13.3
Impact Factor
Theranostics 2022; 12(7):3474-3487. doi:10.7150/thno.72028 This issue Cite
Research Paper
Acidic microenvironment triggered in situ assembly of activatable three-arm aptamer nanoclaw for contrast-enhanced imaging and tumor growth inhibition in vivo
State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Biology, College of Chemistry and Chemical Engineering, Hunan University, Key Laboratory for Bio-Nanotechnology and Molecular Engineering of Hunan Province, Changsha 410082, China.
Received 2022-2-14; Accepted 2022-4-4; Published 2022-4-24
Abstract
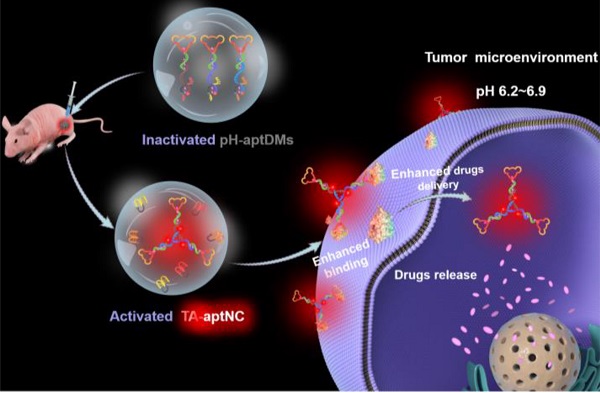
Rationale: Static assembled multivalent DNA nanotheranostics system have encountered some bottleneck problems in cancer imaging and therapy, such as poor penetration and high immunogenicity. Herein, we proposed an acidic tumor microenvironment triggered assembly of activatable multivalent nanodevice, called “three-arm aptamer nanoclaw” (TA-aptNC), assembled from three pH-responsive aptamer-decorated DNA monomers (pH-aptDMs) to facilitate their functions of imaging and therapy.
Methods: The activated TA-aptNC was constructed by acidic microenvironment triggered in situ assembly of three pH-aptDMs. Designer pH-aptDM was established based on the combination of a pH-responsive i-motif switch and an assembly module with a cell membrane anchoring aptamer ligand. Acidic microenvironment-triggered the assembly of the TA-aptNC was characterized by electrophoresis and atomic force microscopic (AFM). The binding affinity and stability of the TA-aptNC, comparing the monovalent pH-aptDM, were studied via the flow cytometry and nuclease resistance assays. Acidic microenvironment-activated contrast-enhanced tumor imaging and significantly antitumor efficiency were evaluated in vitro and in vivo.
Results: At physiological pH environment, the pH-aptDMs with excellent tissue permeability exited as inactivated and monodispersed small monomer. When encountering acidic microenvironment at the tumor site, pH-responsive i-motif switch liberated from the pH-aptDMs, and the three unconstrained DNA modules (DM1, DM2 and DM3) subsequently assembled in situ into the TA-aptNC. Compared with monovalent pH-aptDMs, the spontaneously formed activatable TA-aptNC afforded 2-fold enhanced binding ability via the multivalent effect, which further facilitated the selective tumor cell uptake capability, thus enabling a contrast-enhanced tumor imaging and significantly antitumor efficiency in vivo without systemic toxicity.
Conclusions: The proposed strategy offers valuable insight into excavating an endogenous stimuli-triggered assembly of multivalent nanodevice for accurate diagnosis and efficient tumor therapy.
Keywords: in situ assembly of multivalent nanodevice, in situ enhanced binding affinity, acidic tumor microenvironment, contrast-enhanced tumor imaging, tumor growth inhibition
Introduction
Aptamers termed as 'chemical antibodies', are short oligonucleotide sequences with a folded three-dimensional structure, which can selectively bind to specific small molecules, proteins, or the whole cells [1,2]. Owing to the unique advantages of high selectivity, specificity, easy synthesis and modification, aptamers promise an emerging molecule tool for applications in tumor detection [3,4], imaging [5-7], and therapy [8-10]. As powerful recognition ligands, aptamers have been extensively explored to be labelled with other units (isotopic indicators, radionuclides, fluorescent dyes, and drug molecules) or assemble into aptamer-decorated DNA nanostructures, generating monovalent aptamer probes [9,11]. Despite monovalent aptamer probes showing the potential applications in disease diagnosis and therapy, the unsatisfactory affinity and poor stability in the complex physiological environment inevitably restrict their widespread applications in vivo [6,12].
To meet these requirements, multivalent aptamer probes, by integrating multiple aptamer units into one nanomaterial or DNA nanostructure, have been employed to enhance the binding affinity via the multivalent effect. It has been shown the multivalent aptamer probes strategy increased local ligand density or synergistic interaction [13-15]. In particular, benefiting from the great progress of DNA nanotechnology, series of DNA nanostructures including DNA tetrahedron [10,16,17], DNA nanowire [18], DNA nanohydrogel [19,20], RCA-based DNA nanostructure [21,22], etc., have been exploited and used as scaffolds to incorporate multivalent aptamers. Despite the progress has made, the preassembled DNA nanostructures, also known as “static assembled multivalent nanodevices”, often possess the characteristic of large size, which makes them difficult to penetrate into the entire surface-to-core tumor tissues and prone to be cleared by the reticuloendothelial system in vivo [23,24]. Besides, the multiple aptamers are embedded in various sites of DNA scaffold by an imprecise controlled manner, which inevitably results in higher steric hindrance, thus affording undesired improvement of binding affinity [21,25,26].
Recently, the concept of in situ assembly in specific site has been proposed for cancer imaging and therapy. In situ self-assembly is a process that exogenous small monomers integrating stimulus-responsive primitives and assembly units self-assemble into specific architecture by the stimulation of the pathological abnormalities [27]. Endogenous stimulus-triggered in situ assembly strategy shows some prominent merits: first, they can not only transform the morphology of functional nanostructures, but also facilitate their functions of imaging or therapy [28-30]. Second, this strategy imitates naturally occurring process at physiological environment, which is essential to understand the mechanism of disease occurrence and guide the therapy precisely [31]. Currently, the distinctive tumor microenvironment (TME) of solid tumors, such as acidic extracellular pH, overexpressed proteins/enzymes and hypoxia [32,33], as potential endogenous triggers of in situ assembly methods provides an alternative opportunity to spatiotemporally manipulate biological behaviors at special site. Especially, acidic tumor microenvironment as ubiquitous stimulus induces the conformation transition of pH-responsive units (i-motif, triplex DNA, pH-sensitive amphiphilic polymers and so on) for realizing precise assembly/disassembly [34-36], promising potential applications in construction of in situ assembled multivalent nanodevice for modulation of binding affinity and cellular internalization behavior at tumor site. Thus motivated, our group first reported extracellular pH-powered in situ assembly of multivalent DNA assemblies by the crosslink of aptamer functional DNA monomer on the cell surface, which achieved enhanced binding affinity and drugs delivery [37]. Nevertheless, this strategy employed an “always-on” mode for cancer diagnosis and therapeutic, leading to a long diagnosis time and poor imaging contrast. Subsequently, we continuously developed acid-responsive assembly of multivalent nanotheranostics systems based on activatable strategy and successfully used to realize contrast-enhanced imaging of cancer cells. One is acid-activated reconfiguration of DNAzyme-based multivalent theranostics nanodevice for cancer cells imaging and gene therapy [38]. The other is pH-driven in situ constructed of activatable multivalent theranostics platform for intracellular TK1 mRNA activated cancer cells imaging and killing [39]. Essentially, these encouraging attempts at stimuli-responsive assembly of activatable multivalent nanotheranostics system demonstrated their superiority of contrast-enhanced imaging, enhanced cell killing and lower normal tissue toxicity. However, both reports are inevitably limited by the undefined aptamer density, imprecise controlled spatial orientation and physical dimension, thereby leading to the undesirable improvement of binding. Additionally, these above-mentioned in situ assemblies of multivalent nanotheranostics systems were initiated in a simulated solution. Therefore, it is desirable to excogitate endogenous stimuli-responsive in situ assembly strategy to construct precisely controlled activatable multivalent nanodevice in vivo.
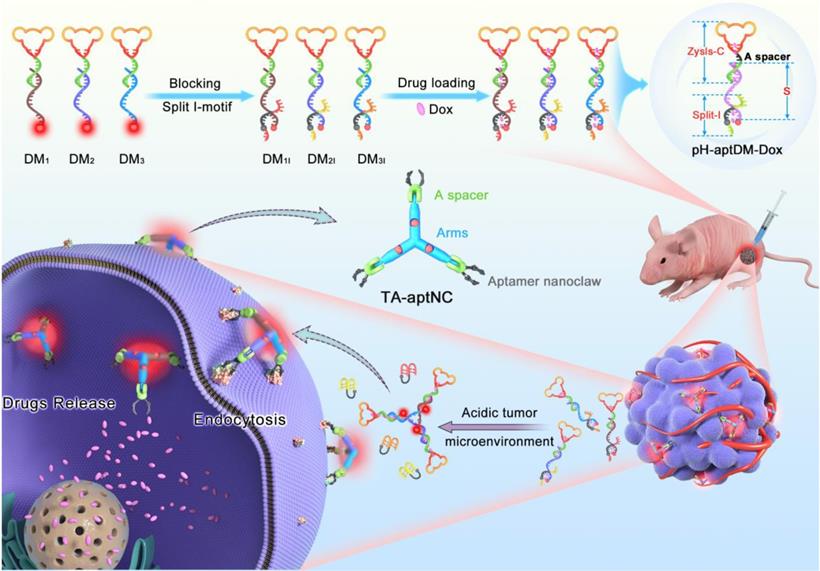
For cancer diagnosis and therapy, there is still a blank area to improve the imaging contrast and therapy using an in situ assembly multivalent nanotheranostics at tumor site. Based on our previous design, we first proposed an acidic tumor microenvironment-powered autonomous in situ assembly of activatable multivalent strategy by designing three pH-responsive and aptamer-decorated DNA monomers (pH-aptDMs). The resultant activated multivalent nanodevice, called “three-arm aptamer nanoclaw” (TA-aptNC), possessed the unique superiorities of the precise controlled spatial orientation, defined ligand density and structure size, which is crucial for the improvement of binding avidity and cellular internalization efficiency. As illustrated in Scheme 1, three DNA modules (DM1, DM2 or DM3) were designed to be partly complementary to each other. Each of them has a sticky-end that was respectively locked by the split i-motif sequence (I1, I2 or I3), generating pH-responsive and aptamer-decorated DNA monomers (pH-aptDMs, including DM1I, DM2I or DM3I). Doxorubicin, a model anticancer drug, embed into the abundant CG base pairs of pH-aptDMs. At physiological condition (pH 7.2-7.4), three pH-aptDMs existed as monodispersed structure, showing excellent tissue permeability and inactivated fluorescence due to the fluorophore in close proximity to the quencher. When encountering the acidic tumor microenvironment (pH 6.2-6.9), the protonated split i-motif (I1, I2 and I3) could form quadruplex structures and then liberate from DNA monomers, thus resulting in fluorescence recovery and partial drugs release. Three DNA modules with the unconstrained sticky ends would then be assembled with each other to form the TA-aptNC via sticky-end hybridization at tumor site. Three aptamers respectively positioned on the three outstretched arms of TA-aptNC, as “recognition claws”, were able to grasp the receptor on the cell membrane. The resultant multivalent effect could accelerate the internalization of the TA-aptNCs into target tumor cell via receptor-mediated internalization pathways, thus improving the drug delivery and greatly inhibiting tumor growth in vivo.
Schematic illustration of acidic tumor microenvironment-triggered in situ assembly of activatable multivalent nanodevice (TA-aptNC) for contrast-enhanced imaging and antitumor growth. In the free state, three pH-responsive aptamer-decorated DNA monomer (pH-aptDMs, including DM1I, DM2I and DM3I) exited as monodispersed small monomer with quenched fluorescence, showing excellent tissue permeability. When the mixture of three pH-aptDMs was administered into SMMC-7721 tumor-bearing nude mice via an intratumoral injection, the mixture responses to the acidic tumor microenvironment (pH 6.2-6.9), leading to the assembly of activatable TA-aptNC. The TA-aptNC with in situ improved binding affinity specifically targeted tumor cell, which further facilitates rapid drug delivery, thereby showing a contrast-enhanced in vivo tumor imaging and a significant antitumor growth efficiency.
As far as we know, this is the first report of endogenous stimulus-powered in situ assembly of precisely controlled multivalent aptamer nanodevice for facilitating their imaging and therapy functions in vivo. Irrespective of the introduction of any exogenous additives, the TA-aptNC-based in situ assembly system not only allowed acidic tumor microenvironment-activated contrast-enhanced in vivo imaging and selective cytotoxicity to target tumor cells, but also afforded significantly antitumor efficiency in vivo. As a modularly molecular design concept, targeting module, stimuli- responsive module and assembling module were intelligently integrated to form a multifunctional pH-aptNC. Furthermore, the striking features of DNA-based multivalent system are biocompatible, biodegradable and no self-toxicity.
Materials and Methods
Preparation of three monomer
The pH-aptDMs (DM1I, DM2I and DM3I) were prepared via an annealing protocol. In the experiment, the DM1I monomer was assembled from three oligonucleotide strands (ZYsls-C, S1 and I1) with a molar ratio of 1:1:1.5 in 10 mM PBS (pH 7.4). Then, the mixture was annealed by using enzyme-free self-assembled procedure: 95 °C for 5 min, 70 °C for 10 min, 60 °C for 10 min, 50 °C for 15 min, 40 °C for 20 min, 30 °C for 30 min and 20 °C for 30 min. Similarly, the self-assembly of DM2I (ZYsls-C, S2 and I2), DM3I (ZYsls-C, S3 and I3) and control probes were synthesized by following the same methods. A slightly excessive blocking sequence (split i-motif, including I1, I2 and I3) was respectively added to sufficiently locking the sticky ends of pH-aptDMs. The formation of pH-aptDMs was finally verified by native Gel Electrophoresis.
Gel electrophoresis analysis
All probes were dissolved in buffer (1 μM), respectively. The stepwise assembly of DM1I, DM2I and DM3I in PBS (pH 7.4) was estimated by 3% agarose gel electrophoresis. To characterization of acidic pH-triggered in situ assembly of three pH-aptDMs, the mixture of DM1I, DM2I and DM3I (1 μM) were incubated in PBS (pH 7.4 or 6.3) at 37 °C for 1 h. Gel electrophoresis analysis were performed in 1× TBE buffer at 4 °C for 1 h under a constant potential of 100 V. For imaging, the gel was then stained in SYBR Gold for 30 min, finally photographed by an Azure C600 Imaging Biosystems (CA).
Flow cytometry assays
To evaluate the acidic pH-powered in situ assembly multivalent system for fluorescent labeling of target cells, equal DM1I, DM2I and DM3I (15 nM) were co-incubated with 1.5 × 105 target tumor cells in binding buffer (pH 7.4 or 6.3) at 37 °C for 1 h. Subsequently, it was determined by a Gallios cytometer (Beckman Coulter, USA) by counting 10,000 events.
To evaluating the binding capacity of TA-aptNC-based in situ assembly strategy at cellular level, different probes were reasonably designed, including TA-CaptNC (the mixture of CDM1I, CDM2I and CDM3I, aptamer sequence replaced by arbitrary sequences), pH-C (the mixture of DM1C, DM2C and DM3C, split i-motif sequence substituted by pH-insensitive sequence), pH-aptDM (the corresponding monovalent probe) and TA-aptNC (the mixture of DM1I, DM2I and DM3I). These probes were respectively treated with the target cells and then determined by a Gallios cytometer (Beckman Coulter, USA).
Laser Scanning Confocal Microscopy imaging
To investigate the acidic tumor microenvironment-triggered fluorescent labeling of TA-aptNC-based strategy, cells were firstly plated in confocal dish and incubated at 37 °C in 5% CO2 for 24 h, then washed three times using the binding buffer before incubation. After SMMC-7721 cells treated with different composition (pH-C, pH-aptDM, TA-aptNC) in binding buffer at pH 6.3 or 7.4 for 1 h in the dark. Then, the cells were washed with PBS 3 times and stained with Hoechst 33342 before observation. Samples were monitored using microscope (Nikon, Japan) with a 100× objective. Note that the concentrations of these probes were regulated to have the same amount of Cy5 and aptamer.
To investigate the specifical binding of in situ assembled TA-aptNC to target SMMC-7721 cells, the mixture of three pH-aptDMs were co-incubated with different cells (target SMMC-7721, HeLa, HepG2, MCF-7 and L02 cells) in different pH at 37 °C for 1 h. After incubation, all samples were washed 2 times and then resuspended in PBS for microscopic observation.
Intracellular delivery of Dox
The localization of Dox and Cy5-TA-aptNC was studied by incubating SMMC-7721 cells with different compositions. Cy5-pH-aptDM or Cy5-TA-aptNC and Dox (mole rations of 1:5) were firstly mixed to prepare Cy5-pH-aptDM-Dox and Cy5-TA-aptNC-Dox as described above. subsequently, different compositions, including free Dox, Cy5-TA-aptNC-Dox (the mixture of Cy5- DM1I-Dox, Cy5- DM2I-Dox and Cy5- DM3I-Dox), Cy5-pH-aptDM-Dox, Cy5-TA-aptNC (the mixture of Cy5- DM1I, Cy5- DM2I and Cy5- DM3I), or Cy5-pH-aptDM, were respectively incubated with SMMC-7721 cells at 37 °C in CO2 incubator for 1 h, then removing the medium. All samples were washed twice with PBS. Hoechst 33342 was added for specific staining of the nucleus for 10 min. The samples were washed with PBS twice, and then resuspended in binding buffer (1000 µL) for cellular fluorescent images by Nikon A1 confocal laser scanning microscopy with a 100× objective. Fluorescent imaging of Hoechst 33342 was detected using 405/430 nm excitation/emission filters. Dox was detected using 488/505 nm. Cy5 was detected using 633/660 nm.
Cytotoxicity assay
MTS assay was employed to evaluate cell viability. 0.2 mL SMMC-7721 cells were seeded in 96-well culture plates at a density of 1 × 104 cells per well for 2 h. Cells were then treated with the corresponding probes at different concentrations for 3 h. After 80% of the supernatant medium removed, 0.2 mL RPMI 1640 medium (12% FBS) was added for further cell growth until 48 h. Last, 20 μL of MTS reagent diluted in 100 μL of cell culture medium was added to incubate with the cells for 2 h at 37 °C. Finally, the absorbance of 490 nm was obtained by using a microplate reader. (Bio Tek Instrument, Inc.). Cell viability was calculated according to manufacturer's description.
In vivo fluorescence imaging
All animal studies were performed in compliance with institutional animal use and care regulations, according to protocol No. SYXK (Xiang) 2018-0006, approved by the Laboratory Animal Center of Hunan Province. Male BALB/c nude mice were purchased from Hunan SJA Laboratory Animal Co., Ltd. All mice used in this experiment are 4-6 weeks old at the start of each experiment and weighed 20-25 g. To establish the mouse xenograft tumor models, male nude mice were subcutaneously injected with 1 × 106 SMMC-7721 cells on the right back of the mouse. The SMMC-7721-implanted mice were then allowed to grow for 3-4 week until the tumor reached 1 cm in diameter. The mice were randomized into 3 groups (five mice each group) and administrated with 1.5 μΜ of pH-C, pH-aptDM and TA-aptNC into the tumor tissue by intratumoral injection and the corresponding normal muscle tissues. The real-time fluorescence images were monitored by an IVIS Lumina II in vivo imaging system (Caliper Life Science, U.S.A.) at 0, 1, 5, 15, 30, 45 and 60 min. The mice were anesthetized with anesthetic before imaging.
Tissue slice imaging
For tissue slice imaging, when the mice were deeply anesthetized, SMMC-7721 tumor-bearing mice were firstly sacrificed. The fresh tumor tissues and normal muscle tissues were excised, immediately embedded in OCT and frozen at -80 °C for several minutes. The frozen tissues were then cut into 8 μm slices using a frozen tissue slicer of CM1850 (Leica, Germany). Subsequently, the frozen tissue slices were quickly adsorbed on glass slides for the assessment of the acidic tumor microenvironment-triggered in situ assembly of three pH-aptDMs into TA-aptNC. 1.5 μM of the corresponding probes were respectively added to the central area of the tissue section. After incubation for 1 h at 37 °C, the slicer fluorescent images were collected on a Nikon confocal microscope with a 60× oil immersion objective.
In vivo tumor growth inhibition
Male BALB/c mice with SMMC-7721 tumor-bearing xenograft, when the tumor volume reached 100 mm3, were randomly allocated into three groups (five mice each group) and administered with PBS, TA-aptNC (the mixture of DM1I, DM2I and DM3I) and TA-aptNC-Dox (the mixture of DM1I-Dox, DM2I-Dox and DM3I-Dox) by intratumoral injection. Dox dose of 0.5 mg/kg-1 were injected every other day for a total of six injections. The same amount of PBS and the mixture of DM1I, DM2I and DM3I were treated with the control group, respectively. The changes of body weight and tumor volumes were monitored and recorded every other day for 14 days. The tumor volume was obtained as the following formula: tumor volume = Length × Width2/2 using a caliper. At the end of the relevant timeline (day 14), the animals were sacrificed by cervical dislocation under general anesthesia. The tumors were harvested for further histological analysis.
Results and Discussion
Fabrication and characterization of the pH-responsiveness of TA-aptNC-based in situ assembly system
In this strategy, the aptamer ZYsls, selected by our group, was used as model function domain against human liver cancer SMMC-7721 cells. To ensure that the whole system can operate as expected, the stepwise assembly of three pH-aptDMs was characterized by 3% agarose gel electrophoresis, showing an obvious band migration and high yield (Figure 1A). In this strategy, acidic pH as a trigger could turn on the pH-switch integrated into three pH-aptDMs, and then power unblocked DNA modules assembled into activated TA-aptNC. The feasibility of our strategy was explored by electrophoresis assays (Figure 1B). Lanes 1 and 2 represented pH-aptDM (DM2I) and TA-aptNC formed by annealing assembly, respectively. Due to the excellent stability of split i-motif sequences at pH 7.4, the mixture of three pH-aptDMs failed to assemble into TA-aptNC, resulting in unnoticeable band movement in lane 3. In comparison, there existed two bands with different gradients representing the activated TA-aptNC and the residual pH-aptDMs, demonstrating successful activation of the assembly of TA-aptNC at pH 6.3. The pH-triggered formation of the TA-aptNC was then characterized by atomic force microscopic (AFM). As expected, a series of three arms-like DNA nanostructures with an average size of ~15 × 2 nm were observed at pH 6.3, indicating pH-triggered the successful assembly of three pH-aptDMs into TA-aptNC (Figure S1). The pH-responsiveness of pH-aptDM was further characterized by circular dichroism (CD) spectroscopy. Compared with free ZYsls-C (Figure S2A), the pH-aptDM monomer and the mixture of three pH-aptDMs, similar to the free split i-motif, exhibited distinct red shift of both positive and negative bands at pH 6.3 owing to base stacking and polynucleotide helicity of unstructured single-stranded DNA (Figure 1C, S2B-C) [40,41]. The results indicated that acidic pH-driven conformational switch went from the random coil to intact i-motif successfully.
Characterization of pH-triggered in situ construction of TA-aptNC. (A) 3% agarose gel electrophoresis analysis of the stepwise assembly of DNA monomers. Lane L, 20 bp DNA ladder; Lane 1, ZYsls-C; Lane 2, ZYsls-C + S1 (DM1); Lane 3, ZYsls-C + S2 (DM2); Lane 4, ZYsls-C + S3 (DM3); Lane 5, ZYsls-C + S1 + I1 (DM1I); Lane 6, ZYsls-C + S2 + I2 (DM2I); Lane 7, ZYsls-C + S3 + I3 (DM3I). (B) 3% agarose gel electrophoresis verification of the formation of TA-aptNC. Lane L, 20 bp DNA ladder; Lane 1, DM2I; Lane 2, TA-aptNC (annealing assembly); Lane 3, DM1I + DM2I + DM3I at pH 7.4; Lane 4, DM1I + DM2I + DM3I at pH 6.3. The CD spectra (C) and fluorescence spectra (D) characterization of the mixture of three pH-aptDMs in the simulated buffer. Flow cytometry analysis of the target SMMC-7721 cells (E) after treated with the mixture under different pH values.
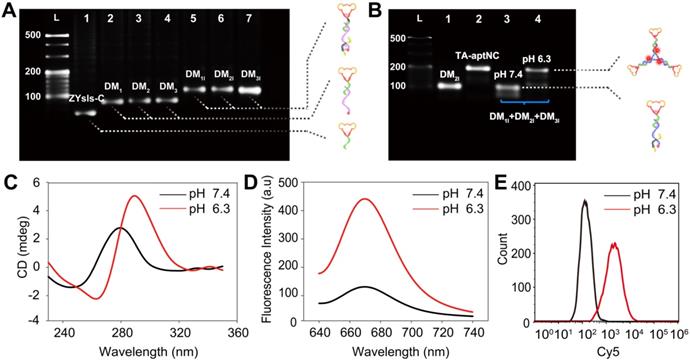
Furthermore, the fluorescence spectra analysis also revealed the pH-response performance of the proposed strategy by employing the pH-insensitive fluorophore/quencher pair (Cy5/BHQ2) (Figure S3). We first optimized the assembly ratio of BHQ2-I to Cy5-S in pH-aptDM (Figure S4). On the basis of the above optimized results, the pH-responsiveness of pH-aptDM was monitored. As shown in Figure 1D, the mixture of three pH-aptDMs could exist as monodispersed small monomer at pH 7.4 and gave a low background fluorescence signal. When adjusting the pH to 6.3, BHQ2-I disassembled from Cy5-S, resulting in a notable activated fluorescence increase. Three monomers also displayed a similar phenomenon (Figure S5), indicating the excellent pH-response performance of pH-aptDMs. Inspired by the results from cell-free system, flow cytometric analyses of target SMMC-7721 cells (Figure 1E) were also employed to investigate the feasibility of pH-triggered in situ assembly of multivalent nanodevice. The analyses indicated that the in situ assembled TA-aptNC exhibited an activatable fluorescence signal response to target SMMC-7721 cells at pH 6.3. Together, these results showed that split i-motif unit integrated into the pH-aptDM as a pH-responsive switch could manipulate the assembly of DNA modules into multivalent nanodevice in response to acidic pH.
Evaluation of the binding performance of the TA-aptNC-based in situ assembly system
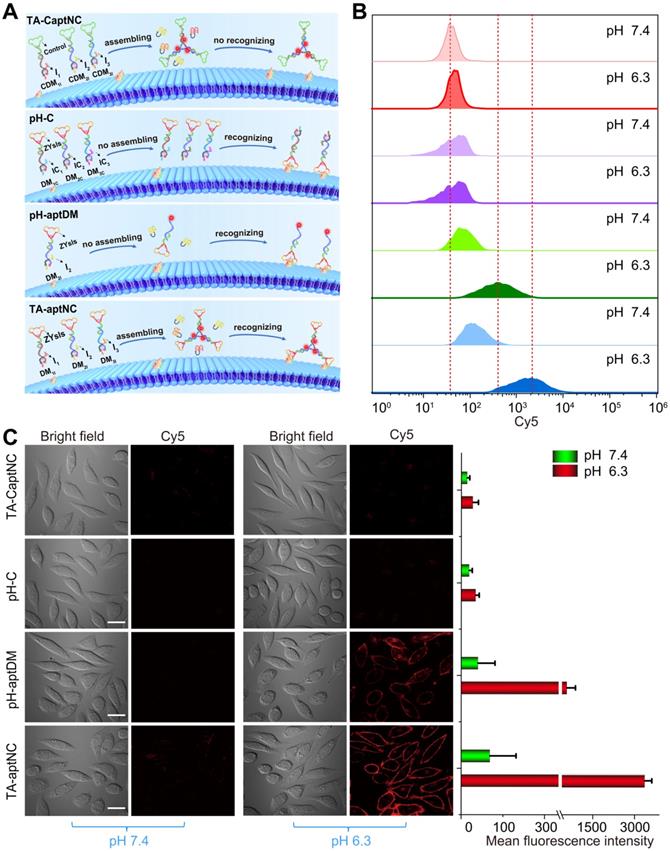
The binding capacity of TA-aptNC-based in situ assembly strategy was investigated at cellular level. To make a reasonable comparison, a variety of control probes were reasonably designed, including TA-CaptNC (the mixture of CDM1I, CDM2I and CDM3I, aptamer sequence replaced by arbitrary sequences), pH-C (the mixture of DM1C, DM2C and DM3C, split i-motif sequence substituted by pH-insensitive sequence) and pH-aptDM (the corresponding monovalent probe). Note that these probes needed to be adjusted to possess the same concentration of targeting ligand ZYsls and Cy5. After treated with different compositions at pH 6.3 (Figure 2A), the flow cytometric analyses demonstrated that the in situ formed TA-aptNC acquired high fluorescence labelling of target cells (Figure S6), exhibiting an excellent SNR that was ~30 times of control compositions (TA-CaptNC and pH-C). While target cells treated with four compositions at different pH, the pH-aptDM and the mixture of three monomers (TA-aptNC group) showed discernible signal in response to target SMMC-7721 cells at pH 6.3 (Figure 2B and S7). The latter established outstanding SBR that was ~8 times of pH-C, which was ~2 times higher than that of the former. In contrast, despite the mixture of CDM1I, CDM2I and CDM3I could be assembled into TA-CaptNC with activated signal at pH 6.3, the poor affinity prevented it from targeting SMMC-7721 cells. As to pH-C, the acid-insensitive performance made it difficult to be activated at pH 6.3, thus generating undistinguished fluorescence labelling. Meanwhile, the confocal laser scanning microscopy images also revealed the binding advantage of in situ formed TA-aptNC to target cell (Figure 2C). An ignorable fluorescence was observed on the membrane of cells under neutral pH, which is due to the probes failing to be activated. When adjusting the pH to 6.3, the mixture of three monomers incubated with SMMC-7721 cells for 1 h displayed twinkle halos around cells, much more than that of the pH-aptDM under the same conditions. The corresponding mean fluorescence intensity of Cy5 labelled SMMC-7721 cells once again highlighted the excellent binding capability of the TA-aptNC-based in situ assembly strategy. Together, these results strongly validate that acidic pH-activated in situ assembly multivalent system could greatly enhance image contrasts.
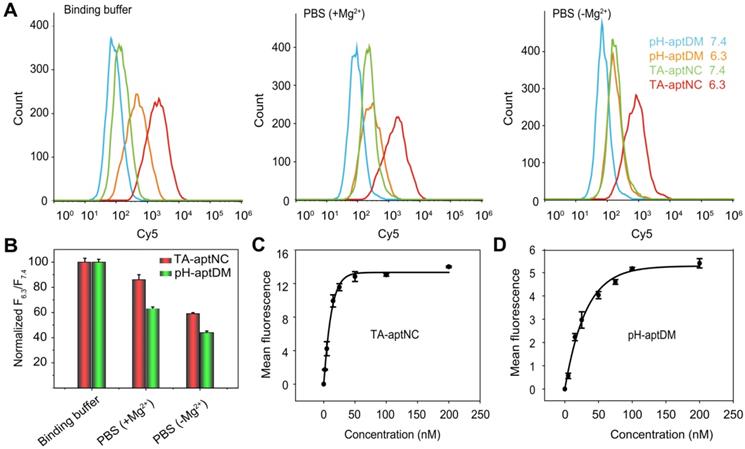
Compared to the pH-aptDM, the resultant TA-aptNC was desired to keep relatively stable affinity to target cells. Subsequently, the binding capacity of TA-aptNC was explored in some typical buffers, including binding buffer (provided yeast-tRNA, Mg2+, glucose and BSA into PBS), PBS with Mg2+ provided (+ Mg2+) and PBS without Mg2+ provided (- Mg2+). The flow cytometry assays indicated that a high binding affinity was in binding buffer, by comparing with that in other buffers, which is due to that the majority of aptamers were selected via cell-SELEX in binding buffer (Figure 3A-B). When removing bivalent cationic, monovalent pH-aptDM displayed an ~60% drop of signal-to-background ratio (SBR). Surprisingly, the binding ability of TA-aptNC could still maintained ~60% of that in binding buffer. We can conclude that the binding stability of TA-aptNC is superior to that of pH-aptDM, which might be due to the multivalent effect and the increase in mass accompanied by the reduction of conformational energy penalty. The stable binding capacity would greatly promote the application of the TA-aptNC in real biological systems.
Binding affinity of aptamers is an important parameter for evaluating the binding performance of diagnostic and/or therapeutic probes to target cells. In this study, the multivalent binding ability of TA-aptNC was dependent on the multiple aptamer nanoclaw embedded in the three outstretched arm of scaffold. The binding affinity of TA-aptNC against target cancer cells was evaluated by flow cytometric analysis, and quantified by the equilibrium dissociation constant (Kd, which is inversely related to the binding affinity). The TA-aptNC activated at acidic pH exhibited a better binding affinity (Kd = 9.00 ± 0.60 nM, Figure 3C), which was 2 times higher than that of pH-aptDM (Kd = 18.00 ± 0.10 nM, in Figure 3D).
Sensitivity and selectivity of the TA-aptNC-based in situ assembly system
In view of the in situ assembled TA-aptNC with superior binding affinity, the sensitivity of the proposed strategy was evaluated by quantitative analysis of SMMC-7721 cells. The samples dispersed with different number of cells ranging from 16 to 160000 were prepared by gradient dilution in 200 μL of binding buffer. The flow cytometry assays indicated that the TA-aptNC-labelled SMMC-7721 cells locating in the upper right region decreased gradually with the decrease of cell number (Figure S8A). An excellent linear relationship was obtained in the range of 16 to 160000 cells. The calibration equation was y = 0.865x - 15.284 with a correlation coefficient R2 = 0.9989 (Figure S8B). The lowest number of SMMC-7721 cell detected by this strategy was 16 in 200 μL of sample, showing the excellent sensitivity of this strategy.
The accurate recognition of tumor has presented a great challenge not only in sensitivity but more importantly in selectivity. To indicated that TA-aptNC-based in situ assembly system could specifically recognize target SMMC-7721 cells, normal L02 cells and three kinds of cancer cells (HepG2, HeLa and MCF-7 cells) were chosen to investigate the specificity of this strategy. As shown in Figure S9, the mixture of three DNA monomers (TA-aptNC group) presented an obvious fluorescence labelling to SMMC-7721 cells at pH 6.3. By contrast, negligible fluorescence signals in response to pH changes were observed for the normal cell and other three tumor cells. To further confirm the above results from flow cytometry, we examined the TA-aptNC for distinguishing of target cell using confocal microscopy (Figure S10). Bright red halos around cells were only observed for target SMMC-7721 cells after incubating with the mixture of three pH-aptDMs at pH 6.3. Whereas there was almost no obvious Cy5 fluorescence in negative control cells, which was in accord with the results of flow cytometry analysis. The great sensitivity and specificity promised the potential application of TA-aptNC-based in situ assembly system in accuracy cancer diagnosis and therapy.
Binding behavior investigation of pH-triggered in situ assembly of multivalent nanodevice. (A) Schematic diagram of pH-triggered the in situ assembly of multivalent nanodevice and binding behavior of various compositions at pH 6.3. Flow cytometry analysis (B) and confocal microscopy assays (C) of SMMC-7721 cells after incubation with different compositions in binding buffer with different pH at 37 oC for 1 h. The corresponding histogram of the four compositions show the mean fluorescence intensity in the images (C) (scale bar: 20 μm). Data are represented as means ± SD (n = 3).
Flow cytometry analysis of the binding performance of multivalent TA-aptNC, in comparison with monovalent pH-aptDM. (A) The binding stability of TA-aptNC in binding buffer, PBS (+ Mg2+ ) and PBS (- Mg2+ ). (B) Histogram of the corresponding normalized signal ratios of SMMC-7721 cells at pH 6.3 to at pH 7.4 for TA-aptNC in (A). Dissociation constant of TA-aptNC (C) and pH-aptDM (D) against target SMMC-7721 cells. Data are represented as means ± SD (n = 3).
Cytotoxicity and cell uptake behavior of the TA-aptNC-based in situ assembly system in vitro
Considering the in situ assembled TA-aptNC with significant increased binding to target cells, we envisioned that it could be evaluated as targeted drug delivery carrier for enhancing cancer therapy. In this study, Dox as a drug model was preferentially inserted into GC base pairs of three pH-aptDMs with quenched fluorescence. Accordingly, the drug payload performance of the DM1I, DM2I and DM3I were evaluated by the fluorescence spectrum, respectively. As shown in Figure S11, the Dox signals were gradually decreased as pH-aptDM concentration increased and reached a saturation at a molar radio of 1:5 (pH-aptDM/Dox). Then, we ascertain the stability and drug release of the Dox-loaded TA-aptNC (TA-aptNC-Dox) at physiological temperature (Figure S12A). The TA-aptNC-Dox and pH-aptDM-Dox both displayed desirable stability in PBS, and a slight drug leakage in 10% fetal bovine serum. However, when both of Dox-loaded probes diffusing in high dose of nuclease, Dox was successfully released with strong fluorescence signals owing to hydrolysis of DNA nanocarriers (Figure S12B). However, TA-aptNC-Dox showed more slower fluorescence restoration than pH-aptDM-Dox, suggesting the excellent stability of TA-aptNC in intracellular drug delivery.
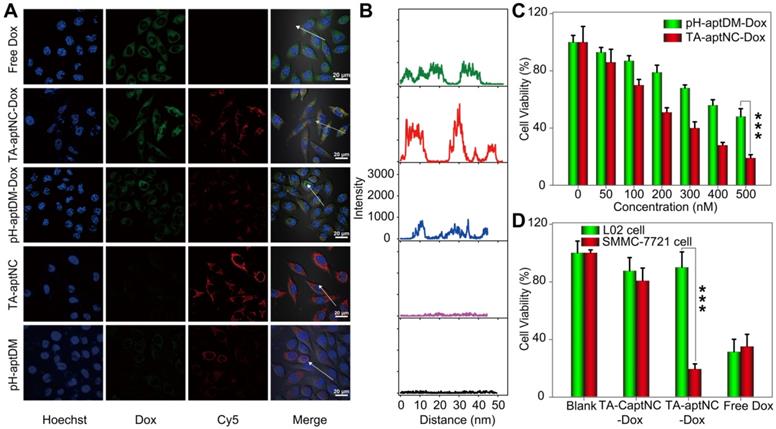
Subsequently, we investigated the time dependent internalization of Cy5-labelled TA-aptNC (Cy5-TA-aptNC) into target cells (Figure S13). The results showed an enhanced intracellular fluorescence signal with the increase of culture time from 0.5 to 4 h and a plateau at about 3 h. Further, we evaluated the drug delivery and release capacity of the TA-aptNC in target cells. As shown in Figure 4A-B, the brighter fluorescence signal of Cy5 and Dox were observed by the TA-aptNC-Dox treated group (the mixture of three Cy5-pH-aptDMs-Dox incubated with SMMC-7721 cells at pH 6.3), in comparison with the pH-aptDM-Dox treated group under the same condition. Presumably, at pH 6.3, the mixture of the three pH-aptDM-Dox was assembled into TA-aptNC-Dox, which could aggregate on the target cell surface, thus facilitating cellular internalization with the aid of surface-anchored multiple aptamer nanoclaw. In the effect of pH, ionic and ubiquitous nucleases present in cytoplasm, Dox was released from the resulting TA-aptNC-Dox with significant fluorescence recovery. The results demonstrated that acidic pH successfully activated the assembly of TA-aptNC-Dox with an improved drug delivery efficiency at tumor site. Meanwhile, the selective drug uptake behavior was studied with target SMMC-7721 cells and notarget L02 cells (Figure S14). An obvious Dox fluorescence signal was observed in SMMC-7721 cells treated with the mixture at pH 6.3 than that of at pH 7.4. It is due to that the acquired multivalent effect could facilitate internalization of TA-aptNCs-Dox into target tumor cells through aptamer-receptor interaction with improved drug delivery efficience. Moreover, when L02 cells were treated with the mixture at pH 6.3, the formation of i-motif led to the release of partial Dox from the pH-aptDMs-Dox. Therefore, a relatively weak Dox fluorescence signal was observed only at pH 6.3 but not at 7.4. It indicates the highly selective drug delivery mediated by TA-aptNC-based in situ assembly system.
Having established that TA-aptNC-Dox could be selectively internalized into cells and realize intracellular drug release, we next assessed the selective cytotoxicity of the TA-aptNC-Dox using MTS assays. As presented in Figure 4C, both the mixture of three Dox-loaded monomer (TA-aptNC-Dox group) and the pH-aptDM-Dox could induce cell apoptosis in a dose-dependent manner. The resulting multivalent effect could accelerate the internalization of the in situ assembled TA-aptNC-Dox into target SMMC-7721 cells, thereby enabling the enhanced cytotoxicity. Furthermore, benefiting from the enhanced affinity and excellent specificity, the TA-aptNC-Dox showed robust cytotoxicity on target cells rather than nontarget cells at pH 6.3 (Figure 4D). In contrast, free Dox displayed indiscriminate cytotoxicity for both SMMC-7721 cells and L02 cells, indicating the excellent selective cytotoxicity of the TA-aptNC. As anticipated, the pure TA-aptNC and pH-aptDM displayed a negligible cytotoxicity for target cells either, suggesting good biocompatibility of the TA-aptNC-based in situ assembly strategy (Figure S15). In general, the selective and potent in vitro therapeutic efficacy of TA-aptNC might pave the way for precise therapy in vivo.
(A) Confocal laser scanning microscopy images of target SMMC-7721 cells after treatment with different compositions, including free Dox, TA-aptNC-Dox (the mixture of DM1I-Dox, DM2I-Dox and DM3I-Dox), pH-aptDM-Dox, TA-aptNC (the mixture of DM1I, DM2I and DM3I) and pH-aptDM in binding buffer at pH 6.3 (scale bar: 20 μm). (B) The corresponding diagrams show the Dox fluorescence intensity on the arrows drawn in the images (A). (C) Cytotoxicity investigation of target tumor cells after incubation with the mixture at pH 7.4 (pH-aptDM-Dox) or pH 6.3 (TA-aptNC-Dox) for 48 h at different doses. (D) The selective cytotoxicity of target tumor cells and normal L02 cells after treated with different formulations. Data are represented as means ± SD (n = 3). The p values were calculated by the Student's t test (***p < 0.001).
In vivo imaging of the TA-aptNC-based in situ assembly system
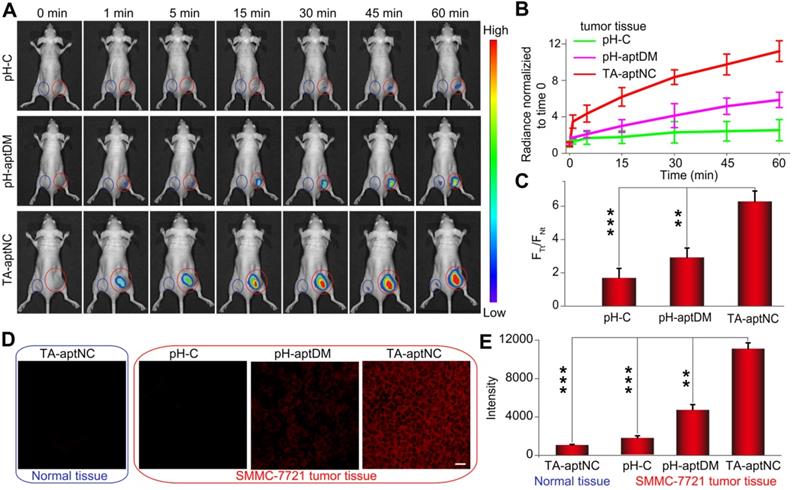
After verifying the TME-activated contrast-enhanced imaging effect and selective cytotoxicity to target SMMC-7721 cells in vitro, we evaluated the utility of the proposed strategy in vivo wherein acidic tumor microenvironment could guide in situ assembly of three pH-aptDMs into TA-aptNC for contrast-enhanced tumor imaging. pH-C, pH-aptDM and the mixture of three monomers (TA-aptNC group), under the same condition, was injected into the tumor site (red circle) of SMMC- 7721 tumor-bearing nude mice, respectively. Simultaneously, the corresponding normal tissues (blue circle) were treated by a same method as negative controls for time-dependent in vivo fluorescence images (Figure 5A). Both the TA-aptNC group and pH-aptDM group established an increased intratumoral fluorescence with the time extended as a result of the continuous activation of fluorescence signals by acidic tumor microenvironment. Significantly, unlike pH-aptDM, TA-aptNC group realized a fast, accuracy and strong fluorescence recovery at tumor site as early as 1 min after intertumoral injection. However, the control groups (pH-C-treated tumor and all the corresponding normal tissues treated with different probes) displayed an ignorable fluorescence enhancement. Quantification analysis indicated that TA-aptNC group displayed outstanding fluorescence contrast (FTt/FNt, the fluorescence signals ratio of the same probe treated with tumor tissues to corresponding normal tissues), which was 3.8-fold higher than that of pH-C group at 60 min (Figure 5B-C). While the fluorescence contrast of monovalent pH-aptDM group was merely 1.8 times of pH-C. We thus deduced that three monomers in the mixture could competitively bind with each other to promote the release of BHQ2 labelled split-I from Cy5 labelled S at complex acidic tumor microenvironment, thus generating significant activatable intratumoral signals. Consistent with the results in vitro, the results in animal model highlighted that this strategy could be high-efficiently activated in TME but remains silence in normal tissues, which is of great importance for sensitive and accurate tumor diagnosis.
The contrast-enhanced tumor imaging was further investigated by imaging frozen tumor sections and normal tissue sections. As presented in Figure 5D, a negligible fluorescence signal was observed for the pH-C treated tumor sections and all of normal tissue sections. Note that TA-aptNC group showed stronger capability to image heterogeneous tumor microenvironments than pH-aptDM-treated group. Taken together, the in situ activated multivalent nanodevice (TA-aptNC) showed accurate and contrast-enhanced imaging capacity in extremely complex tumor environment without any auxiliary additives.
Investigation of acidic tumor microenvironment specificlly activated in vivo tumor imaging by the in situ assembled multivalent nanodevice. (A) Time-dependent in vivo fluorescence imaging monitoring of the SMMC-7721 tumor-bearing nude mice after intratumoral injection of pH-C, pH-aptDM and TA-aptNC (the mixture of three pH-aptDM monomers), respectively. The corresponding probes treated normal tissues were imaged as controls (Normal tissue: blue circle; SMMC-7721 tumor: red circle.). (B) Quantification of the fluorescence intensity at the tumor sites and normal tissues in (A). (C) Analysis of signal intensity ratios of tumor tissues (FTt) to normal tissues (FNt) for different probes in (A) at 1 h. (D) Confocal laser scanning microscopy images of the tumor slices and normal tissue slices after incubation with different probes (scale bar: 200 μm). (E) The corresponding histogram of the four compositions show the mean fluorescence intensity in (D). Data are represented as means ± SD (n = 3). The p values were calculated by the Student's t test (***p < 0.001, **p < 0.01).
In vivo antitumor effect of the pH-triggered TA-aptNC-based in situ assembly system
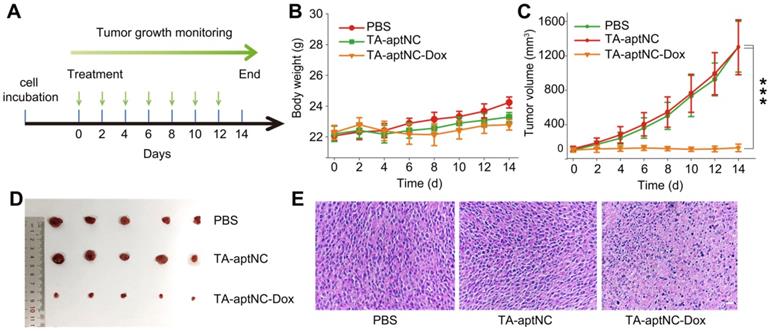
We subsequently evaluated its antitumor performance in vivo using nude mice bearing SMMC-7721 xenograft tumor. All tumor-bearing BALB/c mice were randomly divided into three groups (n = 5), and treated with PBS, the mixture of three monomers (TA-aptNC group) and the mixture of Dox-loaded three monomers (TA-aptNC-Dox group) every two days by intratumoral injection, respectively. The time schedule of the treatment process was shown in Figure 6A. During the animal experiment, the body weight of each nude mice was stable without a significant fluctuation (Figure 6B), demonstrating the limited systemic toxicity and the excellent biocompatibility of in situ assembled TA-aptNC. The real-time variations of tumor volume were also recorded (Figure 6C). The continuous increase of tumor volume was observed for the mixture of three monomers injected mice (TA-aptNC group) in similar with the PBS-treated control group, indicating almost no inhibition of tumor growth, which excluded the systemic toxicity of our proposed strategy. Noteworthy, the TA-aptNC-Dox group displayed a high antitumor efficacy with 90% inhibition. The growth inhibition of tumor was clearly validated by the photographs of dissected tumors for different treatment groups (Figure 6D). In addition, the hematoxylin and eosin (H&E) staining also confirmed that the TA-aptNC group caused a significant disruption of nucleus structures and tumor tissue morphology (Figure 6E), in accordance with the outcome of abovementioned tumor inhibition. Collectively, these results showed that in situ assembly of precisely controlled TA-aptNC afforded superior antitumor efficacy and a negligible systemic toxicity.
In vivo tumor growth inhibition study of the TA-aptNC-based in situ assembly system. (A) The SMMC-7721 tumor-bearing BALB/c nude mice model was established at day 0. The mice were randomly divided into three groups and treated with various formulations (PBS, TA-aptNC, TA-aptNC-Dox (the mixture of three pH-aptDMs)) at days 0, 2, 4, 6, 8, 10 and 12. The body weight curve (B) and the tumor volume curve (C) of nude mice treated with various formulations, respectively. (D) Photographs of dissected tumors in various treatment groups. (E) H&E staining images of tumor tissues in all treatment groups (scale bars: 200 μm). The p values were calculated by the Student's t test (***p < 0.001).
Biocompatibility evaluation
Biocompatibility of drug deliverer is regarded as a requisite in biomedical applications. The hemolysis experiment was performed to investigate the in vivo biocompatibility of this system. First, the important hematological biomarkers of rat red blood cells (RBC) were treated with ultrapure H2O and PBS as the positive control and negative control, respectively. As shown in Figure S16, there was no significant hemolysis after incubating RBCs with different concentrations of the mixture of three pH-aptDMs range from 1 to 150 μM for 12 h at 37 °C, even at high concentrations of the mixture of three pH-aptDMs, indicating the excellent blood compatibility of the TA-aptNC system. The hematological analyses further revealed the exceptional biocompatibility and robust in vivo applicability of this system.
Conclusion
In summary, endogenous stimuli-powered in situ assembly of precisely controlled activatable multivalent nanodevice (TA-aptNC) in an extremely complex tumor microenvironment was engineered for facilitating their imaging function and antitumor efficiency. In comparison with the monovalent DNA monomer or static assembled multivalent DNA nanotheranostics system, the proposed strategy possessed some superiorities: (1) The activated TA-aptNC, possessed the unique superiorities of the precise controlled spatial orientation, defined ligand density and structure size, which allowed 2-fold enhanced binding ability and improved endocytosis efficiency, thus enabling the TME-activated contrast-enhanced in vivo tumor imaging and significant growth inhibition of tumor. TA-aptNC-based in situ assembly system could minimize the background signal in nonspecific site and exclude a risk of irreversible damage to normal tissues. (2) Irrespective of the recruitment of any exogenous biological or nonbiological auxiliary additives, the assembly of TA-aptNC imitated naturally occurring process at a certain location, which might offer unprecedented opportunity to understand the mechanism of disease occurrence and guide the therapy precisely. The strategy could be universally applicable to different tumor by altering targeted identification primitives, which broadened its application scope in various diseases. The proposed strategy will open new avenue for modulating biological behavior in specific site, laying a foundation for the future accurate diagnosis and therapeutics.
Abbreviations
TA-aptNC: three-arm aptamer nanoclaw; pH-aptDMs: pH-responsive aptamer-decorated DNA monomers; DM: DNA module; TA-aptNC-Dox: Dox-loaded TA-aptNC; pH-aptDM-Dox: Dox-loaded pH-aptDM; Cy5-TA-aptNC: Cy5 labeled TA-aptNC; Cy5- pH-aptDM: Cy5 labeled pH-aptDM; PAGE: polyacrylamide gel electrophoresis; AFM: atomic force microscopic atomic force microscopic; RCA: rolling circle amplification; TME: tumor microenvironment; MTT: methyl thiazolyl tetrazolium; SNR: signal-to background ratio; H&E: hematoxylin and eosin.
Supplementary Material
Supplementary materials and methods, figures and table.
Acknowledgements
This work was supported by the Natural Science Foundation of China (21735002, 21874035, 21675046, and 22174044).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Shangguan DH, Li Y Tang ZW, Cao ZH Chen HW, Mallikaratchy P et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci U S A. 2006;103:11838-11843
2. Meng HM, Liu H, Kuai HL, Peng RZ, Mo LT, Zhang XB. Aptamer-integrated DNA nanostructures for biosensing, bioimaging and cancer therapy. Chem Soc Rev. 2016;45:2583-2602
3. Ding CP, Zhang CL, Cheng SS, Xian YZ. Multivalent aptamer functionalized Ag2S nanodots/hybrid cell membrane-coated magnetic nanobioprobe for the ultrasensitive isolation and detection of circulating tumor cells. Adv Funct Mater. 2020;30:1909781-1909781
4. Qin WW, Chen L, Wang ZR, Li Q, Fan CH, Wu MH. et al. bioinspired DNA nanointerface with anisotropic aptamers or accurate capture of circulating tumor cells. Adv Sci. 2020;7:2000647-2000654
5. Ouyang CH, Zhang SB, Xue C, Yu X, Xu H, Wang ZM. et al. Precision-guided missile-like DNA nanostructure containing warhead and guidance control for aptamer-based targeted drug delivery into cancer cells in vitro and in vivo. J Am Chem Soc. 2020;142:1265-1277
6. Kuai HL, Zhao ZL, Mo LT, Liu H, Hu XX, Fu T. et al. Circular bivalent aptamers enable in vivo stability and recognition. J Am Chem Soc. 2017;139:9128-9131
7. Song JB, Lin SL, Yang Z, Zhu R, Zhou ZJ, Li ZW. et al. Self-assembled responsive bilayered vesicles with adjustable oxidative stress for enhanced cancer imaging and therapy. J Am Chem Soc. 2019;141:8158-8170
8. Lei YL, Qiao ZZ, He XX, Shi H, Ye XS, Yan LA. et al. DNA nanotriangle-scaffolded activatable aptamer probe with ultralow background and robust stability for cancer. Theranostics. 2018;8:4062-4071
9. Ren KW, Liu Y, Wu J, Zhang Y, Zhu J, Yang M. et al. A DNA dual lock-and-key strategy for cell-subtype-specific siRNA delivery. Nat Commun. 2016;7:13580
10. Wang D, Li SH, Zhao ZL, Zhang XB, Tan WH. Engineering a second-order DNA logic-gated nanorobot to sense and release on live cell membranes for multiplexed diagnosis and synergistic therapy. Angew Chem Int Ed Engl. 2021;60:15816-15820
11. Kim J, Park W, Kim D, Lee ES, Lee DH, Jeong S. et al. Tumor-specific aptamer-conjugated polymeric photosensitizer for effective endo-laparoscopic photodynamic therapy. Adv Funct Mater. 2019;29:1900084-1900094
12. Andrusishina INJ, Elementol. 2010; 15: 425-433
13. Chen LF, Xing SH, Lei YL, Chen QS. Zou Z, Quan K, et al. A glucose-powered activatable nanozyme breaking pH and H2O2 limitations for treating diabetic infections. Angew Chem Int Ed Engl. 2021;60:1-6
14. Liu BW, Liu JW. Interface-driven hybrid materials based on DNA-functionalized gold nanoparticles. Matter. 2019;1:825-847
15. Geng ZM, Wang L, Liu K, Liu JY, Tan WH. Enhancing anti-PD-1 immunotherapy by nanomicelles self-assembled from multivalent aptamer drug conjugates. Angew Chem Int Ed Engl. 2021;60:15459-15465
16. Li M, Ding HM, Lin MH, Yin FF, Song L, Mao XH. et al. DNA framework-programmed cell capture via topology engineered receptor-ligand interactions. J Am Chem Soc. 2019;141:18910-18915
17. Hu QQ, Li H, Wang LH, Gu HZ, Fan CH. DNA nanotechnology-Enabled drug delivery systems. Chem Rev. 2019;119:6459-6506
18. Xue C, Zhang SB, Yu X, Hu SY, Lu Y, Wu ZS. Periodically ordered, nuclease-resistant DNA nanowires decorated with cell-specific aptamers as selective theranostic agents. Angew Chem Int Ed Engl. 2020;59:17540-17547
19. Lacroix A, Sleiman HF. DNA Nanostructures: current challenges and opportunities for cellular delivery. ACS Nano. 2021;15:3631-3645
20. Heinen L, Heuser T, Steinschulte A, Walther A. Antagonistic enzymes in a biocatalytic pH feedback system program autonomous DNA hydrogel life cycles. Nano Lett. 2017;17:4989-4995
21. Wang J, Wang H, Wang HM, He SZ, Li RM, Deng Z. et al. Nonviolent self-catabolic DNAzyme nanosponges for smart anticancer drug delivery. ACS Nano. 2019;13:5852-5863
22. Zhang ZQ, Ali MM, Eckert MA, Kang DK, Chen YY, Sender LS. et al. A polyvalent aptamer system for targeted drug delivery. Biomaterials. 2013;34:9728-9735
23. Hu QY, Chen Q, Gu Z. Advances in transformable drug delivery systems. Biomaterials. 2018;178:546-558
24. Xiao MS, Lai W, Wang F, Li L, Fan CH, Pei H. Programming drug delivery kinetics for active burst release with DNA toehold switches. J Am Chem Soc. 2019;141:20354-20364
25. Han JP, Cui YC, Gu Z, Yang DY. Controllable assembly/disassembly of polyphenol-DNA nanocomplex for cascade-responsive drug release in cancer cells. Biomaterials. 2021;273:120846
26. Mela I, Vallejo-Ramirez PP, Makarchuk S, Christie G, Bailey D, Henderson RM. et al. DNA nanostructures for targeted antimicrobial delivery. Angew Chem Int Ed Engl. 2020;59:12698-12702
27. Hu X, Li FY, Wang SY, Xia F, Ling DS. Biological stimulus-driven assembly/disassembly of functional nanoparticles for targeted delivery, controlled activation, and bioelimination. Adv Healthc Mater. 2018;7:1800359-1800377
28. Park H, Kim J, Jung S, Kim WJ. DNA-Au Nanomachine equipped with i-Motif and G-Quadruplex for triple combinatorial anti-Tumor therapy. Adv Funct Mater. 2018;28:1705416-1705428
29. Xue C, Zhang SX, Ouyang CH, Chang DR, Salena BJ, Li YF. et al. Target-induced catalytic assembly of Y-shaped DNA and its application for in situ imaging of microRNAs. Angew Chem Int Ed Engl. 2018;57:9739-9743
30. Ren KW, Zhang Y, Zhang XB, Liu Y, Yang M, Ju HX. In situ siRNA assembly in living cells for gene therapy with microRNA triggered cascade reactions templated by nucleic acids. ACS Nano. 2018;12:10797-10806
31. Liu R, Zhang SB, Zheng TT, Chen YR, Wu JT, Wu ZS. Intracellular nonenzymatic in situ growth of three-dimensional DNA nanostructures for imaging specific biomolecules in living cells. ACS Nano. 2020;14:9572-9584
32. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674
33. Jiang SX, Ge ZL, Mou S, Yan H, Fan CH. designer DNA nanostructures for therapeutics. Chem. 2021;7:1156-1179
34. Chen XX, Chen TS, Ren LJ, Chen GF, Gao XH, Li GX. et al. Triplex DNA nanoswitch for pH-sensitive release of multiple cancer drugs. ACS Nano. 2019;13:5959-5967
35. Wang YG, Zhou KJ, Huang G, Hensley C, Huang XN, Ma XP. et al. A nanoparticle-based strategy for the imaging of a broad range of tumours by nonlinear amplification of microenvironment signals. Nat Mater. 2014;13:204
36. Kim J, Jo C, Lim WG, Jung SJ, Lee YM, Lim J. et al. Programmed nanoparticle-loaded nanoparticles for deep penetrating 3D cancer therapy. Adv Mater. 2018;30:1707557-1707564
37. Zou SZ, Lei YL, Ma WJ, Chen B, Cheng H, Jia RC. et al. Extracellular pH-manipulated in situ reconfiguration of aptamer functionalized DNA monomer enables specifically improved affinity, detection and drug delivery. Analyst. 2020;145:2562-2569
38. Lei YL, Tang JL, He XX, Shi H, Zeng Y, Sun HY. et al. In situ modulating DNAzyme activity and internalization behavior with acid-Initiated reconfigurable DNA nanodevice for activatable theranostic. Anal Chem. 2021;93:5629-5634
39. Ma WJ, Chen B, Jia RC, Sun HH, Huang J, Cheng H. et al. In situ hand-in-hand DNA tile assembly: A pH-driven and aptamer targeted DNA nanostructure for TK1 mRNA visualization and synergetic killing of cancer cells. Anal Chem. 2021;93:10511-10518
40. Chen F, Lu QJ, Huang LN, Liu BW, Liu ML, Zhang YY. et al. DNA triplex and quadruplex assembled nanosensors for correlating K+ and pH in lysosomes. Angew Chem Int Ed Engl. 2021;60:5453-5458
41. Kaiser CE, Van Ert NA, Agrawal P, Chawla R, Yang DZ, Hurley LH. Insight into the complexity of the i-motif and G-Quadruplex DNA structures formed in the KRAS promoter and subsequent drug induced gene repression. J Am Chem Soc. 2017;139:8522-8536
Author contact
![]() Corresponding author: xiaoxiaoheedu.cn (X. H.), kmwangedu.cn (K. W.).
Corresponding author: xiaoxiaoheedu.cn (X. H.), kmwangedu.cn (K. W.).
 Global reach, higher impact
Global reach, higher impact