cells and tissues via sialic acid-imprinted nanotags. Chem Commun. 2015;51:17696-9
113. Miller KD, Fidler-Benaoudia M, Keegan TH, Hipp HS, Jemal A, Siegel RL. Cancer statistics for adolescents and young adults, 2020. CA Cancer J Clin. 2020;70:443-59
114. Mereiter S, Balmaña M, Campos D, Gomes J, Reis CA. Glycosylation in the era of cancer-targeted therapy: where are we heading? Cancer Cell. 2019;36:6-16
115. Gielen P, Büll C, Schulte B, Kers-Rebel E, Bossman S, ter Laan M. et al. Expression profiling of the Sialic acid-Siglec axis in patients with glioma. The immunosuppressive network in patients with glioma; focus on the role of myeloid derived suppressor cells. 2016: 113.
116. Gu Z, Dong Y, Xu S, Wang L, Liu Z. Molecularly Imprinted Polymer-Based Smart Prodrug Delivery System for Specific Targeting, Prolonged Retention, and Tumor Microenvironment-Triggered Release. Angew Chem Int Ed. 2021;60:2663-7
117. Sellergren B, Allender CJ. Molecularly imprinted polymers: A bridge to advanced drug delivery. Adv Drug Del Rev. 2005;57:1733-41
118. Canfarotta F, Lezina L, Guerreiro A, Czulak J, Petukhov A, Daks A. et al. Specific drug delivery to cancer cells with double-imprinted nanoparticles against epidermal growth factor receptor. Nano Lett. 2018;18:4641-6
119. Rosenblum D, Joshi N, Tao W, Karp JM, Peer D. Progress and challenges towards targeted delivery of cancer therapeutics. Nat Commun. 2018;9:1-12
120. Wicki A, Witzigmann D, Balasubramanian V, Huwyler J. Nanomedicine in cancer therapy: challenges, opportunities, and clinical applications. J Controlled Release. 2015;200:138-57
121. Yin Y, Guan L, Wang Y, Ma Y, Pan J, Peng Y. et al. Sialic acid-imprinted mesoporous nanocarriers for tumor cell targeted drug delivery. Colloid Interface Sci Commun. 2021;42:100421
122. Najjar A, Karaman R. The prodrug approach in the era of drug design. Expert opinion on drug delivery. 2019;16:1-5
123. Najjar A, Najjar A, Karaman R. Newly developed prodrugs and prodrugs in development; an insight of the recent years. Molecules. 2020;25:884
124. Han S, Su L, Zhai M, Ma L, Liu S, Teng Y. A molecularly imprinted composite based on graphene oxide for targeted drug delivery to tumor cells. J Mater Sci. 2019;54:3331-41
125. Li S, Cao S, Whitcombe MJ, Piletsky SA. Size matters: Challenges in imprinting macromolecules. Prog Polym Sci. 2014;39:145-63
126. Li X, Chen K, Zhao Y. Sequence-Selective Protection of Peptides from Proteolysis. Angew Chem Int Ed. 2021;60:11092-7
127. Palladino P, Minunni M, Scarano S. Cardiac Troponin T capture and detection in real-time via epitope-imprinted polymer and optical biosensing. Biosens Bioelectron. 2018;106:93-8
128. Wang X, Chen G, Zhang P, Jia Q. Advances in epitope molecularly imprinted polymers for protein detection: a review. Anal Methods. 2021
129. Hashemi-Moghaddam H, Zavareh S, Karimpour S, Madanchi H. Evaluation of molecularly imprinted polymer based on HER2 epitope for targeted drug delivery in ovarian cancer mouse model. React Funct Polym. 2017;121:82-90
130. Lakshmi BA, Kim S. Current and emerging applications of nanostructured metal-organic frameworks in cancer-targeted theranostics. Mater Sci Eng C. 2019;105:110091
131. Giménez-Marqués M, Hidalgo T, Serre C, Horcajada P. Nanostructured metal-organic frameworks and their bio-related applications. Coord Chem Rev. 2016;307:342-60
132. Luo Z, Fan S, Gu C, Liu W, Chen J, Li B. et al. Metal-organic framework (MOF)-based nanomaterials for biomedical applications. Curr Med Chem. 2019;26:3341-69
133. Qin Y-T, Feng Y-S, Ma Y-J, He X-W, Li W-Y, Zhang Y-K. Tumor-sensitive biodegradable nanoparticles of molecularly imprinted polymer-stabilized fluorescent zeolitic imidazolate framework-8 for targeted imaging and drug delivery. ACS Appl Mater Interfaces. 2020;12:24585-98
134. Liu S, Bi Q, Long Y, Li Z, Bhattacharyya S, Li C. Inducible epitope imprinting:'generating'the required binding site in membrane receptors for targeted drug delivery. Nanoscale. 2017;9:5394-7
135. Peng H, Qin Y-T, He X-W, Li W-Y, Zhang Y-K. Epitope molecularly imprinted polymer nanoparticles for chemo-/photodynamic synergistic cancer therapy guided by targeted fluorescence imaging. ACS Appl Mater Interfaces. 2020;12:13360-70
136. Yoshida A, Kitayama Y, Kiguchi K, Yamada T, Akasaka H, Sasaki R. et al. Gold nanoparticle-incorporated molecularly imprinted microgels as radiation sensitizers in pancreatic cancer. ACS Appl Bio Mater. 2019;2:1177-83
137. Yin D, Li X, Ma Y, Liu Z. Targeted cancer imaging and photothermal therapy via monosaccharide-imprinted gold nanorods. Chem Commun. 2017;53:6716-9
138. Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE. et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707-12
139. Barthelemy P, Leblanc J, Goldbarg V, Wendling F, Kurtz J-E. Pertuzumab: development beyond breast cancer. Anticancer Res. 2014;34:1483-91
140. Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463-75
141. Dong Y, Li W, Gu Z, Xing R, Ma Y, Zhang Q. et al. Inhibition of HER2-positive breast cancer growth by blocking the HER2 signaling pathway with HER2-glycan-imprinted nanoparticles. Angew Chem Int Ed. 2019;58:10621-5
142. Nishino M, Ramaiya NH, Hatabu H, Hodi FS. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nature reviews Clinical oncology. 2017;14:655-68
143. Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161:205-14
144. Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275-87
145. Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350-5
146. Phuengkham H, Song C, Lim YT. A designer scaffold with immune nanoconverters for reverting immunosuppression and enhancing immune checkpoint blockade therapy. Adv Mater. 2019;31:1903242
147. Zhou Z-R, Wang X-Y, Jiang L, Li D-W, Qian R-C. Sialidase-Conjugated “NanoNiche” for Efficient Immune Checkpoint Blockade Therapy. ACS Appl Bio Mater. 2021;4:5735-41
148. Wang H-Y, Cao P-P, He Z-Y, He X-W, Li W-Y, Li Y-H. et al. Targeted imaging and targeted therapy of breast cancer cells via fluorescent double template-imprinted polymer coated silicon nanoparticles by an epitope approach. Nanoscale. 2019;11:17018-30
Author contact
![]() Corresponding author: E-mail addresses: duzxbuct.edu.cn; lianghaihuedu.cn
Corresponding author: E-mail addresses: duzxbuct.edu.cn; lianghaihuedu.cn
 Global reach, higher impact
Global reach, higher impact