13.3
Impact Factor
Theranostics 2023; 13(10):3188-3203. doi:10.7150/thno.80435 This issue Cite
Research Paper
Integration of metabolomics and peptidomics reveals distinct molecular landscape of human diabetic kidney disease
1. Institution of Analytical Chemistry, Department of Chemistry, Zhejiang University, Hangzhou, 310058, China.
2. Department of Nephrology, The First Affiliated Hospital of Zhejiang Chinese Medical University, Zhejiang Provincial Hospital of Traditional Chinese Medicine, Hangzhou, 310006, China.
3. Department of Nephrology, The First People's Hospital of Hangzhou Lin'an District, Affiliated Lin'an People's Hospital of Hangzhou Medical College, Hangzhou, 311300, China.
4. Well-healthcare Technologies Co., Hangzhou, 310051, China.
Abstract
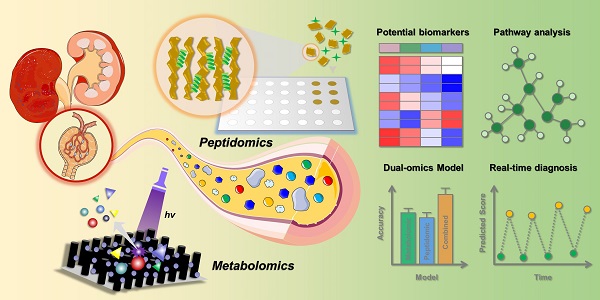
Diabetic kidney disease (DKD) is the most common microvascular complication of diabetes, and there is an urgent need to discover reliable biomarkers for early diagnosis. Here, we established an effective urine multi-omics platform and integrated metabolomics and peptidomics to investigate the biological changes during DKD pathogenesis.
Methods: Totally 766 volunteers (221 HC, 198 T2DM, 175 early DKD, 125 overt DKD, and 47 grey-zone T2DM patients with abnormal urinary mALB concentration) were included in this study. Non-targeted metabolic fingerprints of urine samples were acquired on matrix-free LDI-MS platform by the tip-contact extraction method using fluorinated ethylene propylene coated silicon nanowires chips (FEP@SiNWs), while peptide profiles hidden in urine samples were uncovered by MALDI-TOF MS after capturing urine peptides by porous silicon microparticles.
Results: After multivariate analysis, ten metabolites and six peptides were verified to be stepwise regulated in different DKD stages. The altered metabolic pathways and biological processes associated with the DKD pathogenesis were concentrated in amino acid metabolism and cellular protein metabolic process, which were supported by renal transcriptomics. Interestingly, multi-omics significantly increased the diagnostic accuracy for both early DKD diagnosis and DKD status discrimination. Combined with machine learning, a stepwise prediction model was constructed and 89.9% of HC, 75.5% of T2DM, 69.6% of early DKD and 75.7% of overt DKD subjects in the external validation cohort were correctly classified. In addition, 87.5% of grey-zone patients were successfully distinguished from T2DM patients.
Conclusion: This multi-omics platform displayed a satisfactory ability to explore molecular information and provided a new insight for establishing effective DKD management.
Keywords: Multi-omics, Diabetic kidney disease, Type 2 diabetes, Non-invasive diagnosis, Laser desorption/ionization mass spectrometry
 Global reach, higher impact
Global reach, higher impact