13.3
Impact Factor
Theranostics 2022; 12(6):2758-2772. doi:10.7150/thno.67661 This issue Cite
Review
From engineered heart tissue to cardiac organoid
1. Department of Medicine, Division of Cardiology, Emory University School of Medicine, Atlanta, GA 30322, USA.
2. Severance Biomedical Science Institute, Yonsei University College of Medicine, Seoul, Republic of Korea.
3. Division of Cardiology, Department of Medicine, Yonsei University College of Medicine, Seoul, Republic of Korea.
4. Brain Korea 21 PLUS Project for Medical Science, Yonsei University College of Medicine, Seoul, South Korea.
5. Karis Bio Inc., Seoul, Republic of Korea
* Equal contributions
Received 2021-10-1; Accepted 2022-3-1; Published 2022-3-14
Abstract
The advent of human pluripotent stem cells (hPSCs) presented a new paradigm to employ hPSC-derived cardiomyocytes (hPSC-CMs) in drug screening and disease modeling. However, hPSC-CMs differentiated in conventional two-dimensional systems are structurally and functionally immature. Moreover, these differentiation systems generate predominantly one type of cell. Since the heart includes not only CMs but other cell types, such monolayer cultures have limitations in simulating the native heart. Accordingly, three-dimensional (3D) cardiac tissues have been developed as a better platform by including various cardiac cell types and extracellular matrices. Two advances were made for 3D cardiac tissue generation. One type is engineered heart tissues (EHTs), which are constructed by 3D cell culture of cardiac cells using an engineering technology. This system provides a convenient real-time analysis of cardiac function, as well as a precise control of the input/output flow and mechanical/electrical stimulation. The other type is cardiac organoids, which are formed through self-organization of differentiating cardiac lineage cells from hPSCs. While mature cardiac organoids are more desirable, at present only primitive forms of organoids are available. In this review, we discuss various models of hEHTs and cardiac organoids emulating the human heart, focusing on their unique features, utility, and limitations.
Keywords: Human Pluripotent Stem Cell, Engineered Heart Tissue, Organoid, Disease Modeling, Drug Screening
1. The need for three-dimensional culture systems for mimicking human hearts
Despite long-term and extensive investment of time and funds, about 90% of new drugs fail during phase 1 clinical trials [1]. The success rate of drug candidates for cardiovascular disease and oncology is the lowest, mainly due to cardiotoxicity [2-4]. Cardiotoxicity, such as drug-induced QT prolongation, can cause torsades de pointes (TdP), a rapid polymorphic ventricular tachyarrhythmia leading to sudden cardiac death [5]. A broad range of drugs including anti-arrhythmics [6, 7], anti-cancer [8-10], anti-histamines [11], anti-psychotics [12], and anti-virus [13, 14] were reported to provoke cardiac side effects. Since early detection of cardiotoxicity has the foremost significance in new drug development, various model systems of drug screening have been introduced with their potential to detect cardiotoxicity. Drugs for cardiovascular diseases are usually given chronically, and therefore ensuring low toxicity is essential for preclinical development.
The ideal cells for examining cardiotoxicity are human adult cardiomyocytes (CMs) isolated from the patient's cardiac biopsy. However, the limited availability of cardiac biopsy necessitates the use of animal models or nonmyocyte cell lines. Animal models and non-CM cell lines; however, have innate drawbacks for predicting drug efficacy and cardiotoxicity [15, 16]. Animal model systems are low throughput, time-consuming, and relatively expensive compared to other preclinical experiments [15]. More problematic is their low predictability due to inter-species differences. A non-CM cell-line overexpressing a voltage-dependent potassium channel (hERG) was widely used as a drug testing system to evaluate the risk of QT prolongation and TdP for non-cardiovascular drugs on the suggestion of the Committee for Proprietary Medicinal Products in 1997 [17]. However, estimation of QT prolongation in these cells could not correctly predict the response of human CMs [16]. For example, verapamil and ketoconazole were predicted as potentially lethal drugs, but no cases of TdP were reported in the recipients [18], suggesting a potential attrition of valuable drugs from the market.
The emergence of human PSCs, including embryonic stem cells (hESCs) and induced PSCs (hiPSCs), presented a new opportunity for using a more physiological system for drug screening and toxicity testing because they allow generation of human CMs. hPSCs can produce an unlimited number of functional and patient-specific CMs harboring unique genetic signatures. Accumulating studies demonstrated an unprecedented variety of genetic heart disease models and corresponding drug testing results with hiPSC-CMs. Patient-derived hiPSC-CMs are known to recapitulate pathological phenotypes incurred by gene mutations and are especially useful for identifying disease-specific drug candidates and cardiotoxicity. However, hPSC-CMs differentiated in 2D monolayer culture exhibited immature CM phenotypes, restricting their utility. Excitation-contraction coupling (requiring transverse tubules (T-tubules)), positive force frequency relationship (requiring mature calcium handling), slow action potential conduction, efficient energy conversion (requiring oxidative metabolism), and the CM size were notably underdeveloped in hPSC-CMs in 2D monolayer culture [19].
Under 3D conditions, cells are cultured in a more physiological and dynamic microenvironment mimicking in vivo status [19]. Cardiomyocytes cultured in 2D and 3D conditions showed differences in cellular morphology, expression of myofibrils, and junctional proteins [19, 20]. Cells in 3D cultures exhibited less sensitivity to drugs [21] and mechanical stimuli [22], and resistance to apoptotic signaling [23], suggesting the need for 3D culture of hPSC-CMs for appropriate drug testing [24-27]. Accordingly, studies have attempted to differentiate and culture hPSC-CMs in 3D culture systems [28, 29]. For 3D culture, cardiomyocytes are seeded with or without other cardiac cell types embedded in a solidified gel, allowing subsequent tissue formation and mimicking the native physiological state. hPSC-CMs cultured in a 3D environment showed different electrophysiological and mechanical responses compared to those cultured in 2D systems [30, 31], and also allow non-invasive and repeated measurements of contractility [31, 32]. In addition, 3D cultures of hPSC-CMs can better induce CM maturation, which is a critical challenge in regenerative medicine and drug testing. Various approaches have been developed to improve hPSC-CM maturity. Co-culture with non-cardiac cells was the most frequently used method for CM maturation[33-35]. Soluble factors secreted from human mesenchymal stem cells (hMSCs) co-cultured with hiPSC-CMs were employed to impact hiPSC-CM maturation [35]. Extended culture periods also increased iPSC-CM maturity [36, 37]. Electromechanical stress was also reported for better CM maturation. Mechanical stress [38-40] improved cardiac maturation via stretching CMs [41]. Electrical stimulation [38, 39] with gradually increasing frequency over weeks also matured hiPSC-CMs. For example, physical conditioning with increasing intensity allowed hiPSC-CMs to have a transcriptionally and structurally advanced mature identity. The use of biochemical cues including changing the energy source from glucose to fatty acids [42, 43], treatment with humoral factors such as T3 [44], insulin-like growth factor-1, or corticosteroids [45] were shown to induce CM maturation. Moreover, approaches combining the above methods were also developed [38]. Although CM maturation characteristics such as T-tubule and Z-disks with A- and I-band were achieved through maturation of hEHT, the overall degree of maturity was far below the level of adult cardiomyocytes.
Using 3D culture platforms, two types of artificial tissues have been developed, which can emulate some of the function and structure of native heart with various sizes and shapes. Human engineered heart tissue (hEHT) is the most representative type and can be constructed by combining different types of cardiac cells and biomaterials [38, 46, 47]. Another more recently developed type are human cardiac organoids, which are generated by self-organization of the differentiating hPSCs. Unlike hEHTs, organoids are induced by self-organization [48]. While each model mimics a few aspects of native heart, all currently available hEHTs or organoids are dissimilar to the human heart in structure, function, and cellular components. For example, no models include native cardiac elements such as inflow and outflow tracts, each cardiac chamber, and nerves. In this review, we address the progress and applications of hEHT and cardiac organoids generated with hPSCs.
2. Models of human engineered heart tissue (hEHT)
The heart is a sophisticated muscular engine constantly pumping blood via the circulatory system. Efforts to emulate cardiac muscle-like tissue constructs have yielded various types of hEHTs for different purposes. Representative hEHT types include strip [26, 31, 38, 46, 49-52], ring [28, 32], patch [53-55], film [56, 57], heart-on-a-chip [58, 59], spheroid [60-65], hollow spherical chamber [66, 67] and tube types [68]. Various tissue engineering technologies were employed to assemble them in three dimensions. While current hEHTs do not contain the sophisticated structure of the heart, they can represent more than one functional feature of the heart. These hEHTs are classified by production methods and characteristics, and each type has distinct advantages and disadvantages (Table 1). Representative cell culture conditions and compositions of hEHTs are summarized in Table 2.
Various types of hEHTs
| Type | Shape | Generation | Characteristics | Applications |
|---|---|---|---|---|
| Strip |  | Compaction of cells and hydrogel around two parallel wires or posts[26, 31, 38, 46, 49-52] | Drug testing; No vasculature; Limited diffusion | Drug screening |
| Ring |  | Condensation of hydrogel with cells in circular casting mold and transfer onto silicon passive stretcher[28, 32] | Applicable for electrical pacing studies; No vasculature; Arrhythmogenic | Disease Modeling; Drug screening |
| Patch |  | Accumulation of cell layers on coated plates[53-55] | Regenerative therapies; Higher number of cells required; Low throughput; Risk of breaking; Unequal distribution of the cells | Cardiac Regeneration; Disease modeling |
| Film |  | Seeding of cells onto coated film [56, 57] | Limited cell-cell and cell-matrix interactions; Embedded electrode | Disease modeling |
| Microfluidic Chips |  | Seeding of cells onto chip (MPS)[58, 59] | Easy manipulation of the microenvironment; Realtime on-chip analysis; Limited cell-cell and cell-matrix interactions; Limited force measurement; Requiring multi-step fabrication; Unintended drug absorption | Drug screening |
| Spheroid |  | Assembly of cell mixture and hydrogel[60-65] | No requirement for expensive instruments; High-throughput drug testing; Necrosis in the core (>250) (limitation of diffusion) Not feasible for electromechanical stimulation | Disease modeling; Drug screening |
| Chamber |  | Cell seeding in agarose mold around balloon Foley catheter[66, 67] | Recapitulating 3D structure of the heart; Low throughput; No chamber specification; Catheter related complications | Disease modeling; Drug screening |
| Tube |  | Wrapping of cell sheets (CM, fibroblasts) around a hollow column[68] | Emulating the multi-layered cardiac wall; Low throughput; Medium leakage leading to the shrinkage of the tube; Requiring high percent of fibroblasts for wall stiffness | Cell-based cardiac pump |
Representative cell compositions and culture conditions of hEHTs
| Type | Paper | Cell composition | Culture condition |
|---|---|---|---|
| Strip | Mills et al. [26] | hESC-CMs (5 × 104) | α-MEM with 10% fetal bovine serum (FBS) |
| bovine collagen I and Matrigel | |||
| electrical stimulation | |||
| Huebsch et al. [49] | hiPSC-CMs: fibroblast final density: 2 x 107 cells/mL | maintained in EB20 media | |
| Tulloch et al. [40] | hESC-CMs (2 x 106) HUVEC (1 x 106) MSCs or MEFs (1 x 106) | RPMI medium with B27 supplement | |
| collagen type I | |||
| uniaxial mechanical stress conditioning | |||
| Ronaldson-Bouchard et al. [38] | hiPSC-CMs 75% Fibroblast 25% | DMEM supplemented with 10% FBS | |
| human fibrinogen and human thrombin | |||
| electrical stimulation | |||
| Zhao et al. [46] | hESC-CMs: cardiac fibroblasts atrial 10:1.5 ventricle 10:1 final density: 5.75 x 104 cells/mL | DMEM, 10% FBS | |
| rat tail collagen and Matrigel | |||
| electrical stimulation | |||
| Ring | Goldfracht et al. [32] | hESC-derived atrial/ventricular cells (2 x 106) | IMDM |
| bovine collagen | |||
| electrical stimulation | |||
| Patch | Gao et al.[105] | hPSC-CMs: SMCs: ECs (2:1:1) | DMEM containing 10% fetal calf serum, B27+, Ɛ-aminocaproic acid, and ROCK inhibitor |
| fibrinogen, Matrigel, and thrombin solution | |||
| mechanical stimulation | |||
| Ye et al.[133] | hiPSC-CMs (3.5 x 105) hiPSC-ECs (4 x 105) hiPSC-SMCs (3.5 x 105) | 1:1 mixture of medium collected from hiPSC-ECs and hiPSC-SMCs that had been cultured in serum- and glucose-free MEM medium | |
| fibrin with thrombin | |||
| Stevens et al.[99] | hESC-CM (2 x 106) HUVEC (2 x 106) MEF (1 x 106) | RPMI medium with B27 supplement | |
| electrical pacing | |||
| Amano et al.[55] | hiPSC-CMs (1 x 106) cardiac microvascular endothelial cells (1 x 106) cardiac fibroblasts (1 x 106) | DMEM | |
| fibronectin and gelatin | |||
| Film | Lind et al.[134] | hiPSC-CMs (2.2 x 106) NRVMs (1 x 106) | M199 containing 2% FBS |
| fibronectin | |||
| electrical point stimulation | |||
| Chip | Matuhr et al.[59] | hiPSC-CMs (4~5 × 105) | maintained in EB20 media |
| Spheroid | Archer et al.[63] | hiPSC-CMs: primary human cardiac microvascular endothelial cells: primary human cardiac fibroblasts (4:2:1) final density: 8 x 104 cell/mL | cardiomyocyte maintenance medium: endothelial basal medium 2 (1:1) |
| Chamber | MacQueen et al. [66] | hiPSC-CMs (1 x 106) | M199 plus vitamin B12 |
| human fibronectin | |||
| Tubular | Tsuruyama et al. [68] | hiPSC-CMs (1 x 106) NHDF (1 x 106) | DMEM |
| fibrin and collagen | |||
| electrical stimulation |
The strip model comprised of elongated muscle fiber and shaped like a femur, is the most studied form of hEHT. The strip type is generated by the assembly of hPSC-CMs with or without supporting cardiac cells in a hydrogel including collagen, Matrigel, fibronectin or fibrin [26, 31, 38, 49]. Generally, differentiated CMs on day 14 are enzymatically dissociated into single cells and mixed with supporting cells including cardiac fibroblasts. The cell mixture is suspended in a mold and maintained for more than a week for compaction, thereby forming a strip [26, 38]. The blunt ends of the strip are connected to a wire or column which provides mechanical stress for cardiomyocyte maturation [38]. For example, Mills et al. developed a 96-well device for functional screening of small molecules using hPSC-derived cardiac organoids (hCOs) [26]. For each hCO, cardiac cells were mixed with collagen I and Matrigel and cultured in serum-free conditions in DMEM. This study demonstrated the utility of a hEHT for pro-regenerative drug development, identification of the biological mechanisms of CM maturation, and minimization of adverse side effects. The potential for mass production of hEHT from a small number of cells makes it attractive for high-throughput drug screening [49]. Yumi Zhao et al. reported a heteropolar strip type with atrial and ventricular sides and chamber-specific drug responses [46]. However, there is a risk of necrosis due to the lack of vasculature for diffusion, and cardiac pathological conditions such as pressure overload or myocardial infarction are not induced in this model.
The ring model is engineered by pipetting a cell mixture into circular casting molds [28, 32]. The ring type differs from the strip type by a hole in the middle of the hEHT, reducing the risk of necrosis [28, 32]. Additionally, the hEHT rings can be hooked around a silicon exerciser for transducing force and measuring their functions in response to electrical pulses [32, 38]. The utility of the ring-type hEHTs was shown in pharmaceutical testing [32], a cryoinjury model [28], and cardiac regeneration studies [28]. For example, Goldfracht et al. established ring-shaped hEHTs by differentiating hPSCs into ventricular or atrial cardiomyocytes, and then embedding these cardiomyocytes in a collagen-hydrogel to create chamber-specific, ring-shaped hEHTs [32]. Ring-shaped hEHTs were transferred onto a silicon passive stretcher and cultured in IMDM Medium. The potential of this chamber-specific cardiac tissue model was demonstrated in physiologic studies, disease modeling, and drug testing. However, the maturity of the CMs was still far lower than adult human CMs [69-71].
The patch type is a flat shape of tissue and can be generated with or without scaffold. Matrigel, fibrin, or decellularized organ was used as scaffold [53-55], and temperature-sensitive dishes were utilized for scaffold-free generation of hEHT [72, 73]. The patch type is mainly applied for regenerating injured hearts when transplanted onto the outer cardiac wall [53-55]. For example, Gao et al. generated human cardiac muscle patches of clinically relevant dimensions (4 cm × 2 cm × 1.25 mm) by suspending cardiomyocytes, smooth-muscle cells, and endothelial cells that were differentiated from hiPSCs in a fibrin scaffold. When transplanted into a porcine infarct model, this patch-shaped tissue was shown to reduce infarct size. However, the patch type requires a large number of cells, and the risk of tissue destruction during the culture period was raised [74].
The spheroid type is a small and simple form of hEHT. Spheroids can be generated by culturing cell mixtures of hPSC-CMs, human cardiac fibroblasts, and hPSC-ECs in a non-adhesive and round-bottom 96-well plate [60-65]. Archer et al. fabricated spheroids using hiPSC-CMs, human cardiac microvascular endothelial cells, and human cardiac fibroblasts (4:2:1). After 14 days, spontaneously beating microtissues were formed [63]. Due to the availability of multiple spheres produced simultaneously from a relatively small number of cells, spheroids were claimed to be suitable for high-throughput drug testing [75]. However, their utility is limited, since the spheroids lack vasculature and cell necrosis can occur inside spheroids larger than 250µm [76].
The heart-on-a-chip type is a module-based heart tissue connected to a microfluidic system [58, 59]. Heart-on-a-chip can precisely control the microenvironment and monitor the real-time drug response of CMs [77, 78]. A primitive example is a cardiac microphysiological system (MPS) developed by Maher et al. [59]. By aligning CMs in the microtissue and creating separate fluid transport channels mimicking vasculature and continuous nutrient exchange, this MPS was able to keep hiPSC-derived cardiac tissue viable and functional over multiple weeks. This system allowed multiple modes of cardiac functional analysis and drug screening. However, the need for sophisticated equipment limits high-throughput drug screening. Furthermore, the microfluidic channel could absorb test drugs, preventing accurate prediction of drug response [79, 80].
The film type is a biomaterial coated sheet or low-adhesive tape with a monolayer of cardiomyocytes [56, 57]. A strain sensor embedded in the thin film enables non-invasive and continuous readout. Lind et al. introduced an instrumented cardiac microphysiological device with a monolayer of hiPSC-CMs or commercially available Cor4U cell line seeded on the fibronectin coated film [124]. They established high-throughput cardiotoxicity screening through flexible sensors, allowing fabrication of a multi-well platform with continuous readout of the contractile stress and beating rate. Compared to the heart-on-a-chip type, the film type can be made relatively simply with general materials in the lab [57]. The forward and backward movements of the film type can be measured as the contractile capacity of the hEHT. The film type was utilized for the modeling of Barth syndrome and the cardiotoxicity drug test [57].However, CM immaturity, lack of capacity for long-term culture, and low reproducibility were claimed as disadvantages of the film models [57].
There have been attempts to generate a hollow, 3D, fluid-ejecting hEHT to mimic the native heart. The following two types have such characteristics, having a cardiac cavity enabling functional analyses such as PV loop analysis, developed pressure, cardiac output, and ejection fraction [66-68]. The hollow chamber type is a fluid-pumping cardiac tissue made of either catheter or nanofibrous scaffolds [66, 67]. In the catheter type, a mixture of differentiated CMs and collagen was solidified surrounding a silicon Foley catheter [67]. After gel polymerization, the hollow cardiac chamber was carefully removed and incubated in a bioreactor containing cell culture medium. MacQueen et al. created tissue-engineered ventricles by using ventricle-shaped scaffold through nanofiber spinning and nanofibrous scaffolds [66]. These scaffolds were incubated with fibronectin and CMs (rat ventricular CMs or hiPSC-CMs) at high density. The hollow chamber had a cavity of 500 µl of diastolic volume, or about 1/250 of the native human heart, and a very thin wall (~0.1 mm) to maintain cell viability in the diffusion-limited condition without vessels. This engineered tissue allowed measurements of multiscale in vitro cardiac assays including tissue coverage and alignment, calcium-transient propagation, and pressure-volume loops in the presence or absence of test compounds [66]. The tube type was generated by serially wrapping custom-made tubes with hiPSC-CM cell sheets and fibroblast sheets [68]. Fibrin and collagen gels were applied as glue between the attached sheets and the tube. Measurement of inner pressure was available using a catheter, and the multi-layered structure of this type is distinct from the simple combination of cells in other models. However, the tube type carries the risks of necrosis in multi-layered cell sheets without vasculature, and tube shrinkage caused by medium leakage [68].
3. Applications of hEHT
hEHT can present a more relevant human disease model than animal models. hEHT enables experiments that would have been impossible with hPSC-CMs in conventional 2D systems. In 2D cultures of hPSC-CMs, invasive electrophysiological experiments are the major analytical tools; however, in hEHT, noninvasive and repeated measurements of contractile force and electrophysiological activities are possible. In addition, owing to a drug resistance generally exhibited in 3D cell culture, hEHT is expected to recapitulate in vivo responses better than hPSC-CMs in 2D culture [81-83]. Furthermore, technical advances enabled the generation of atrial [84], ventricular [67] or bipolar (atrial and ventricular ends) [46] hEHT, allowing more sophisticated modeling of chamber-specific cardiac diseases. With the aforementioned advantages, hEHT can be applied for disease modeling including genetic and non-genetic cardiac diseases, drug screening, cardiac regeneration, and cell-based cardiac pumps.
3.1 Disease modeling
Cardiac disease models were investigated with 2D-cultured hiPSC-CMs carrying genetic mutation(s) or induced pathological cardiac conditions [85-87]. Diseased hiPSC-CMs and electrophysiological experiment systems are appropriate to investigate disorders caused by abnormal ion channel activities but are not suitable for cardiac maladaptations caused by mechano-structural problems. Recent advances in hEHT technologies enabled the investigation of various disease models overcoming the limitations of CMs cultured in 2D systems. Here, we discuss representative genetic and nongenetic cardiac diseases which can be modeled by hEHT.
3.1.1 Genetic disease
Barth syndrome (BTHS) patients exhibit mutations in the TAZ gene and abnormal sarcomerogenesis in hiPSC-CMs [57]. The contractile pathophysiology caused by abnormal sarcomerogenesis was investigated with the film type of hEHT by seeding purified BTHS hiPSC-CMs on a thin elastic film, which was called a muscular thin film tissue (MTF). MTF generated with BTHS hiPSC-CMs (BTHS-MTF) showed significantly lower twitch and peak systolic stress compared to controls, and introduction of modified TAZ mRNA reversed the myopathic phenotypes, demonstrating that the BTHS phenotypes were driven by a mutation in the TAZ gene [57].
Duchenne muscular dystrophy (DMD) is caused by mutations in the X-linked dystrophin gene. The majority of DMD patients eventually develop dilated cardiomyopathy (DCM) [88]. A DMD disease model was made with hiPSC-CMs from a DMD patient (DMD hiPSC-CMs), and a recovered model was generated after genome editing of the dystrophin gene using CRISPR-Cas9 technology (corrected DMD) [89]. hEHT generated with corrected DMD hiPSC-CMs exhibited improved contractile function over DMD hEHT but lower contractility than normal hEHT. Subsequently, corrected DMD hiPSC-CMs were mixed with DMD hiPSC-CMs in the range of 10-100% to identify the percentage of corrected CMs needed to rescue the DMD phenotype, and the gene correction required to restore the cardiac function was found to be 30 to 50% of CMs [89].
Hypertrophic cardiomyopathy (HCM) is a polygenic disease that is strongly influenced by environmental factors and usually associated with mutations in contractile components of the sarcomere [90]. Increased expression of hypertrophic markers, aberrant calcium handling, and thickening of myocardium are characteristics of HCM patients [90]. hEHT models of HCM were generated with hiPSC-CMs containing BRAF [91], or PRKAG2 mutations, or electrical stimulation on hiPSC-CMs derived from a hypertension patient. BRAF encodes a serine/threonine kinase regulating the RAS/MARK pathway, which has diverse roles in cell cycle, cell growth, differentiation, and senescence [92, 93]. hEHT generated with hiPSC-CMs with a BRAF mutation exhibited hypertrophic characteristics including a trend of shorter twitch duration and higher passive Young's modulus, indicating tissue stiffness. However, the pathological phenotypes were diminished only after 11 days from the hEHT formation, suggesting the need for extra stimulations or further sophisticated development of hEHT to recapitulate the cardiac hypertrophy shown in patients [94]. PRKAG2 mutations can cause inherited autosomal dominant left ventricular hypertrophy [95]. HCM-hEHT with a PRKAG2 mutation exhibited HCM phenotypes with increased AMPK activity and reduced adverse remodeling and arrhythmia with AMPK agonist [96]. In addition, hypertrophic hEHT was generated with hiPSC-CMs derived from hypertension patients and application of electrical stimulation for up to 8 months [46]. The hypertrophic hEHT showed enriched gene expression related to pathological remodeling, cardiac enlargement and dysfunction, heart failure, and cardiac hypertrophy. Chronic electrical stimulation and a long period of hEHT culture might be essential for generation of human hypertrophic heart in a dish.
Inherited arrhythmogenic syndromes, such as short QT syndrome (SQTS), were modeled using patient-specific hiPSCs. hiPSC-derived cardiac cell sheets (hiPSC-CCSs) were generated from a symptomatic SQTS patient carrying the N588K mutation in the KCNH2 gene [97] and were used as a tool for studying conduction and arrhythmogenesis. Optical mapping reported shortened APD, impaired APD-rate adaptation, abbreviated wavelength of excitation, and increased inducibility of sustained spiral waves. Phase-mapping analysis showed accelerated and stabilized rotors. Antiarrhythmic agents including quinidine, disopyramide, and sotalol were shown to rescue the arrhythmic phenotype.
3.1.2 Non-genetic disease
hEHT can be used to emulate cardiac injury and cardiac response to drugs. Upon severe ischemic damage caused by coronary artery occlusion, the adult human heart undergoes pathological changes due to the limited regenerative potential of CMs, although the fetal/neonatal heart undergoes a full functional recovery through CM proliferation [98]. The distinct cardiac repair responses according to CM maturation were investigated with a cryoinjury model of hEHT [28]. Cryoinjury caused CM death in a localized area and high CM proliferation rather than CM hypertrophy and fibrosis, suggesting hEHT as a disease model of the immature human heart [28]. This immature heart model was used for screening drugs and identifying pathways for CM proliferation, and led to the discovery of a synergistic activation of the mevalonate pathway and a cell-cycle network during CM proliferation [26].
hEHT has been proposed as a myocardial infarction (MI) model. For inducing MI-like conditions through low-oxygen supply conditions, small spheroids with a radius of ~150 µm were fabricated by the self-assembly of cardiac cells (iCell cardiomyocytes: human cardiac ventricular fibroblasts: human umbilical vein endothelial cells: human adipose-derived cells = 7:4:2:1) [62]. Due to the lack of vessel formation, oxygen gradients were naturally formed in a normoxic condition (20% oxygen) and severe oxygen deprivation was created in the center of the spheroids. Necrotic core, which is considered a common problem among hEHT [99], was exacerbated by hypoxic conditions (10% oxygen). A shell of fibroblasts was found in spheroids together with cell apoptosis in the core, loss of contractile function and unsynchronized CM contractions. An anti-fibrotic reagent, JQ1 (bromodomain inhibitor) reduced the number of fibroblasts and enhanced synchronized contractions of CMs. In addition, doxorubicin exacerbated the pathological conditions [62]. However, due to the severe cell death without CM regeneration and fibrosis in the core, replacing dead CMs with fibroblasts, the MI spheroid model has limitations to be addressed before deeming it an appropriate MI model.
A heart failure model was inducible in the matured hEHT by chronic catecholamine overstimulation [39, 97]. A ring type of hEHT formed by a mixture of cells (hPSC-CMs and fibroblasts) and matrix (collagen and Matrigel) was induced to mature under mechanical load and conditioned media containing several growth factors including IGF and FGF. After treatment with catecholamine, hEHT exhibited contractile dysfunction, CM hypertrophy, and increased cell apoptosis [39]. Heart failure model of hEHT is characterized by transcriptional profiling in 6-week engineered human myocardium (EHM) in agreement with the structural and functional data. The limitations of the heart failure model of hEHT come from the immaturity (equivalent to fetal human heart at 13 weeks of gestation).
hEHTs are also used for mimicking arrhythmias. Using linear and circular hEHTs, the dynamics of activation propagation was shown to depend on geometry [100]. Linear hEHT showed the normal propagation pattern across the distal ends. However, spontaneous infinite reentrance of activation propagation was seen in the circular shape, mimicking tachycardia in a model of arrhythmogenic cardiomyopathy. Defibrillation through electrical field-stimulation reversed arrhythmias in the circular hEHT to a normal rhythm state. Consequently, the circular hEHT was suggested as an arrhythmic disease model and a screening platform for antiarrhythmic drugs. Another group also established a circular cell sheet using hiPSC-CMs for studying arrhythmogenesis via optogenetic stimulation [101]. hEHTs were transduced with lentivirus expressing channelrhodopsin-2 (H134R) and stimulated by bursts of blue light separated by no pacing for 3 weeks. This optical pacing induced chronic tachycardia in the hEHTs, showing shortening of action potential duration 90 (APD90) and reduction of L-type Ca2+ current. This induced tachycardia was terminated by ryanodine receptor stabilization, or sodium, or hERG potassium channel inhibitor. Furthermore, arrhythmia modeled in ring-shaped atrial hEHT showed a large single circular re-entry wave propagating around the ring and multiple spiral-wave reentrant loops observed in rhythm disorders [32]. Particularly, this model proposed a chamber specific, atrial-hEHT for atrial arrhythmias. Anti-arrhythmic agents vernakalant and flecainide converted the arrhythmic hEHTs to normal rhythm.
3.2 Drug testing and pharmacotoxicity using hEHT
hEHTs composed of major cardiac cells including CMs, fibroblasts, and endothelial cells are suggested to be an optimal drug testing platform as they can better emulate functional and structural changes of CMs in a tissue environment and have higher sensitivity to cardiotoxins compared to 2D-monolayer CMs [63]. Metrics of cardiac tissue function such as contraction rate, conduction properties, and mechanical motion can be recorded in a non-invasive manner and analyzed in real-time. Chip (MPS system), strip, and spheroid types which are compatible with computer devices are generally suitable for high-throughput drug screening platforms.
Mathur et al., performed the pharmacological studies using the MPS system to test the cardiac response to four drugs (isoproterenol, metoprolol, E-4031, verapamil) [59]. The motion tracking was recorded with a microscope and analyzed by automated video-optical recording. Data indicated that the half-maximal inhibitory/effective concentration values (IC50/EC50) were consistent with the data on tissue-scale references compared to cellular-scale studies. A strip type, fibrin-based hEHT in a 24-well format was established as a simple in vitro model for cardiac research [102, 103]. Spontaneous contractions of hEHT were analyzed by automated video-optical recording. Chronotropic responses of the β-adrenergic agonist isoprenaline were observed in the presence of calcium. Also, concentration-dependent irregular beating, and reversible decreases in relaxation velocity were induced by the proarrhythmic compounds [102]. Another strip type of hEHT was established with 5 commercial and 5 academic hPSC-CM lines for suitability for drug screening. Spontaneous and stimulated contractions were induced in these hEHTs for evaluating baseline contractile force, kinetics, and beating rate. Those parameters varied depending on the lines, while canonical drug responses were observed in most hEHTs. This study suggested that this hEHT may not be relevant for drug screening, while it could be used for disease modeling with the addition of isogenic controls [103]. Milles et al. performed functional screening of 105 small molecules with pro-regenerative potential with their previously developed strip type platform [26]. The study revealed discordance between their hEHT and conventional 2D assays for many known pro-regenerative compounds. Using their 3D hEHT, they identified two pro-proliferative small molecules that did not have detrimental effects on cardiac function. High-throughput proteomics of these two compounds revealed synergistic activation of the mevalonate pathway and a cell-cycle network, suggesting the utility of this platform for identifying biological mechanisms as well as drug screening.
hEHT was found to be useful for investigating structural changes of CMs induced by various drugs. This structural cardiotoxicity can be evaluated by morphological damage such as CM degeneration, necrosis, fibrosis, and progressive pathological changes in subcellular organelles of CMs. FDA-approved structural cardiotoxins (https://www.pharmapendium.com) were evaluated using the spheroid type of hEHT (less than 100 µm radius) generated by co-culturing of CMs, fibroblasts, and endothelial cells in an ultra-low adhesion plate [63]. Structural cardiotoxicity is usually assessed for non-cardiac drugs such as anti-cancer agents. For example, lapatinib, an anti-cancer drug, was found to induce ATP depletion in endothelial cells but not in CMs and fibroblasts, suggesting that lapatinib caused the structural cardiotoxicity through the damage of endothelial cells.
3.3 Other applications of hEHT
Other applications of hEHTs include in vivo cardiac regeneration and cell based cardiac pump. A patch type of hEHT was generally used for in vivo cardiac regeneration. For example, this hEHT was generated with a cell mixture of human embryonic stem cell-derived CMs (hESC-CMs), human umbilical vein endothelial cells (HUVECs), and fibroblasts with [104] or without [99] a biodegradable porous scaffold. When transplanted into the injured hearts of animals, this patch, which included a mixture of CMs, ECs, and fibroblasts, substantially increased the survival of engrafted hESC-CMs, and the preexisting vessels were anastomosed with host vasculature inside the engrafted hEHT [99, 104]. Generation of hEHT of a clinically relevant size was attempted through an increase in scaffold size (up to 4 x 4 cm). However, the increased scaffold size was not associated with a dramatic increase in cell number in the patch [25, 105]. Regardless of the scaffold size, the total cell number was at most 8 ~ 10 million cells, suggesting the limitation of cell density in clinical application.
hEHT can also be used as a cardiac pump. Left ventricular assist devices (LVADs) could be one therapeutic option for patients with end-stage heart failure [106]. However, mechanical circulatory support devices such LVADs have limitations including thrombogenicity, power transmission, and infection [107]. With the development of hEHTs, a cell-based cardiac pump could be used instead of a LVAD. Such a cell-based LVAD is expected to offer a high degree of immune tolerance and create a bypass blood flow from the apex of the heart directly to the aorta [107]. An optimal cell-based cardiac pump would be composed of a hollow chamber wrapped by contracting cardiac cells, one-way valves to ensure unidirectional flow, and electrical detectors and stimulators embedded as part of the device [107]. The currently available cell-based pump, which is at the early developmental stage, is a tube-type hEHT (Table 1) consisting of a long hollow column wrapped by CM/fibroblast cell sheets without cardiac valves or electrical devices [68].
4. Human cardiac organoids
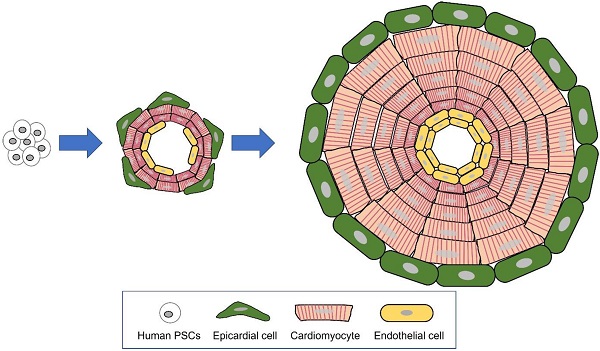
An organoid is an in vitro 3-dimensional miniature organ which is generated with organ-specific adult stem cells or pluripotent stem cells (PSCs). According to Lancaster and Knoblich, an organoid is defined by three characteristics: 1) constitution of multiple organ-specific cell types, 2) capability to recapitulate some specific function of the organ, and 3) sorted multi-cell populations grouped together and organized similar to an organ [48]. Although organoids and spheroids are both cultured in 3 dimensions and the terminology has been ambiguously used, there are distinct differences between organoids and spheroids [108, 109]. Self-organization, which consists of cell sorting out and spatially restricted lineage commitment of precursor cells, is the key mechanism differentiating between organoid and spheroid [48] (Figure 1). Cell sorting out is a general capacity of cells to reorganize and classify to form structures with much the same histogenic properties. In spatially restricted lineage commitment, precursor cells give rise to more differentiated progeny by spatial constraints of the tissue and/or division orientation [48]. Being self-organized, the growing organoid mimics the major processes in development. These novel tissue-patterning mechanisms offer the organoid higher complexity, having in vivo-like physiological features and long-term maintenance (Figure 2).
Although self-organizing organoids have been reported for almost all major organs for over a decade [110-112], human cardiac organoids were more recently developed [113, 114]. Beginning with the first organoid for optic cup developed with hESCs in 2008 [115], various types of organoids including brain [116], retina [117, 118], intestine [119], kidney [120-122], and liver [123] have been generated with hPSCs. For cardiac organoid-like structures, dissociated neonatal chicken [124] and rat cardiac cells [125] showed innate capacity to re-aggregate and construct a “mini-heart” with a cavity. However, a mixture of individually differentiated CMs, ECs, fibroblasts, and smooth muscle cells from human PSCs did not self-organize or grow into a “mini-heart” [114, 126, 127].
Unlike hEHTs, which use mixtures of individually differentiated cardiac cells, cardiac organoids are formed by self-organization of stem cells during cardiac differentiation (Table 3). Spontaneously contracting human cardiac organoids with cavities have been generated by several investigators. Human cardiac organoids recapitulate developmental events and model genetic malformation by specific mutations in genes or modification of the culture conditions (Table 4). Thus, cardiac organoid is a favorable model for studying cardiac development and developmental disorders. Since human cardiac organoids include vessels, [126], long-term culture is possible, which can induce more mature CMs.
Comparisons of hEHT and cardiac organoid
| hEHT | Cardiac organoid | |
|---|---|---|
| Cell source | hPSC-CM hPSC-CM with non-cardiac cells | Differentiating hPSC |
| Use of a mold during tissue formation | O | X |
| Connection with an instrument for real-time assessment | O (except spheroid) | X |
| Developmental study | Unfeasible | Feasible |
| Functional assay | - Contractility/force generation - Intraventricular pressure/volume - Action potential - calcium transient | - Contractility/beating rate - Action potential - Calcium transients |
| Applications | - Cardiotoxicity test - Drug efficacy test - Disease modeling - Cardiac regeneration - Cardiac pump | - Developmental studies - Disease modeling ▪ congenital cardiac defect ▪ cryoinjury |
Organoid generated through self-organization of differentiating pluripotent stem cells (PSCs). The major mechanisms of organogenesis are self-organization, consisting of cell sorting out (gathering of similar cell types) and spatially restricted lineage commitment.
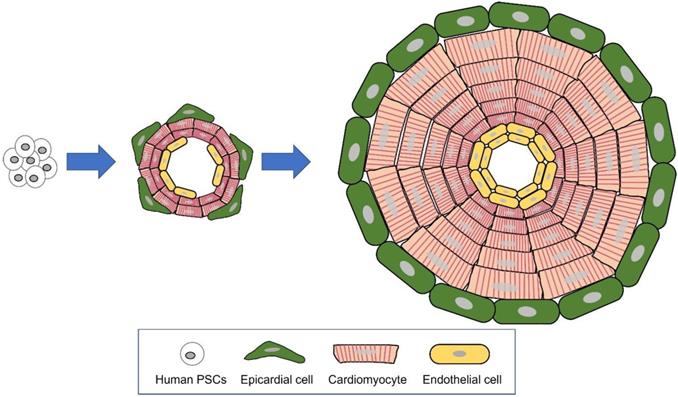
Major differences between hEHT and cardiac organoid. A Strip type hEHT, a most common form of hEHT. B Cardiac organoid resembling a native heart in cell composition (multi-cardiac cells), macro- (a hollow chamber) and micro- (multi-specialized layers composing a wall) structure, repeated systolic and diastolic contractions, and volume-pressure relationship.
Drakhlis et al. generated heart-forming organoids (HFO) with hPSC aggregates embedded in Matrigel via biphasic WNT pathway modulation [128]. HFOs are composed of a myocardial layer lined by endocardial-like cells surrounded by septum-transversum-like anlagen. HFOs also possess spatially and molecularly distinct anterior versus posterior foregut endoderm tissues and a vascular network. The architecture of HFOs recapitulates early cardiomyogenesis, which requires an interplay between cardiac mesoderm and foregut endoderm. Disease models using HFOs with a NKX2.5-knockout (KO) hESC reporter line showed a phenotype previously observed in the same gene knockout mice (less organized, larger cardiomyocytes, and decreased cell adhesion).
Hofbauer et al. established hPSC-derived self-organizing cardioids that intrinsically specify, pattern, and morph into chamber-like structures containing a cavity, reminiscent of the early human left ventricular heart chamber. Beating organoids with small homogeneous sizes were developed within the dish without exogenous ECM and using a high-throughput approach to reach optimal signaling conditions [126]. The average diameter of cardioids at CM specification (day 5.5~7.5) was about 2,000 μm. The cardioid corresponded to the first heart field lineage and the early stages of cardiogenesis. The study further demonstrated that this cardioid can model mechanisms underlying development of the three major components of cardiac architecture CMs, endothelial layers and epicardial lining as well as response to cardiac injury (cryoinjury). While this paper explores the potential for generating an organoid with a cavity, events at a late developmental stage are lacking, such as cardiac structure formation, maturity of cardiac cells, or compaction of myocardium. To examine the response to cryoinjury, cardioids were cultured with epicardial cells; however, co-cultures do not meet the concept of organoid as it is not an intrinsic organogenesis process.
Lewis-Israeli et al. generated human heart organoids (hHOs) using a two-step canonical Wnt signaling modulation strategy using growth factors and chemical inhibitors [129]. hHOs mimic human cardiac development and are similar to age-matched fetal heart tissues at the transcriptional, cellular, and structural levels. hHOs develop internal cardiac chambers, with multiple lineage cells having regional identities reminiscent of the heart fields and the atrial and ventricular chambers, epicardium, endocardium, and coronary vasculature. These hHOs were shown to model congenital heart disease induced by pregestational diabetes, suggesting the utility of this model for emulating the effects of complex metabolic disorders on cardiac development. However, the maturation level of hHO is restricted to embryonic fetal hearts, and hHOs tend to deviate from their normal developmental pathway as a function of time, becoming less relevant over time.
Song et al. fabricated cardiac mesoderm cell-derived cardiac organoids (CMC-COs) and CM-derived COs (CM-COs) to investigate the effect of differential self-organizing capacity of mesoderm-derived cells or CMs on maturation of CMs in organoids [130]. hPSC-derived CMCs and -CMs were dissociated at day 4 or 11 of cardiac differentiation and then plated on poly (2-hydroxyethyl methacrylate) (poly-HEMA)-coated plates. CMC-COs exhibited structural differences compared with CM-COs such as more organized sarcomere structures and mitochondria, well-arranged T-tubule structures, evenly distributed intercalated discs, and increased expression of ventricular CM and junctional markers. CMC-COs showed mature ventricular-like function including faster motion vector speed, decreased beats per min, increased peak-to-peak duration, and prolonged APDs. This study further revealed that LEFTY-PITX2 signaling plays a crucial role for CM maturation and specification into ventricular-like CMs.
Human Cardiac Organoids
| Model | Formation | Characteristics | Applications | Multilayered cardiac chamber | |
|---|---|---|---|---|---|
| Chamber | Sponta-neous beating | ||||
| Heart-forming organoids (HFOs) | Differentiating hPSC aggregates embedded in Matrigel via biphasic WNT pathway modulation [128] | Stage: pre-heart tube-like stage at 2-3 weeks of human gestation | Cardiac development, cardiac malformation in HFOs generated with NKX2.5-knockout hESCs | X | |
| Anterior-posterior endoderm patterning, EC-lined vessel-like structures, distinct foregut endoderm tissues | X | O | |||
| Cardioid | hPSCs differentiated with chemically defined medium [126] | Stage: early human ventricular heart chamber | Cardiac development, Cryoinjury to model myocardial infarction | O | |
| Self-organization of CMs and endothelial cells | O | O | |||
| Engineered heart organoid; Addition of epicardial cells | |||||
| Human heart organoid (hHO) | Differentiation of hPSC-EB with three-step WNT signaling modulation[129] | Stage: embryonic fetal heart | Cardiac development, Modulation of glucose and insulin level to examine the effect of pregestational diabetes on cardiac development | X | |
| Containing major cardiac cells without spatially restricted lineage commitment; Vascularization | O | O | |||
| CMC or CM -derived cardiac organoid (CMC or CM-COs) | hPSC-CMs cultured with B27 without insulin (CMC-COs) | Stage: early cardiac developmental stage | Cardiac development | X | |
| B27 without vitamin A (CM-COs) +thiazovivin and repeated size selection(>70µm) [130] | Spatially and metabolically matured CMC-COs compared to CM-COs | X | O | ||
| Multi lineage organoid | hiPSCs-derived mesendoderm progenitor aggregates differentiated with cardiac-permissive medium + ascorbic acid [131] | Stage: early human embryonic heart (specifically atrial tissue) | Cardiac development | O | |
| Formation of the epicardial layer and primitive endoderm epithelial cystic structure | O | O | |||
| Extensive tissue growth during >1 year culture | |||||
HFO: heart-forming organoids CMCs, Cardiac mesoderm cells, CO: cardiac organoid
Recently, Silva et al. used hiPSCs to produce multi lineage organoids that recapitulate cooperative cardiac and gut development and maturation [131]. Mesendoderm progenitor aggregates were differentiated in cardiac-permissive medium supplemented with ascorbic acid. Multilineage organoid progenitor cells have a transcriptomic profile that supports the co-development of cardiac and gut tissues. The cardiac and gut organoid was maintained for a long period of time (>1 year) in culture and exhibited a millimeter size-scale and improved physiological maturation of cardiac tissue. However, there are technological hurdles to image large size organoids at later stages of culture due to significant light scattering during light microscopy imaging.
5. Remaining challenges and future perspectives
While a number of papers claimed a hEHT model as a cardiac organoid, those hEHTs fell short of the original definition of an “organoid”. This confusion might have arisen from a misunderstanding of the main defining principle of organoids, i.e., self-organization of cells. Thus, there is a need for a uniform definition in this field. Our review attempts to distinguish these two entities by the criteria of “self-organization” of stem cells.
Various methods of hEHT generation were reported and their utility was highlighted for drug screening, disease modeling, and cardiac regeneration. In general, hEHTs are constructed by combining cardiac cells including CMs with natural or artificial matrix using engineering technologies. hEHTs do not mimic the gross structure of the heart and represent one or several functions of the heart (Table 1). Since each model has a specific merit, it would be better to understand their utility in the context of a specific purpose. Among them, the main utility focuses on drug screening and disease modeling. The advantage of hEHTs over pure CMs for such purposes is their similarity to the in vivo environment and their superiority for CM maturation. To guarantee validity of drug testing and disease modeling when using in vitro cell or tissue systems, use of mature CMs is crucial. While CM maturity is improved with hEHTs, most hEHTs fall short of reaching the neonatal state of the heart. To mature the CMs in hEHTs, not one but a combination of biochemical, electrical, mechanical, and tissue engineering technologies would be required. Moreover, most hEHTs lack vasculature, limiting the size and culture period of hEHT without cell death. This is important because CM maturation needs long-term culture. For cardiac regeneration, a patch type hEHT has been widely used. At present, most patches do not show scalability and have a low cell density per unit volume. Cardiac cells need to have close or direct contact with neighbors to function properly; however, engineering technologies to organize cells at high density and of sufficient size (a few centimeters in length and width) without inducing cell necrosis are undeveloped. A minimum of 1-10 billion cells were suggested for clinically meaningful regeneration purposes [132]. Optimal technologies for hEHTs need to organize a large number of hPSC-derived cardiac cells at high density, with vasculature, into a micro-structure mimicking the heart.
Such challenges have been in part addressed by organoids generated from differentiated hPSCs. The most sophisticated currently developed cardiac organoids take primitive heart forms having a cavity and cardiac walls composed of thin layers of epicardial cells and cardiomyocytes, with or without vasculature (Table 4). At the structural level, cardiac organoids better represent the structure of the native heart than hEHTs. Despite their structural closeness to the native heart, no cardiac organoids were shown to generate flow by contraction, although one of the major functions of the heart is its pumping function. Thus far, cardiac organoids have been mostly applied to cardiac developmental studies and disease modeling, while one study reported their utility for studying the response to cryoinjury [126]. Due to the difficulties of monitoring the function of organoids, which are complex in structure, their utility for drug testing has yet to be reported. At present, these self-organized cardiac organoid models lack a system to monitor or control their function in real-time. In addition, the maturity of CMs in the organoids is at best at the level of fetal heart. Therefore, it would help to incorporate bioengineering technologies to equip monitoring and further induce CM maturation. For example, a bioengineering technology to control the flow in and out of organoids would help regulate drug concentrations in drug testing systems. In addition, accessibility to two-way cameras to monitor voltage/calcium transients or ultrasound probes for echocardiography would enhance the utility of organoids. A combination of self-organizing cardiac organoids together with controllable bioengineering technologies could advance and expand their utility for drug screening, disease modeling, developmental studies, cardiac regeneration, and cardiac pump. In the future, a more physiological cardiac organoid emulating native heart is required, which would be a multilayered organ having thick myocardium together with endothelium and epithelium, atrial and ventricular chambers with their own cavities, one-way valves between chambers to ensure unidirectional flow inside the organ, vascularization throughout the organoid, and innervation.
Acknowledgements
This work was supported by National Research Foundation of Korea (NRF) funded by the Korean government (MSIT) (No. 2020R1A2C3003784, No.2020M3A9I4038454), the Faculty Research Assistance Program of Yonsei University College of Medicine (6-2020-0184), the Parts/Materials Development Project in 2021 (20016564) funded by the Ministry of Trade, Industry & Energy (MOTIE, Korea), the Brain Korea 21 Project for Medical Science, Yonsei University and by grants from NHLBI (R01HL150877, R01HL156008), American Heart Association Transformational Project Award (20TPA35490282).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Horvath P, Aulner N, Bickle M, Davies AM, Nery ED, Ebner D. et al. Screening out irrelevant cell-based models of disease. Nat Rev Drug Discov. 2016;15:751-69
2. Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Clinical development success rates for investigational drugs. Nat Biotechnol. 2014;32:40-51
3. Liang P, Lan F, Lee AS, Gong T, Sanchez-Freire V, Wang Y. et al. Drug screening using a library of human induced pluripotent stem cell-derived cardiomyocytes reveals disease-specific patterns of cardiotoxicity. Circulation. 2013;127:1677-91
4. Frommeyer G, Eckardt L. Drug-induced proarrhythmia: risk factors and electrophysiological mechanisms. Nat Rev Cardiol. 2016;13:36-47
5. Kannankeril PJ, Roden DM. Drug-induced long QT and torsade de pointes: recent advances. Curr Opin Cardiol. 2007;22:39-43
6. Tang CPS, McMullen J, Tam C. Cardiac side effects of bruton tyrosine kinase (BTK) inhibitors. Leuk Lymphoma. 2018;59:1554-64
7. Yonemizu S, Masuda K, Kurata Y, Notsu T, Higashi Y, Fukumura K. et al. Inhibitory effects of class I antiarrhythmic agents on Na(+) and Ca(2+) currents of human iPS cell-derived cardiomyocytes. Regen Ther. 2019;10:104-11
8. Burridge PW, Li YF, Matsa E, Wu H, Ong SG, Sharma A. et al. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat Med. 2016;22:547-56
9. Von Hoff DD, Layard MW, Basa P, Davis HL Jr, Von Hoff AL, Rozencweig M. et al. Risk factors for doxorubicin-induced congestive heart failure. Ann Intern Med. 1979;91:710-7
10. Lefrak EA, Pitha J, Rosenheim S, Gottlieb JA. A clinicopathologic analysis of adriamycin cardiotoxicity. Cancer. 1973;32:302-14
11. Woosley RL. Cardiac actions of antihistamines. Annu Rev Pharmacol Toxicol. 1996;36:233-52
12. Glassman AH, Bigger JT Jr. Antipsychotic drugs: prolonged QTc interval, torsade de pointes, and sudden death. Am J Psychiatry. 2001;158:1774-82
13. Johnson RA, Palacios I. Dilated cardiomyopathies of the adult (second of two parts). N Engl J Med. 1982;307:1119-26
14. Matsumori A, Matoba Y, Sasayama S. Dilated cardiomyopathy associated with hepatitis C virus infection. Circulation. 1995;92:2519-25
15. Gintant GA, Limberis JT, McDermott JS, Wegner CD, Cox BF. The canine Purkinje fiber: an in vitro model system for acquired long QT syndrome and drug-induced arrhythmogenesis. J Cardiovasc Pharmacol. 2001;37:607-18
16. Kramer J, Obejero-Paz CA, Myatt G, Kuryshev YA, Bruening-Wright A, Verducci JS. et al. MICE models: superior to the HERG model in predicting Torsade de Pointes. Sci Rep. 2013;3:2100
17. Food Drug Administration HHS. International Conference on Harmonisation; guidance on S7B Nonclinical Evaluation of the Potential for Delayed Ventricular Repolarization (QT Interval Prolongation) by Human Pharmaceuticals; availability. Notice. Fed Regist. 2005;70:61133-4
18. Redfern WS, Carlsson L, Davis AS, Lynch WG, MacKenzie I, Palethorpe S. et al. Relationships between preclinical cardiac electrophysiology, clinical QT interval prolongation and torsade de pointes for a broad range of drugs: evidence for a provisional safety margin in drug development. Cardiovasc Res. 2003;58:32-45
19. Pontes Soares C, Midlej V, de Oliveira ME, Benchimol M, Costa ML, Mermelstein C. 2D and 3D-organized cardiac cells shows differences in cellular morphology, adhesion junctions, presence of myofibrils and protein expression. Plos One. 2012;7:e38147
20. Abbott A. Cell culture: biology's new dimension. Nature. 2003;424:870-2
21. Bokhari M, Carnachan RJ, Cameron NR, Przyborski SA. Culture of HepG2 liver cells on three dimensional polystyrene scaffolds enhances cell structure and function during toxicological challenge. J Anat. 2007;211:567-76
22. Li Y, Huang G, Li M, Wang L, Elson EL, Lu TJ. et al. An approach to quantifying 3D responses of cells to extreme strain. Sci Rep. 2016;6:19550
23. Li CL, Tian T, Nan KJ, Zhao N, Guo YH, Cui J. et al. Survival advantages of multicellular spheroids vs. monolayers of HepG2 cells in vitro. Oncol Rep. 2008;20:1465-71
24. Jabs J, Zickgraf FM, Park J, Wagner S, Jiang X, Jechow K. et al. Screening drug effects in patient-derived cancer cells links organoid responses to genome alterations. Mol Syst Biol. 2017;13:955
25. Shadrin IY, Allen BW, Qian Y, Jackman CP, Carlson AL, Juhas ME. et al. Cardiopatch platform enables maturation and scale-up of human pluripotent stem cell-derived engineered heart tissues. Nat Commun. 2017;8:1825
26. Mills RJ, Parker BL, Quaife-Ryan GA, Voges HK, Needham EJ, Bornot A. et al. Drug Screening in Human PSC-Cardiac Organoids Identifies Pro-proliferative Compounds Acting via the Mevalonate Pathway. Cell Stem Cell. 2019;24:895-907 e6
27. Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernandez-Mateos J, Khan K. et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920-6
28. Voges HK, Mills RJ, Elliott DA, Parton RG, Porrello ER, Hudson JE. Development of a human cardiac organoid injury model reveals innate regenerative potential. Development. 2017;144:1118-27
29. Giacomelli E, Bellin M, Sala L, van Meer BJ, Tertoolen LG, Orlova VV. et al. Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development. 2017;144:1008-17
30. Uzun AU, Mannhardt I, Breckwoldt K, Horvath A, Johannsen SS, Hansen A. et al. Ca(2+)-Currents in Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes Effects of Two Different Culture Conditions. Front Pharmacol. 2016;7:300
31. Mannhardt I, Breckwoldt K, Letuffe-Breniere D, Schaaf S, Schulz H, Neuber C. et al. Human Engineered Heart Tissue: Analysis of Contractile Force. Stem Cell Reports. 2016;7:29-42
32. Goldfracht I, Protze S, Shiti A, Setter N, Gruber A, Shaheen N. et al. Generating ring-shaped engineered heart tissues from ventricular and atrial human pluripotent stem cell-derived cardiomyocytes. Nat Commun. 2020;11:75
33. Giacomelli E, Meraviglia V, Campostrini G, Cochrane A, Cao X, van Helden RWJ. et al. Human-iPSC-Derived Cardiac Stromal Cells Enhance Maturation in 3D Cardiac Microtissues and Reveal Non-cardiomyocyte Contributions to Heart Disease. Cell Stem Cell. 2020;26:862-79 e11
34. Varzideh F, Mahmoudi E, Pahlavan S. Coculture with noncardiac cells promoted maturation of human stem cell-derived cardiomyocyte microtissues. J Cell Biochem. 2019;120:16681-91
35. Yoshida S, Miyagawa S, Fukushima S, Kawamura T, Kashiyama N, Ohashi F. et al. Maturation of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes by Soluble Factors from Human Mesenchymal Stem Cells. Mol Ther. 2018;26:2681-95
36. Lundy SD, Zhu WZ, Regnier M, Laflamme MA. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells Dev. 2013;22:1991-2002
37. Kamakura T, Makiyama T, Sasaki K, Yoshida Y, Wuriyanghai Y, Chen J. et al. Ultrastructural maturation of human-induced pluripotent stem cell-derived cardiomyocytes in a long-term culture. Circ J. 2013;77:1307-14
38. Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song L, Sirabella D. et al. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature. 2018;556:239-43
39. Tiburcy M, Hudson JE, Balfanz P, Schlick S, Meyer T, Chang Liao ML. et al. Defined Engineered Human Myocardium With Advanced Maturation for Applications in Heart Failure Modeling and Repair. Circulation. 2017;135:1832-47
40. Tulloch NL, Muskheli V, Razumova MV, Korte FS, Regnier M, Hauch KD. et al. Growth of Engineered Human Myocardium With Mechanical Loading and Vascular Coculture. Circulation Research. 2011;109:47-U195
41. Abilez OJ, Tzatzalos E, Yang HX, Zhao MT, Jung GH, Zollner AM. et al. Passive Stretch Induces Structural and Functional Maturation of Engineered Heart Muscle as Predicted by Computational Modeling. Stem Cells. 2018;36:265-77
42. Yang X, Rodriguez ML, Leonard A, Sun L, Fischer KA, Wang Y. et al. Fatty Acids Enhance the Maturation of Cardiomyocytes Derived from Human Pluripotent Stem Cells. Stem Cell Reports. 2019;13:657-68
43. Correia C, Koshkin A, Duarte P, Hu D, Teixeira A, Domian I. et al. Distinct carbon sources affect structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Sci Rep. 2017;7:8590
44. Yang X, Rodriguez M, Pabon L, Fischer KA, Reinecke H, Regnier M. et al. Tri-iodo-l-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells. J Mol Cell Cardiol. 2014;72:296-304
45. Birket MJ, Ribeiro MC, Kosmidis G, Ward D, Leitoguinho AR, van de Pol V. et al. Contractile Defect Caused by Mutation in MYBPC3 Revealed under Conditions Optimized for Human PSC-Cardiomyocyte Function. Cell Rep. 2015;13:733-45
46. Zhao Y, Rafatian N, Feric NT, Cox BJ, Aschar-Sobbi R, Wang EY. et al. A Platform for Generation of Chamber-Specific Cardiac Tissues and Disease Modeling. Cell. 2019;176:913-27 e18
47. Forsythe SD, Devarasetty M, Shupe T, Bishop C, Atala A, Soker S. et al. Environmental Toxin Screening Using Human-Derived 3D Bioengineered Liver and Cardiac Organoids. Front Public Health. 2018 6
48. Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125
49. Huebsch N, Loskill P, Deveshwar N, Spencer CI, Judge LM, Mandegar MA. et al. Miniaturized iPS-Cell-Derived Cardiac Muscles for Physiologically Relevant Drug Response Analyses. Sci Rep. 2016;6:24726
50. Keung W, Chan PKW, Backeris PC, Lee EK, Wong N, Wong AOT. et al. Human Cardiac Ventricular-Like Organoid Chambers and Tissue Strips From Pluripotent Stem Cells as a Two-Tiered Assay for Inotropic Responses. Clin Pharmacol Ther. 2019;106:402-14
51. Schulze ML, Lemoine MD, Fischer AW, Scherschel K, David R, Riecken K. et al. Dissecting hiPSC-CM pacemaker function in a cardiac organoid model. Biomaterials. 2019;206:133-45
52. Lee EK, Tran DD, Keung W, Chan P, Wong G, Chan CW. et al. Machine Learning of Human Pluripotent Stem Cell-Derived Engineered Cardiac Tissue Contractility for Automated Drug Classification. Stem Cell Reports. 2017;9:1560-72
53. Iyer RK, Odedra D, Chiu LL, Vunjak-Novakovic G, Radisic M. Vascular endothelial growth factor secretion by nonmyocytes modulates Connexin-43 levels in cardiac organoids. Tissue Eng Part A. 2012;18:1771-83
54. Nakane T, Masumoto H, Tinney JP, Yuan F, Kowalski WJ, Ye F. et al. Impact of Cell Composition and Geometry on Human Induced Pluripotent Stem Cells-Derived Engineered Cardiac Tissue. Sci Rep. 2017;7:45641
55. Amano Y, Nishiguchi A, Matsusaki M, Iseoka H, Miyagawa S, Sawa Y. et al. Development of vascularized iPSC derived 3D-cardiomyocyte tissues by filtration Layer-by-Layer technique and their application for pharmaceutical assays. Acta Biomater. 2016;33:110-21
56. Lind JU, Busbee TA, Valentine AD, Pasqualini FS, Yuan H, Yadid M. et al. Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat Mater. 2017;16:303-8
57. Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A. et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med. 2014;20:616-23
58. Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Mousavi Shaegh SA. et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc Natl Acad Sci U S A. 2017;114:E2293-E302
59. Mathur A, Loskill P, Shao K, Huebsch N, Hong S, Marcus SG. et al. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci Rep. 2015;5:8883
60. Forsythe SD, Devarasetty M, Shupe T, Bishop C, Atala A, Soker S. et al. Environmental Toxin Screening Using Human-Derived 3D Bioengineered Liver and Cardiac Organoids. Front Public Health. 2018;6:103
61. Arai K, Murata D, Verissimo AR, Mukae Y, Itoh M, Nakamura A. et al. Fabrication of scaffold-free tubular cardiac constructs using a Bio-3D printer. PLoS One. 2018;13:e0209162
62. Richards DJ, Li Y, Kerr CM, Yao J, Beeson GC, Coyle RC. et al. Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity. Nat Biomed Eng. 2020;4:446-62
63. Archer CR, Sargeant R, Basak J, Pilling J, Barnes JR, Pointon A. Characterization and Validation of a Human 3D Cardiac Microtissue for the Assessment of Changes in Cardiac Pathology. Sci Rep. 2018;8:10160
64. Richards DJ, Coyle RC, Tan Y, Jia J, Wong K, Toomer K. et al. Inspiration from heart development: Biomimetic development of functional human cardiac organoids. Biomaterials. 2017;142:112-23
65. Devarasetty M, Forsythe S, Shupe TD, Soker S, Bishop CE, Atala A. et al. Optical Tracking and Digital Quantification of Beating Behavior in Bioengineered Human Cardiac Organoids. Biosensors (Basel). 2017 7
66. MacQueen LA, Sheehy SP, Chantre CO, Zimmerman JF, Pasqualini FS, Liu X. et al. A tissue-engineered scale model of the heart ventricle. Nat Biomed Eng. 2018;2:930-41
67. Li RA, Keung W, Cashman TJ, Backeris PC, Johnson BV, Bardot ES. et al. Bioengineering an electro-mechanically functional miniature ventricular heart chamber from human pluripotent stem cells. Biomaterials. 2018;163:116-27
68. Tsuruyama S, Matsuura K, Sakaguchi K, Shimizu T. Pulsatile tubular cardiac tissues fabricated by wrapping human iPS cells-derived cardiomyocyte sheets. Regen Ther. 2019;11:297-305
69. Robertson C, Tran DD, George SC. Concise review: maturation phases of human pluripotent stem cell-derived cardiomyocytes. Stem Cells. 2013;31:829-37
70. Yang X, Pabon L, Murry CE. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Res. 2014;114:511-23
71. Itzhaki I, Rapoport S, Huber I, Mizrahi I, Zwi-Dantsis L, Arbel G. et al. Calcium Handling in Human Induced Pluripotent Stem Cell Derived Cardiomyocytes. Plos One. 2011 6
72. Shimizu T, Yamato M, Isoi Y, Akutsu T, Setomaru T, Abe K. et al. Fabrication of pulsatile cardiac tissue grafts using a novel 3-dimensional cell sheet manipulation technique and temperature-responsive cell culture surfaces. Circ Res. 2002;90:e40
73. Sekine H, Shimizu T, Sakaguchi K, Dobashi I, Wada M, Yamato M. et al. In vitro fabrication of functional three-dimensional tissues with perfusable blood vessels. Nat Commun. 2013;4:1399
74. Zuppinger C. 3D Cardiac Cell Culture: A Critical Review of Current Technologies and Applications. Front Cardiovasc Med. 2019;6:87
75. Burnham MP, Harvey R, Sargeant R, Fertig N, Haddrick M. A Scalable Approach Reveals Functional Responses of iPSC Cardiomyocyte 3D Spheroids. SLAS Discov. 2021;26:352-63
76. Bartosh TJ, Ylostalo JH, Mohammadipoor A, Bazhanov N, Coble K, Claypool K. et al. Aggregation of human mesenchymal stromal cells (MSCs) into 3D spheroids enhances their antiinflammatory properties. Proc Natl Acad Sci U S A. 2010;107:13724-9
77. Lee P, Klos M, Bollensdorff C, Hou L, Ewart P, Kamp TJ. et al. Simultaneous voltage and calcium mapping of genetically purified human induced pluripotent stem cell-derived cardiac myocyte monolayers. Circ Res. 2012;110:1556-63
78. Vunjak-Novakovic G, Bhatia S, Chen C, Hirschi K. HeLiVa platform: integrated heart-liver-vascular systems for drug testing in human health and disease. Stem Cell Res Ther. 2013;4(Suppl 1):S8
79. van Meer BJ, de Vries H, Firth KSA, van Weerd J, Tertoolen LGJ, Karperien HBJ. et al. Small molecule absorption by PDMS in the context of drug response bioassays. Biochem Biophys Res Commun. 2017;482:323-8
80. Shirure VS, George SC. Design considerations to minimize the impact of drug absorption in polymer-based organ-on-a-chip platforms. Lab Chip. 2017;17:681-90
81. Walker DM, Boey G, McDonald LA. The pathology of oral cancer. Pathology. 2003;35:376-83
82. Loessner D, Stok KS, Lutolf MP, Hutmacher DW, Clements JA, Rizzi SC. Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials. 2010;31:8494-506
83. Uchida Y, Tanaka S, Aihara A, Adikrisna R, Yoshitake K, Matsumura S. et al. Analogy between sphere forming ability and stemness of human hepatoma cells. Oncol Rep. 2010;24:1147-51
84. Lemme M, Ulmer BM, Lemoine MD, Zech ATL, Flenner F, Ravens U. et al. Atrial-like Engineered Heart Tissue: An In vitro Model of the Human Atrium. Stem Cell Reports. 2018;11:1378-90
85. Drawnel FM, Boccardo S, Prummer M, Delobel F, Graff A, Weber M. et al. Disease modeling and phenotypic drug screening for diabetic cardiomyopathy using human induced pluripotent stem cells. Cell Rep. 2014;9:810-21
86. McKeithan WL, Savchenko A, Yu MS, Cerignoli F, Bruyneel AAN, Price JH. et al. An Automated Platform for Assessment of Congenital and Drug-Induced Arrhythmia with hiPSC-Derived Cardiomyocytes. Front Physiol. 2017;8:766
87. Sharma A, Burridge PW, McKeithan WL, Serrano R, Shukla P, Sayed N. et al. High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Sci Transl Med. 2017 9
88. Fairclough RJ, Wood MJ, Davies KE. Therapy for Duchenne muscular dystrophy: renewed optimism from genetic approaches. Nat Rev Genet. 2013;14:373-8
89. Long C, Li H, Tiburcy M, Rodriguez-Caycedo C, Kyrychenko V, Zhou H. et al. Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci Adv. 2018;4:eaap9004
90. Arad M, Seidman JG, Seidman CE. Phenotypic diversity in hypertrophic cardiomyopathy. Hum Mol Genet. 2002;11:2499-506
91. Cashman TJ, Josowitz R, Johnson BV, Gelb BD, Costa KD. Human Engineered Cardiac Tissues Created Using Induced Pluripotent Stem Cells Reveal Functional Characteristics of BRAF-Mediated Hypertrophic Cardiomyopathy. Plos One. 2016;11:e0146697
92. Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S. et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949-54
93. Dhomen N, Marais R. New insight into BRAF mutations in cancer. Curr Opin Genet Dev. 2007;17:31-9
94. Ovchinnikova E, Hoes M, Ustyantsev K, Bomer N, de Jong TV, van der Mei H. et al. Modeling Human Cardiac Hypertrophy in Stem Cell-Derived Cardiomyocytes. Stem Cell Reports. 2018;10:794-807
95. Gollob MH, Green MS, Tang AS, Gollob T, Karibe A, Ali Hassan AS. et al. Identification of a gene responsible for familial Wolff-Parkinson-White syndrome. N Engl J Med. 2001;344:1823-31
96. Hinson JT, Chopra A, Lowe A, Sheng CC, Gupta RM, Kuppusamy R. et al. Integrative Analysis of PRKAG2 Cardiomyopathy iPS and Microtissue Models Identifies AMPK as a Regulator of Metabolism, Survival, and Fibrosis. Cell Rep. 2016;17:3292-304
97. Shinnawi R, Shaheen N, Huber I, Shiti A, Arbel G, Gepstein A. et al. Modeling Reentry in the Short QT Syndrome With Human-Induced Pluripotent Stem Cell-Derived Cardiac Cell Sheets. J Am Coll Cardiol. 2019;73:2310-24
98. Haubner BJ, Schneider J, Schweigmann U, Schuetz T, Dichtl W, Velik-Salchner C. et al. Functional Recovery of a Human Neonatal Heart After Severe Myocardial Infarction. Circ Res. 2016;118:216-21
99. Stevens KR, Kreutziger KL, Dupras SK, Korte FS, Regnier M, Muskheli V. et al. Physiological function and transplantation of scaffold-free and vascularized human cardiac muscle tissue. Proc Natl Acad Sci U S A. 2009;106:16568-73
100. Thavandiran N, Dubois N, Mikryukov A, Masse S, Beca B, Simmons CA. et al. Design and formulation of functional pluripotent stem cell-derived cardiac microtissues. Proc Natl Acad Sci U S A. 2013;110:E4698-707
101. Lemme M, Braren I, Prondzynski M, Aksehirlioglu B, Ulmer BM, Schulze ML. et al. Chronic intermittent tachypacing by an optogenetic approach induces arrhythmia vulnerability in human engineered heart tissue. Cardiovasc Res. 2020;116:1487-99
102. Schaaf S, Shibamiya A, Mewe M, Eder A, Stohr A, Hirt MN. et al. Human Engineered Heart Tissue as a Versatile Tool in Basic Research and Preclinical Toxicology. Plos One. 2011 6
103. Mannhardt I, Saleem U, Mosqueira D, Loos MF, Ulmer BM, Lemoine MD. et al. Comparison of 10 Control hPSC Lines for Drug Screening in an Engineered Heart Tissue Format. Stem Cell Reports. 2020;15:983-98
104. Lesman A, Habib M, Caspi O, Gepstein A, Arbel G, Levenberg S. et al. Transplantation of a tissue-engineered human vascularized cardiac muscle. Tissue Eng Part A. 2010;16:115-25
105. Gao L, Gregorich ZR, Zhu W, Mattapally S, Oduk Y, Lou X. et al. Large Cardiac Muscle Patches Engineered From Human Induced-Pluripotent Stem Cell-Derived Cardiac Cells Improve Recovery From Myocardial Infarction in Swine. Circulation. 2018;137:1712-30
106. Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K. et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110:1465-73
107. Khait L, Birla RK. Cell-based cardiac pumps and tissue-engineered ventricles. Regen Med. 2007;2:391-406
108. Sakalem ME, De Sibio MT, da Costa FAD, de Oliveira M. Historical evolution of spheroids and organoids, and possibilities of use in life sciences and medicine. Biotechnol J. 2021 16
109. Gunti S, Hoke ATK, Vu KP, London NR Jr. Organoid and Spheroid Tumor Models: Techniques and Applications. Cancers (Basel). 2021 13
110. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262-U147
111. Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME. et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373 -+
112. Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat Cell Biol. 2016;18:246-54
113. Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364:952-5
114. Lancaster MA, Huch M. Disease modelling in human organoids. Dis Model Mech. 2019 12
115. Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M. et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519-32
116. Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME. et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373-9
117. Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S. et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51-6
118. Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K. et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771-85
119. Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K. et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105-9
120. Taguchi A, Kaku Y, Ohmori T, Sharmin S, Ogawa M, Sasaki H. et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell. 2014;14:53-67
121. Xia Y, Nivet E, Sancho-Martinez I, Gallegos T, Suzuki K, Okamura D. et al. Directed differentiation of human pluripotent cells to ureteric bud kidney progenitor-like cells. Nat Cell Biol. 2013;15:1507-15
122. Takasato M, Er PX, Becroft M, Vanslambrouck JM, Stanley EG, Elefanty AG. et al. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat Cell Biol. 2014;16:118-26
123. Huch M, Dorrell C, Boj SF, van Es JH, Li VS, van de Wetering M. et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247-50
124. D. A. Fischman AAM. Cardiac Hypertrophy, N. K. Alpert, Ed.. Academic Press, New York. 1971: pp. 125-39.
125. Halbert SP, Bruderer R, Lin TM. In vitro organization of dissociated rat cardiac cells into beating three-dimensional structures. J Exp Med. 1971;133:677-95
126. Hofbauer P, Jahnel SM, Papai N, Giesshammer M, Deyett A, Schmidt C. et al. Cardioids reveal self-organizing principles of human cardiogenesis. Cell. 2021;184:3299-317 e22
127. Schutgens F, Clevers H. Human Organoids: Tools for Understanding Biology and Treating Diseases. Annu Rev Pathol. 2020;15:211-34
128. Drakhlis L, Biswanath S, Farr CM, Lupanow V, Teske J, Ritzenhoff K. et al. Human heart-forming organoids recapitulate early heart and foregut development. Nat Biotechnol. 2021;39:737-46
129. Lewis-Israeli YR, Wasserman AH, Gabalski MA, Volmert BD, Ming Y, Ball KA. et al. Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease. Nat Commun. 2021;12:5142
130. Song MH, Choi SC, Noh JM, Joo HJ, Park CY, Cha JJ. et al. LEFTY-PITX2 signaling pathway is critical for generation of mature and ventricular cardiac organoids in human pluripotent stem cell-derived cardiac mesoderm cells. Biomaterials. 2021;278:121133
131. Silva AC, Matthys OB, Joy DA, Kauss MA, Natarajan V, Lai MH. et al. Co-emergence of cardiac and gut tissues promotes cardiomyocyte maturation within human iPSC-derived organoids. Cell Stem Cell. 2021;28:2137-52 e6
132. Murry CE, Reinecke H, Pabon LM. Regeneration gaps: observations on stem cells and cardiac repair. J Am Coll Cardiol. 2006;47:1777-85
133. Ye L, Chang YH, Xiong Q, Zhang P, Zhang L, Somasundaram P. et al. Cardiac repair in a porcine model of acute myocardial infarction with human induced pluripotent stem cell-derived cardiovascular cells. Cell Stem Cell. 2014;15:750-61
134. Lind JU, Busbee TA, Valentine AD, Pasqualini FS, Yuan HY, Yadid M. et al. Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nature Materials. 2017;16:303 -+
Author contact
![]() Corresponding author: Young-sup Yoon, MD, PhD, Department of Medicine, Division of Cardiology, Emory University School of Medicine, Atlanta, GA 30322, USA. E-mail: yyoon5edu (Y.-s. Y.)
Corresponding author: Young-sup Yoon, MD, PhD, Department of Medicine, Division of Cardiology, Emory University School of Medicine, Atlanta, GA 30322, USA. E-mail: yyoon5edu (Y.-s. Y.)
 Global reach, higher impact
Global reach, higher impact