13.3
Impact Factor
Theranostics 2025; 15(17):8952-8963. doi:10.7150/thno.111807 This issue Cite
Research Paper
Role of MiR-204 in controlling metabolic functions of the subretinal microglia
1. Department of Biochemistry & Physiology, University of Oklahoma Health Sciences Center, Oklahoma City, OK 73104, USA.
2. Department of Ophthalmology, Dean McGee Eye Institute, University of Oklahoma Health Sciences Center, Oklahoma City, OK 73104, USA.
3. Department of Neurobiology and Anatomy, University of Texas Health Science Center at Houston, Houston, TX 77030, USA.
4. Department of Pharmaceutical Sciences, College of Pharmacy, University of Oklahoma Health Sciences Center, Oklahoma City, OK 73117, USA.
Received 2025-2-9; Accepted 2025-7-29; Published 2025-8-11
Abstract
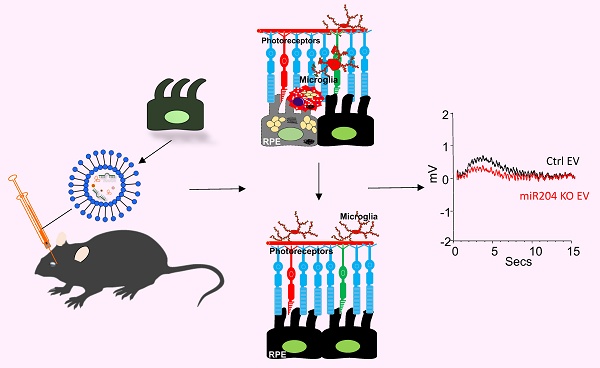
Rationale: MicroRNA-204 (miR-204) is one of the most abundant miRNA species in the retinal pigment epithelium (RPE) and RPE-derived extracellular vesicles (EVs). Knockout (KO) of miR-204 leads to dysfunction and degeneration of both the RPE and the retina. In addition to previously reported retinal pathologies, we observed the accumulation of lipid-laden subretinal microglia in miR-204 KO mice. This study aimed to identify potential molecular targets of miR-204 involved in microglia lipid processing and to determine whether RPE-derived EVs can improve the function of miR-204-deficient retinal microglia.
Methods: Lipid accumulation in microglia was detected by staining with LipidTox, a fluorescent dye specific for neutral lipids, followed by either flow cytometry analysis or direct visualization on RPE/choroid flat mounts. MiRNA database and target prediction tools, such as miRWalk and TargetScan, were used to search for potential target genes of miR-204 in microglia. The identified target mRNA was validated with a miRNA reporter assay. RPE EVs were prepared from ex vivo cultured mice eye cups and administered via retro-orbital injection in miR-204 knockout (KO) mice. RPE integrity was assessed by ERG c-wave measurement.
Results: KO of miR-204 resulted in the accumulation of neutral lipids in subretinal microglia. MiR-204 targeted the TGF-β receptor 2 gene in microglia. TGF-β markedly suppressed the expression of genes related to microglia lipid clearance. Eyes injected with RPE-derived EVs showed improved ERG c-wave responses compared to the fellow eyes injected with saline.
Conclusions: This study supports that TGF-β/TGF-β receptor 2 regulates microglia lipid metabolism primarily by suppressing lipid clearance. By modulating TGF-β signaling, miR-204 in RPE-derived EVs likely enhances the lipid metabolic activities of subretinal microglia, which are crucial for the structural integrity and proper function of the outer retina and RPE. RPE-derived EVs and their delivery of miRNAs represent a potential therapeutic approach for treating retinal diseases, such as age-related macular degeneration, which involve dysregulated lipid metabolism in subretinal microglia.
Keywords: microglia, metabolism, extracellular vesicles, RPE, inflammation
Introduction
MiR-204 is one of the most abundant miRNA species in the retinal pigment epithelium (RPE) [1]. Genetic knockout of miR-204 in the RPE reduced the phago-lysosomal activity and suppressed the turnover of phagocytosed photoreceptor outer segments (POS), thereby leading to the progressive RPE degeneration and loss of retinal function [2]. Mutation in miR-204 gene causes inherited retinal degeneration and coloboma [3].
In addition to the RPE, retinal microglia also perform phagocytosis function [4]. Microglia in the central nervous system can adapt to the microenvironment, displaying plasticity in markers and functions associated with either homeostatic steady-state or activation [5-7]. Once in the subretinal space, microglia facilitate the RPE in POS removal [8]. The uniqueness of the subretinal microenvironment is an enrichment of the immune regulatory factors released by the RPE [9]. RPE-derived extracellular vesicles (EVs), and their associated proteins and microRNAs (miRNAs), can potentially reprogram the subretinal microglia [10]. The underlying mechanisms, however, remain elusive. Whether miR-204 can impact microglial function has not been determined.
In the current study, we characterized the retinal microglia in miR-204 KO mice and identified deficiencies in their functions related to lipid clearance. Subretinal microglia in miR-204 KO mice exhibited an accumulation of neutral lipids. We further determined that the TGF-β receptor 2 gene was a target of miR-204. TGF β signaling has profound impact on cell metabolism [11]. In microglial cells, TGF β inhibited fatty acid utilization and downregulated genes associated with lipid metabolism. Previously, we reported that miRNAs can be transferred from the RPE to retinal microglia [12]. Notably, we found that RPE-derived EVs can improve RPE barrier function in miR-204 KO mice when delivered retro-orbitally. Collectively, our findings suggest that miR-204 plays important roles in regulating retinal microglia functions by modulating their responses to TGF β. RPE-derived EVs have the potential to rejuvenate the protective functions of subretinal microglia, by delivering cargos such as miR-204.
Material and Methods
Animals
miR-204 KO mice were provided by Dr. Sheldon Miller's laboratory at the NIH Intramural Research Program [2]. Common genetic mutations associated with inherited retinal degeneration, such as rd1 and rd8 [13], were screened for and confirmed to be absent in the KO strain. Animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Oklahoma Health Sciences Center (OUHSC), and adhered to the Statement for the “Use of Animals in Ophthalmic and Vision Research” from the Association for Research in Vision and Ophthalmology (ARVO). Mice were housed in the OUHSC animal care facility and maintained on standard rodent chow ad libitum under a 12h dark/light cycle.
Culture of primary retinal microglia
Microglia were isolated from neonatal mice (postnatal days 3-5) and cultured as previously described [12]. Cells were cultured in Dulbecco's Modified Eagle/F12 medium (1:1 mixture of DMEM:F12; Mediatech, Manassas, VA) containing 10% fetal bovine serum and 50 ng/mL granulocyte-macrophage colony stimulation factor (GM-CSF) (R&D Systems, Minneapolis, MN). The enrichment of microglia was validated by CD11b staining and flow cytometry, with a typical purity greater than 90% [12].
Isolation of RPE-derived EVs
Mouse eyes were enucleated from euthanized animals, and extraocular muscles and connective tissues were removed by microdissection. Following retina removal, RPE/sclera/choroid explants were cultured ex vivo in serum-free DMEM medium for 4 h. EVs in the conditioned medium were isolated using size exclusion chromatography on a qEV column (Izon Science, Medford, MA), with phosphate-buffered saline (PBS) as the elution buffer. The eluted fractions were examined for the exosome marker proteins TSG101 and flotillin-2 (Flot2) [12, 14]. Nanoparticle tracking analyses of particle size and density were performed using a NanoSight NS300 (Malvern Panalytical, Westborough, MA) [12]. Ten eye cups were pooled, yielding an EV concentration of approximately 5 x108/mL per eye cup.
EV treatment and Electroretinography (ERG) c-wave measurement
Under anesthesia, miR-204 KO mice received retro-orbital injection of 100 μL of EV (5x108 particles) per eye. A second injection was administrated 7 days after the initial treatment. The contralateral eye served as a control and received 100 μL of saline at both time points. Seven days following the second injection, animals underwent ERG c-wave measurement [15].
ERG recording was conducted as previously described [16]. Briefly, mice were dark-adapted overnight before recording. Anesthesia was induced using ketamine (80-100 mg/kg) /xylazine (5-16 mg/kg). Pupil dilation was achieved with 0.5% Tropicamide (Covetrus, Portland, ME), followed by the application of hydroxypropyl methylcellulose ophthalmic solution. Gold wire electrodes were placed on the corneal surface. ERG a, b and c-wave responses were recorded using a Diagnosys Espion E2 system (Diagnosys LLC, MA, USA) [15].
To track the uptake of EVs by retinal cells, EVs were engineered with the fluorescent BODIPY dye for tracing [17]. Poly(ethylene glycol) (PEG) spacers were conjugated to BODIPY-anchors to prevent the EVs from aggregation [17]. After retro-orbital injection, the co-localization of BODIPY and IBA-1 staining was examined on RPE/choroid flat mounts.
RPE/choroid flat mount
Enucleated mouse eyes were fixed in 4% paraformaldehyde (PFA) for 10 min, and extraocular muscles were carefully removed by microdissection. RPE/choroid flat mounts were prepared as previously described [15] and stained with an anti-IBA-1 antibody (Fujifilm Wako, Tokyo, Japan). The total number of subretinal microglia, including both activated and homeostatic resting morphologies, was quantified across the entire wholemount.
In vivo mouse eye examination
Animals were anesthetized by an intraperitoneal injection of a mixture of ketamine (50-75 mg/kg) and xylazine (5-10 mg/kg). The pupil was fully dilated with Tropicamide, and covered with Gonak™ hypromellose ophthalmic demulcent solution (Akorn, Lake Forest, IL, USA). Fundus photographs were taken with a Micron-IV camera (Phoenix Research Laboratories, Pleasanton, CA) with cornea-contacting lens [18]. Spectral-domain OCT was performed on a Bioptigen Envisu system (Leica Microsystems Inc, Buffalo Grove, IL, USA) with a mouse retina lens [19].
3' UTR reporter assay
The 3′ untranslated region (UTR) target clones for mouse Tgfbr2 gene, either wild type or mutated, were purchased from GeneCopoeia (Rockville, MD). The construct contains the 3′ UTR of Tgfbr2 gene spanning positions 2031-4728 (GeneBank sequence ID NM_029575), inserted downstream of the Gaussia luciferase reporter gene [20]. The mutant construct had nucleotides 2499 and 2500 changed from AA to CC. Primary retinal microglia cultured in 6-well plates were co-transfected with 2.5 μg of the reporter plasmid and 3 pmol of miR-204 mimics or scrambled RNA (Dharmacon, Lafayette, CO), using Lipofectamine 2000 (Invitrogen). Dual-luciferase assays were performed 2 days post-transfection, with Renilla luciferase activity, cloned into the same reporter construct [20], used for normalization.
Cytokine array assay
Conditioned medium from primary retinal microglia were collected from both WT and miR-204 KO cells. Cell debris were removed by centrifugation at 1,000 g for 5 min. Samples were applied to membranes in the Proteome Profiler Mouse Cytokine Array Panel A (R&D systems) kit, which can detect 40 different cytokines. Signals were detected with streptavidin-conjugated fluorescent secondary antibody following manufacture's guide.
Flow cytometry
Mouse RPE/choroid/sclera tissues were collected from enucleated eyes after the removal of the anterior segment and retina. Tissues from six mouse eyes were pooled and digested with 0.2 mg/mL collagenase D (Sigma) and 5 u/mL dispase (Stemcell, Cambridge, MA) at 37 °C for 30 min [15]. Cell clumps were removed by passing through a 40-μm cell strainer. The cells were then blocked with an FcγR blocker and stained for cell surface markers using fluorochrome-labeled antibodies: PE-anti-CD45 and Alexa Fluor 700-anti-CD11b (BioLegend, San Diego, CA). Intracellular staining was used for CD68 with PerCP/Cyanine 5.5-conjugated antibody (BioLegend) [15]. For neutral lipids staining, cells were fixed in 4% PFA for 30 min and stained with HSC LipidTox Green (Invitrogen, Waltham, MA) at room temperature for 30 min. Samples were analyzed on an Attune NxT acoustic focusing flow cytometer (Invitrogen by Thermo Fisher Scientific, MA). Compensation was performed according to the manufacturer's instructions using UltraComp Beads (Thermo Fisher). Flow cytometry data were processed with AttuneTM NxT, FCS Express (DeNovo Software, Pasadena, CA) and FlowJo (Ashland, OR, USA) softwares.
Seahorse Metabolic assay
Primary mouse retinal microglia were used for the metabolic assays to measure the rates of oxygen consumption and extracellular acidification in the presence of glucose or glucose and palmitate-bovine serum albumin (BSA) [15]. 2x105 cells per well were seeded in Seahorse 96-well microplates (Agilent, Santa Clara, CA) and treated with 10 ng/mL TGF-β1 for 16 h. MitoStress and Palmitate Oxidative Stress tests were performed following manufacturer's guides. Normalization was performed with the number of cells per well counted by Cytation 1 (Agilent) after nuclei staining. Data were analyzed in Wave software [15].
Statistics
Statistical analyses were performed with GraphPad Prism software. Data were analyzed using GraphPad Prism software (Boston, MA, USA). Normality of the data distribution and homogeneity of variance were assessed using the Kolmogorov-Smirnov test and either F-test (two-group comparisons) or the Brown-Forsythe test (for multiple-group comparisons). Between-group differences were assessed by Student's t-test or the Mann-Whitney test, with the level of significance presented as P values. For multiple group comparison, if data passed the normality test (P >0.05), one-way analysis of variance (ANOVA) was used, followed by Dunnett's post-tests (everyone vs ctrl), Turkey's (every group) or Sidak's Multiple comparisons Test (specific pairs). If the data were not normally distributed, the Kruskal-Wallis test (a non-parametric alternative to ANOVA) was used, followed by Dunn's multiple comparisons tests. All data were presented as the means ± SEM.
Results
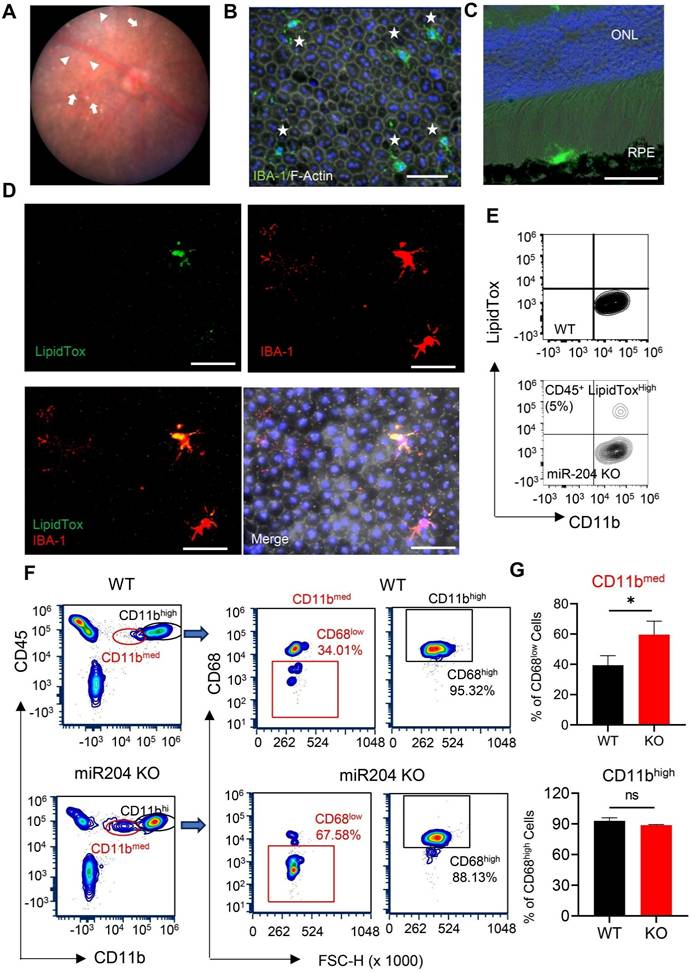
Consistent with previous publications [2], fundus photographs of miR-204 KO mice showed RPE pigmentary changes and retinal cell infiltration at 3 months of age (Figure 1A). Despite these alterations, OCT examination revealed no significant structural changes or retinal thinning in miR-204 KO mice up to 6 months of age (Figure S1A-B). ERG c-wave measurements, an indicator of RPE function, demonstrated an approximately 50% reduction in c- wave amplitude in miR-204 KO mice compared to age-matched controls at 3 months of age (Figure S1C-D). IBA-1+ cells with activated morphology were readily detected on the apical side of the RPE on both RPE flat-mounts and cryosections of posterior eyes (Figure 1B-D). Subretinal cells were also observed on H&E-stained paraffin sections (Figure S1E-F). To further investigate lipid accumulation, and we stained microglia with HSC LipidTox dye which measures neutral lipids, including cholesterol and triacylglycerol [21]. With flow cytometry assay [22], CD45med cells isolated from RPE/choroid preparations of miR-204 KO mice showed a subset of cells with elevated lipid content (Figure 1E), consistent with findings from the flat-mount imaging. These data suggest that, in addition to its established roles in RPE biology [2], miR-204 KO led to lipid accumulation in subretinal microglia. Lipid accumulation in the RPE and choroid, and the presence of subretinal cells are commonly observed in AMD eyes [23-25]. The CD45med cells in miR-204 KO mice showed a higher percentage of cells with lower Cd11b and lower CD68, further support their defects in phagocytosis function (Figure 1F-G) [26].
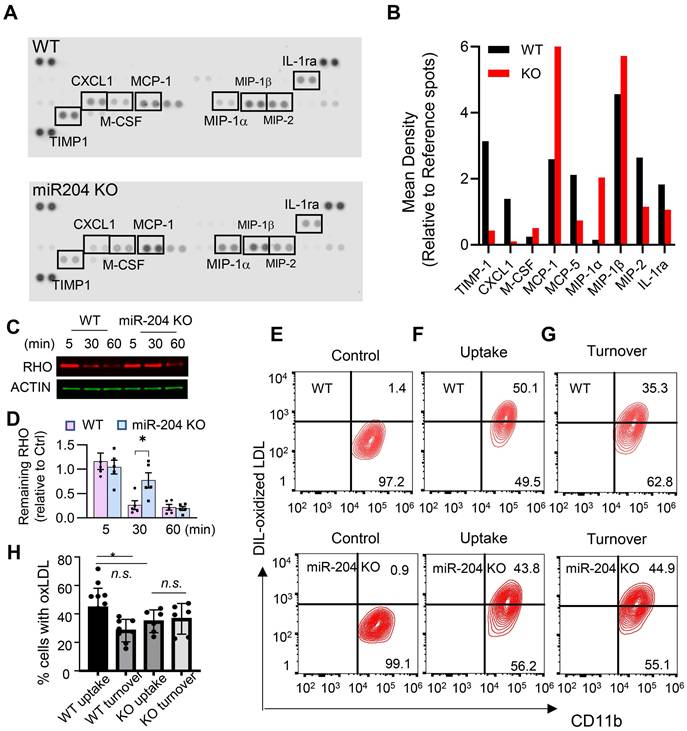
To investigate lipid metabolism-related functions, cultured primary retinal microglia were used. These cells mainly secreted macrophage chemotactic chemokines (e.g. MCP-1, MIP-1a, and MIP-1b) (Figure 2A and B), and deficiency of miR-204 led to their increased production. Microglia can phagocytose and degrade lipid-enriched POS [4]. As shown in Figure 2C and D, WT microglia degraded over 60% of ingested POS within 30 min, as indicated by a reduction in rhodopsin levels (a major protein in POS) to 34 ± 21% of their initial level at 5 min post-loading. In contrast, miR-204-deficient microglia were significantly less effective in degrading POS, with rhodopsin levels remaining at 74 ± 36% of the baseline after 30 min. The differences in POS degradation rates between WT and KO microglia were statistically significant (P < 0.05, Mann-Whitney test, N = 5). We also assessed the ability of miR-204 KO microglia to turn over oxidized LDL (oxLDL), another lipid species microglia can encounter in vivo [27]. Although both WT and KO microglia exhibited comparable initial uptake of oxLDL (Figure 2E and 2F), KO microglia demonstrated a marked deficiency in oxLDL degradation. Approximately 40% of both WT and miR-204 KO microglia initially contained oxLDL after exposure, the percentage of oxLDL+ cells in WT microglia decreased to 28 ± 8% after 16 h of incubation. In contrast, no reduction in oxLDL+ cells was observed in miR-204 KO microglia (Figure 2G). The difference in oxLDL turnover between WT and KO microglia was statistically significant (P < 0.05, N = 6, one-way ANOVA, Dunnett's multiple comparison test) (Figure 2H). Together, these results suggest compromised lipid handling ability in miR-204 KO microglia.
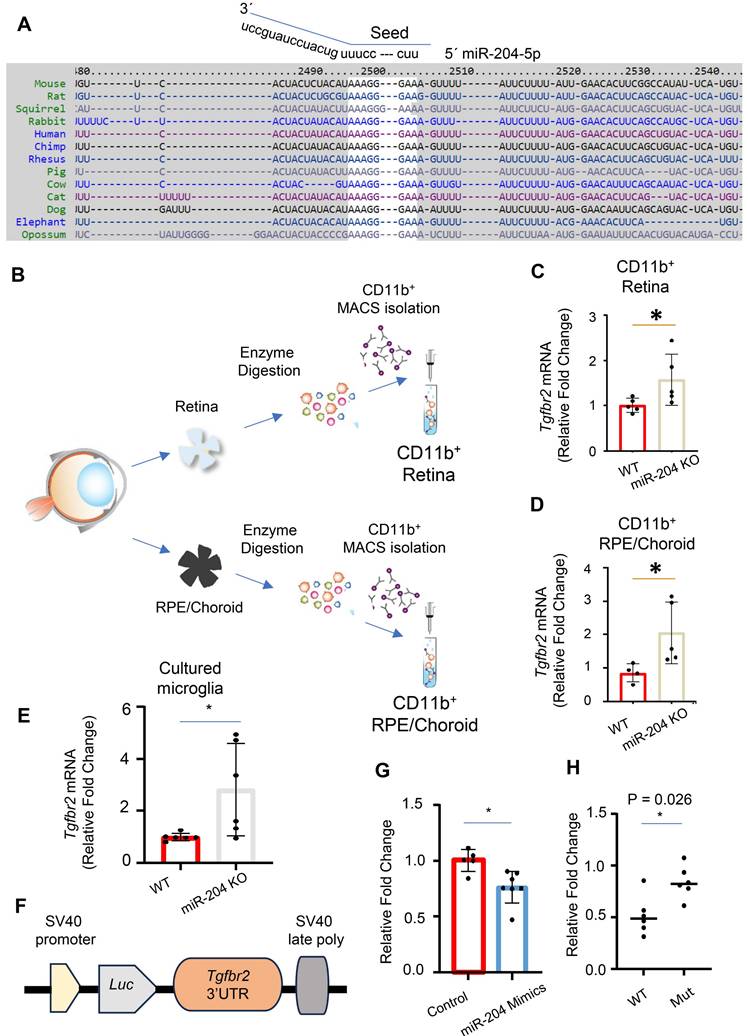
We used miRNA database and target prediction tools, including miRWalk [28] and TargetScan [29], to search for potential target genes of miR-204 in microglia. The 3´ UTR of Tgfbr2 gene was predicted to have a perfect match with nucleotides 1 - 8 of the miR-204 seed sequence, located at positions 2498 to 2505 (Figure 3A). The pairing is conserved among vertebrates including mouse, rat and primates. To experimentally validate Tgfbr2 gene as a target of miR-204, we isolated CD11b+ cells from WT and miR-204 KO retina or RPE/choroid tissues using magnetic-activated cell sorting (MACS) beads (Miltenyi) (Figure 3B), and used quantitative RT-PCR to measure the level of Tgfbr2. The mRNA levels of Tgfbr2 gene were significantly higher in KO CD11b+ microglia and macrophages (Figure 3B -D), as well as in cultured retinal microglia established from miR-204 KO mice (Figure 3E). To confirm the direct interaction between miR-204 and Tgfbr2 mRNA, we co-transfected microglia with miR-204 mimics or a scrambled control, along with a reporter plasmid containing the 3' UTR of the Tgfbr2 gene downstream of a luciferase gene (Figure 3F). Binding of miR-204 to the 3' UTR resulted in downregulation of the reporter gene and lowered the luciferase activity (Figure 3G). To validate the specificity of this interaction, we generated a mutant reporter construct by replacing the predicted miR-204 binding site sequence in the 3' UTR (AAAGGGAAAGTTTA) with a mutated sequence (ACCGGGAAAGTTTA) using site-directed mutagenesis. As shown in Figure 3H, the mutant reporter did not respond to miR-204 mimics co-transfection, indicating that the canonical binding site is required for repression. Collectively, these results support that miR-204, the most abundant miRNA species in RPE-derived EVs (Figure S2), directly targets and downregulates Tgfbr2 in microglia.
Lipid accumulation in miR-204-KO subretinal microglia. (A) Representative fundus photograph of a 3-month-old miR-204 KO mouse showing RPE pigmentary changes (arrowheads) and retinal deposits (arrows). (B-C) Immunostaining of RPE flat mounts (B) and cryosections of posterior eye tissue (C) from miR-204 KO showing infiltrated microglia at the RPE layer. Subretinal microglia were labeled with IBA1 (Green), and phalloidin (white) was used to delineate the boundaries of the RPE in whole-mount tissues. Blue: Nuclei. ONL: outer nuclear layer. (D) Co-staining of IBA-1+ (red) subretinal microglia with LipidTox dye (green) on RPE flat mount prepared from miR-204 KO mouse. (E) Flow cytometry contour plots showing LipidTox and CD11b double-positive cells in RPE/choroid preparations from control and miR-204 KO mice. Scale bar: 50 µm. (F) Flow cytometry of microglia markers, CD11b and CD68, in cells prepared from choroid/RPE fractions of WT and miR-204 KO mice. Quantitation data are presented in (G). Data presented are the averages of 4 separate experiments. * P < 0.05, Student's t-test.
Deficiency in lipid metabolites turnover in miR-204 KO microglia. (A) Cytokine arrays of conditioned media from primary retinal microglia from wild type (WT) and miR-204 knockout mice. (B) Quantitative analyses of cytokine arrays. (C) Western blot analysis of rhodopsin turnover in cultured primary retinal microglia fed with POS. Cells were loaded with POS for 30 min, and rhodopsin levels were monitored at the indicated time points. (D) Quantitative data of rhodopsin turnover normalized to β-actin. (* P < 0.05, one-way ANOVA with Dunnett's multiple comparison post-test, N = 5). (E-G) Flow cytometry analysis of microglia fed with oxLDL. Cells were treated with oxLDL for 30 min (uptake), followed by a 16 h incubation to assess turnover. Quantitative data were presented in (H). Data presented are averages from 4 different experiments (P < 0.05, one-way ANOVA, Dunnett's multiple comparison test).
To further characterize the effects of TGF-β pathway on microglia metabolic activities, we measured glucose- and fatty acid-dependent oxygen consumption rates in cells treated with TGF-β. The results (Figure 4A and 4B) showed that TGF-β treatment reduced fatty acid oxidation (FAO) rates while moderately increased mitochondrial oxidative phosphorylation of glucose. Quantitative RT-PCR analyses showed that TGF-β treatment downregulated the expression of genes crucial for FAO: Cpt1a and Lpl (Figure 4C). These genes encode proteins that facilitate the transport of long-chain fatty acids into mitochondria [30] and the hydrolysis of triglycerides into fatty acids [31, 32], respectively. The effects of TGF-β on lipid metabolism were selective; Cd36, which is involved in lipid intake and lipoprotein production and transport, was downregulated by POS treatment (Figure 4C) [33], but remained unaffected by TGF-β treatment (Figure 4C). MiR-204 KO microglia showed reduced basal expression of both cpt1a and lpl, but their levels were not further decreased in response to TGF-β treatment (Figure 4C). Functionally, TGF-β treatment decreased the turnover rates of rhodopsin in WT microglia (Figure 4D).
miR-204 directly targets Tgfbr2 mRNA. (A) Sequence alignment of miR-204 seed sequence with 3′ UTR of Tgfbr2 mRNA across different species. (B) Schematic representation of the purification of CD11b+ cells from retinal and RPE/choroid tissues using MACS isolation. (C)-(E) Quantitative RT-PCR analysis of Tgfbr2 expression in CD11b+ retinal microglia, Cd11b+ choroidal myeloid cells, and cultured primary retinal microglia. (F) Schematic illustration of the miRNA reporter assay, with 3′ UTR of Tgfbr2 mRNA cloned downstream of a luciferase reporter gene. (G) Dual luciferase reporter assay showing reduced luciferase activity following co-transfection of miR-204 mimics with the reporter construct in primary retinal microglia. Data presented are averages from 3 independent experiments. * P < 0.05, Student's t-test. (H) Dual luciferase reporter assay following co-transfection with miR-204 mimics and reporter plasmids containing either the wile-type (WT) or mutated (Mut) Tgfbr2 3′ UTR. Data presented are averages from 3 independent experiments. * P < 0.05, Mann Whitney test.
Microglial metabolic shift in response to TGF-β. (A) Control or microglia treated with 10 ng/mL TGF-β for 24 h were subjected to Seahorse Fatty Acid Oxidation assay with palmitate-BSA as substrate for mitochondrial oxygen consumption. (B) Quantitative analysis of oxygen consumption rates from the FAO or Mitostress assay of cells treated with or without treatment of TGF-β. (C) quantitative PCR measurement of cpt1a, lpl, and cd36 gene expression in primary retinal microglia, with or without TGF-β treatment. Data presented are averages from 3 independent experiments (P < 0.05, one-way ANOVA, Dunnett's multiple comparison test). (D) Western blot analysis of rhodopsin turnover in cultured primary retinal microglia fed with POS in the presence or absence of TGF-β. (* P < 0.05, Student's t-test. N = 6).
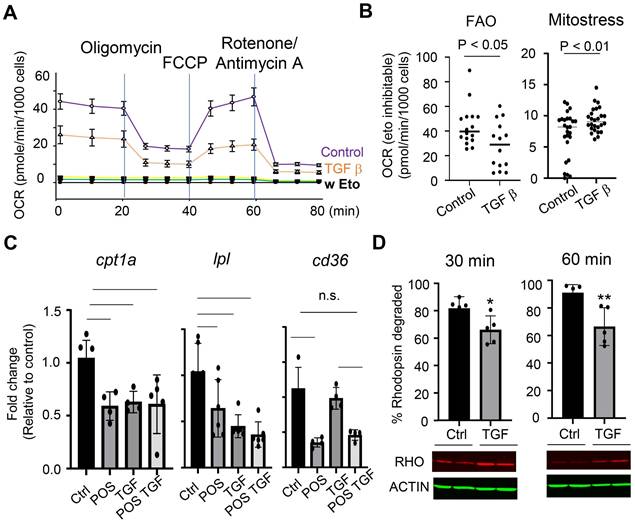
To validate the functional links, we knocked down Tgfbr2 in primary microglial cells using siRNA. Transfection with Tgfbr2 siRNA effectively suppressed TGF-β signaling, as indicated by decreased SMAD2 phosphorylation (Figure 5A). To assess functional consequences, we measured the degradation of phagocytosed POS. Tgfbr2 knockdown led to an accelerated rate of rhodopsin degradation (Figure 5B), indicating enhanced POS turnover. Consistently, miR-204 KO microglia had heightened response to TGF-β, as reflected by increased SMAD2 phosphorylation (Figure 5C). Together with the impaired POS degradation observed in miR-204 KO microglia (Figure 2), these findings suggest that TGF-β signaling suppresses microglial lipid metabolism. MiR-204 can mitigate the inhibitory effects of TGF-β via targeting Tgfbr2 gene.
We have previously demonstrated that EV can mediate the transfer of miRNAs between the RPE and microglia [12], potentially serving as a signaling mechanism between these cells. MiR-204 is the most abundant miRNA species in RPE-derived EVs (Figure S2). To confirm the transfer of the miR-204 from the RPE to microglia via EVs, we employed an ex vivo RPE culture that rapidly released EVs into the conditioned medium (CM) [12, 34, 35]. The RPE/eyecup-CM was fractionated by size exclusion chromatography on a qEV column (Izon Science), and the eluted fractions were examined for the presence of exosome marker proteins TSG101 and FLOT2 [36, 37]. A total of 28 fractions were collected, with EV-enriched fractions identified from 9 to 15 (Figure 6A). Using nanoparticle tracking analyses [13], we quantified that the concentration of EVs was approximately 5x108/mL per eye cup preparation (Figure 6B). These purified RPE EVs were then applied to cultured retinal microglia. Quantitative RT-PCR (Figure 6C) was used to measure the relative level of miR-204 in microglia received EVs. Endogenous level of miR-204 in microglia was significantly elevated by RPE EVs treatment.
To assess the impacts of RPE-derived EVs on retinal function and structural integrity, we performed retro-orbital injections of EVs into miR-204 KO mice. Each eye received 5x108 EV particles in a 100 µL volume, while the contralateral eye received a saline injection as a control. The procedure was repeated one week after the initial injection. The dose is consistent with previously reported dosages used for intraocular injection [38, 39]. ERG c-wave measurement was carried out 7 days after the second injection. The results showed that EV injection effectively improved c-wave responses in eyes when contralateral control eyes had c-wave amplitudes below 400 µV (Figure 7B). However, in instances where control eyes were relatively healthier with c-wave amplitudes exceeding 400 µV, EV treatment reduced the RPE barrier function (Figure 7C). The protective effects were diminished in animals receiving EVs prepared from miR-204 KO mice (Figure 7D and E), supporting the essential roles of miR-204s for the protective functions of RPE-derived EVs. Although EVs delivered by retro-orbital injection to one eye can reach the retina of the contralateral side, their targeting of subretinal microglia was restricted to the injected side (Figure 7A). The outer retina is avascular. The delivery of EV to the subretinal microglia was likely achieved by local diffusion system. This is similar to selective targeting of a specific population of retinal cells reported with retro-orbital injection of adeno-associated virus [40].
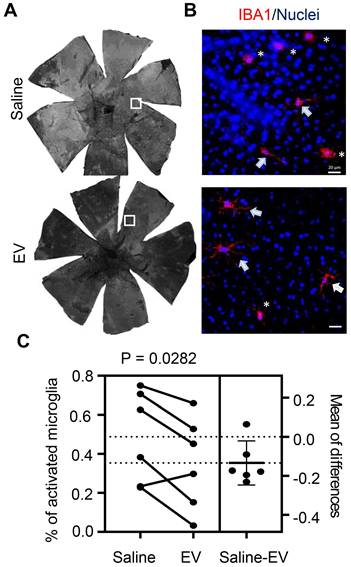
Upon examining the effects of EV treatment on subretinal microglia, we observed a decrease in the percentage of cells with activated morphology compared to those in saline-treated contralateral eyes (Figure 8). Histopathological examination found no difference in eyes with and without EV treatment (Figure S3A). Scotopic and photopic ERG measurements did not identify differences in photoreceptor cell functions after EV treatment (Figure S3B). Retinal thicknesses, as revealed by OCT scan, were not affected by EV (Figure S3C). Thus, EV treatment appears particularly beneficial for eyes with pre-existing RPE injury, likely by promoting inflammation resolution through enhanced lipid metabolic functions in subretinal microglia.
Discussion
In retinal diseases with chronic and low-grade inflammation, such as AMD [41-43], parenchymal microglia migrate from plexiform layers into subretinal space to assist in the removal of debris from the photoreceptor cells and RPE [4]. Subretinal microglia are exposed to a high load of lipid metabolites. However, the mechanisms that preserve their physiological and protective functions of microglia while preventing their overactivation remain largely elusive. The RPE actively participates in immune modulation [44]. MiRNAs encapsulated within RPE-derived EVs can be transferred to microglia [12], acting as a signaling mechanism that regulates gene expression in subretinal microglia.
TGF-β signaling modulates POS degradation in retinal microglia. (A) Western blot analysis of SMAD2 phosphorylation in primary microglia transfected with control (Scr) or Tgfbr2 siRNA and treated with TGF-β (1 ng/mL) (** P < 0.01, Student's t-test. N = 3) (B) Western blot analysis of rhodopsin turnover in primary microglia transfected with Scr or Tgfbr2 siRNA. Rhodopsin (RHO) degradation was assessed 30 and 60 min after POS uptake (* P < 0.05, Student's t-test. N = 6). (C) SMAD2 phosphorylation in primary microglia prepared from miR-204 knockout (KO) and wild-type (WT) mice treated with increasing doses of TGF-β (0, 1, or 10 ng/mL). (*p < 0.05, **p < 0.01; *p < 0.001; one-way ANOVA with Sidak's multiple comparison test; N = 3).
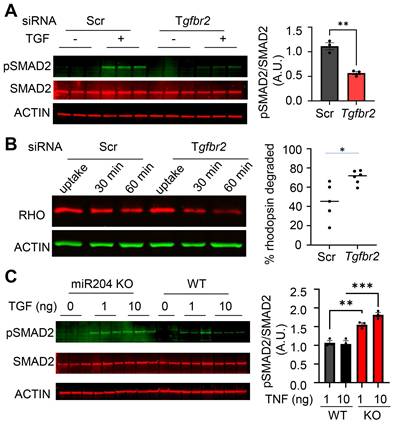
Transfer of miR-204 via RPE-derived EVs. (A) EVs were purified from RPE-conditioned medium through size fractionation. Western blot analysis shows enrichment of EVs markers in selected fractions. (B) Nanosight tracking analysis measured the density and size distribution of purified EVs. (C) Quantitative RT-PCR assessed the level of miR-204 in WT microglia treated with indicated amounts of RPE-produced EVs. Data presented are averages from 3 independent experiments (P < 0.05, one-way ANOVA, Dunnett's multiple comparison test).
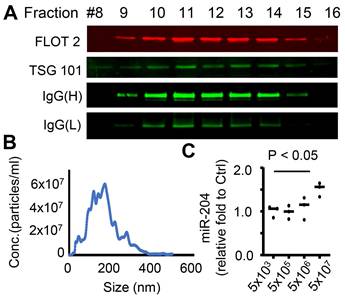
In vivo delivery of RPE EVs by retro-orbital injection. (A) Immunostaining of IBA1+ cells in retina or RPE flat mount from mice with RPE-derived EV injections. White arrows mark EV-containing microglia. (B) and (C) ERG c-wave responses in eyes of miR-204 mice after retro-orbital injection of RPE-derived EVs from wild-type animals. Data presented are comparisons between contralateral eyes from the same animal receiving EVs or Saline. Paired t-tests were used to assess the differences in c-wave responses after EV treatment, with the number of tested animals indicated on the figure. (D) Representative ERG c-wave traces from an animal receiving WT RPE-derived EVs (Ctrl) and miR204 KO RPE-derived EV (miR204 KO) in contralateral eyes. (E) Estimation plot of ERG c-wave amplitudes in animals receiving WT and miR204 KO RPE-derived EVs in contralateral eyes. Paired t-tests, N = 7.
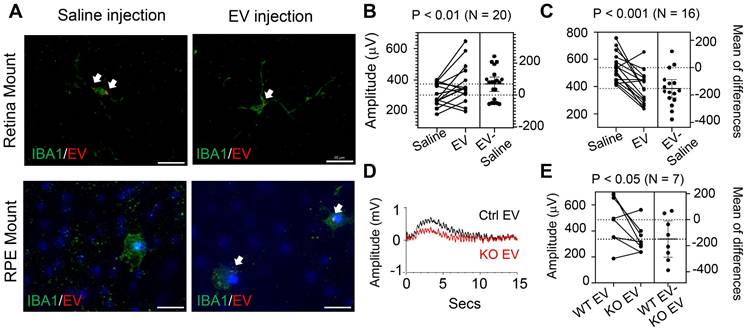
Quantitation of IBA1-positive subretinal microglia after treatment with RPE-derived EVs. (A) Representative images of IBA1-positive cells in whole RPE/choroid mounts from a miR-204 KO mouse at 4 months of age, after receiving EV injection in one eye and saline solution in the contralateral eye. (B) Higher resolution images of the areas boxed in panel A. Arrows indicate resting microglia; asterisks denote activated microglia. Red: IBA1; Blue: Nuclei. Scale bar: 20 µm. (C) Estimation plot showing quantitative results of the percentage of activated microglia under each treatment condition. P value was determined by paired Student's t-test (N = 6).
The intraocular environment is known for its immunosuppressive nature [45], where growth factors and cytokines like TGF-β can dampen both adaptive and innate immune responses [46]. Subretinal microglia, critical for clearing lipid metabolites from the RPE and photoreceptor cells [47], encounter inhibitory factors as they migrate from the inner retina. These factors suppress their metabolic activities, resulting in ineffective inflammation resolution. MiR-204, the most abundant miRNA species in the RPE and RPE-derived EVs [1], increased in microglia following the treatment with RPE-derived EVs (Figure 6C). One of the targets of miR-204 is Tgfbr2. By downregulating this receptor, RPE EVs can alleviate the inhibitory impact of TGF-β on subretinal microglia, which impedes lipid turnover (Figure 2) and utilization (Figure 4). The effects of TGF-β on cellular metabolic functions have been studied extensively in various types of cells and usually are context dependent. In cancer cells and epithelial cells, TGF-β often induces epithelial-mesenchymal transition (EMT) and mitochondrial dysfunction [48]. TGF-β can also regulate many glucose metabolizing enzymes via SMAD-independent non-canonical pathways [11]. In macrophages and dendritic cells, TGF-β promotes lipid droplets (LD) accumulation [49, 50]. The detailed molecular pathways underlying these metabolic effects have not been established. One of our future studies will be to overexpress TGFBR2 in microglia cells and explore the downstream signaling events. With our flow cytometry assays and LipidTox staining, we observed a relatively small percentage of the CD11b+ cells with lipid accumulation (Figure 1E). Such findings suggest that subretinal microglia, rather than the homeostatic macrophages in the choroid [51], are responsible for handling the lipid waste generated by the photoreceptor cells.
When delivered by retro-orbital injection, EVs can quickly reach the retina [52] and are taken up by microglia (Figure 7A). EVs injected into one side of the eye can traverse the other side via systemic circulation, a phenomenon also demonstrated with AAV-mediated, cell type-specific gene expression [40]. However, subretinal microglia, which presumably do not receive supplies from systemic circulation, can uptake these injected EVs through local perfusion. The response between the two eyes were observed to be independent (Figure 7B). Using the contralateral eye as a control, we found that EVs effectively improved ERG c-wave responses only in eyes with existing defects in RPE barrier. Activated myeloid cells have upregulated Fcγ receptors [53]. Similar to previously reported IgG+ exosomes from other sources [54, 55], RPE-produced EVs contain IgG (Figure 6A), which can facilitate the binding to microglia and improves the specificity of cargo delivery. In addition to microglia, RPE can also engulf the EVs. Our study cannot rule out the contribution of RPE engulfed EVs to the protective effects.
A significant challenge in developing EV- and miRNA-based therapies arises from the intricate nature of miRNA-mRNA interactions. A single miRNA species can target several hundred different mRNAs [56]; and it is common for multiple miRNAs to cooperatively target the same gene or genes within the same pathways [57, 58]. EVs in RPE-CM carry numerous miRNA species [12]. Effective interpretation of these “Big Data” requires the development of sophisticated computational methods to study the effects of miRNAs at the genome-wide level.
In conclusion, our studies revealed an important role of miR-204 in regulating retinal microglia lipid metabolism functions. Transferring miR-204 via EVs represents a potential mechanism for intercellular communication between the RPE and subretinal microglia, contributing to a protective immune response following RPE injury.
Abbreviations
CM: Conditioned Medium; DMEM: Dulbecco's Modified Eagle Medium; ERG: Electroretinography; EV: Extracellular Vesicle; FAO: Fatty Acid Oxidation; Flot2: Flotillin-2; KO: Knockout; miRNA: microRNA; POS: Photoreceptor Outer Segments; RPE: Retinal Pigment Epithelium; 3′ UTR: 3′ Untranslated Region.
Supplementary Material
Supplementary figures.
Acknowledgements
The authors wanted to thank Dr. Shelton Miller for providing the miR-204 KO mice. This work was supported by NIH grants R01EY026999 (YC), R01EY 035855 (JC and YL), EY 034742 (JC). The authors would like to acknowledge the Oklahoma University Health Science Center (OUHSC) Vision Research Facilities for the Live Animal Imaging Core, Genotyping Core, Cellular Imaging Core, and Immunology Core services. The core is supported by NIH/NEI grant P30EY027125 to Dr. Michelle C. Callegan and an unrestricted grant from Research to Prevent Blindness to the Dean McGee Eye Institute.
Author Contributions
Y. Chen and J. Cai conceived and designed the research; Y. Chen, SE Bounds, N Kundu, JR Karmoker, D Kim and J. Cai performed the research and acquired the data. Y. Chen, Y Liu and J. Cai analyzed and interpreted the data. Y. Chen and J. Cai drafted the manuscript. All authors edited the manuscript and approved for publication.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Wang FE, Zhang C, Maminishkis A, Dong L, Zhi C, Li R. et al. MicroRNA-204/211 alters epithelial physiology. FASEB J. 2010;24:1552-71
2. Zhang C, Miyagishima KJ, Dong L, Rising A, Nimmagadda M, Liang G. et al. Regulation of phagolysosomal activity by miR-204 critically influences structure and function of retinal pigment epithelium/retina. Hum Mol Genet. 2019;28:3355-68
3. Conte I, Hadfield KD, Barbato S, Carrella S, Pizzo M, Bhat RS. et al. MiR-204 is responsible for inherited retinal dystrophy associated with ocular coloboma. Proc Natl Acad Sci U S A. 2015;112:E3236-45
4. Zhao L, Zabel MK, Wang X, Ma W, Shah P, Fariss RN. et al. Microglial phagocytosis of living photoreceptors contributes to inherited retinal degeneration. EMBO Mol Med. 2015;7:1179-97
5. Gao C, Jiang J, Tan Y, Chen S. Microglia in neurodegenerative diseases: mechanism and potential therapeutic targets. Signal Transduct Target Ther. 2023;8:359
6. Lannes N, Eppler E, Etemad S, Yotovski P, Filgueira L. Microglia at center stage: a comprehensive review about the versatile and unique residential macrophages of the central nervous system. Oncotarget. 2017;8:114393-413
7. Langmann T. Microglia activation in retinal degeneration. J Leukoc Biol. 2007;81:1345-51
8. Yu C, Lad EM, Mathew R, Shiraki N, Littleton S, Chen Y. et al. Microglia at sites of atrophy restrict the progression of retinal degeneration via galectin-3 and Trem2. J Exp Med. 2024 221
9. Taylor AW, Hsu S, Ng TF. The Role of Retinal Pigment Epithelial Cells in Regulation of Macrophages/Microglial Cells in Retinal Immunobiology. Front Immunol. 2021;12:724601
10. Chatterjee A, Singh R. Extracellular vesicles: an emerging player in retinal homeostasis. Front Cell Dev Biol. 2023;11:1059141
11. Liu H, Chen YG. The Interplay Between TGF-beta Signaling and Cell Metabolism. Front Cell Dev Biol. 2022;10:846723
12. Morris DR, Bounds SE, Liu H, Ding WQ, Chen Y, Liu Y. et al. Exosomal MiRNA Transfer between Retinal Microglia and RPE. Int J Mol Sci. 2020;21:3541
13. Mattapallil MJ, Wawrousek EF, Chan CC, Zhao H, Roychoudhury J, Ferguson TA. et al. The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes. Invest Ophthalmol Vis Sci. 2012;53:2921-7
14. Klingeborn M, Dismuke WM, Skiba NP, Kelly U, Stamer WD, Bowes Rickman C. Directional Exosome Proteomes Reflect Polarity-Specific Functions in Retinal Pigmented Epithelium Monolayers. Sci Rep. 2017;7:4901
15. Chen Y, Bounds SE, Ma X, Karmoker JR, Liu Y, Ma JX. et al. Interleukin-17-mediated protective cytokine signaling against degeneration of the retinal pigment epithelium. Proc Natl Acad Sci U S A. 2023;120:e2311647120
16. Shin Y, Moiseyev G, Petrukhin K, Cioffi CL, Muthuraman P, Takahashi Y. et al. A novel RPE65 inhibitor CU239 suppresses visual cycle and prevents retinal degeneration. Biochim Biophys Acta Mol Basis Dis. 2018;1864:2420-9
17. Kang C, Han P, Lee JS, Lee D, Kim D. Anchor, Spacer, and Ligand-Modified Engineered Exosomes for Trackable Targeted Therapy. Bioconjug Chem. 2020;31:2541-52
18. Zhao Z, Liang Y, Liu Y, Xu P, Flamme-Wiese MJ, Sun D. et al. Choroidal gammadelta T cells in protection against retinal pigment epithelium and retinal injury. FASEB J. 2017;31:4903-16
19. Go YM, Zhang J, Fernandes J, Litwin C, Chen R, Wensel TG. et al. MTOR-initiated metabolic switch and degeneration in the retinal pigment epithelium. FASEB J. 2020;34:12502-20
20. Kaurani L, Islam MR, Heilbronner U, Kruger DM, Zhou J, Methi A. et al. Regulation of Zbp1 by miR-99b-5p in microglia controls the development of schizophrenia-like symptoms in mice. EMBO J. 2024;43:1420-44
21. Usoltseva AS, Litwin C, Lee M, Hill C, Cai J, Chen Y. Role of LIPIN 1 in regulating metabolic homeostasis in the retinal pigment epithelium. FASEB J. 2024;38:e70249
22. O'Koren EG, Mathew R, Saban DR. Fate mapping reveals that microglia and recruited monocyte-derived macrophages are definitively distinguishable by phenotype in the retina. Sci Rep. 2016;6:20636
23. Curcio CA, Messinger JD, Sloan KR, McGwin G, Medeiros NE, Spaide RF. Subretinal drusenoid deposits in non-neovascular age-related macular degeneration: morphology, prevalence, topography, and biogenesis model. Retina. 2013;33:265-76
24. Curcio CA, Johnson M, Rudolf M, Huang JD. The oil spill in ageing Bruch membrane. Br J Ophthalmol. 2011;95:1638-45
25. Curcio CA, Johnson M, Huang JD, Rudolf M. Aging, age-related macular degeneration, and the response-to-retention of apolipoprotein B-containing lipoproteins. Prog Retin Eye Res. 2009;28:393-422
26. Jurga AM, Paleczna M, Kuter KZ. Overview of General and Discriminating Markers of Differential Microglia Phenotypes. Front Cell Neurosci. 2020;14:198
27. Gnanaguru G, Choi AR, Amarnani D, D'Amore PA. Oxidized Lipoprotein Uptake Through the CD36 Receptor Activates the NLRP3 Inflammasome in Human Retinal Pigment Epithelial Cells. Invest Ophthalmol Vis Sci. 2016;57:4704-12
28. Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS One. 2018;13:e0206239
29. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015 4
30. Ma X, Wu W, Hara M, Zhou J, Panzarin C, Schafer CM. et al. Deficient RPE mitochondrial energetics leads to subretinal fibrosis in age-related neovascular macular degeneration. Commun Biol. 2024;7:1075
31. Moon JH, Kim K, Choi SH. Lipoprotein Lipase: Is It a Magic Target for the Treatment of Hypertriglyceridemia. Endocrinol Metab (Seoul). 2022;37:575-86
32. Loving BA, Tang M, Neal MC, Gorkhali S, Murphy R, Eckel RH. et al. Lipoprotein Lipase Regulates Microglial Lipid Droplet Accumulation. Cells. 2021 10
33. Terry AR, Nogueira V, Rho H, Ramakrishnan G, Li J, Kang S. et al. CD36 maintains lipid homeostasis via selective uptake of monounsaturated fatty acids during matrix detachment and tumor progression. Cell Metab. 2023;35:2060-76 e9
34. Sanjiv N, Osathanugrah P, Fraser E, Ng TF, Taylor AW. Extracellular Soluble Membranes from Retinal Pigment Epithelial Cells Mediate Apoptosis in Macrophages. Cells. 2021;10:198
35. Locke CJ, Congrove NR, Dismuke WM, Bowen TJ, Stamer WD, McKay BS. Controlled exosome release from the retinal pigment epithelium in situ. Exp Eye Res. 2014;129:1-4
36. Wei D, Zhan W, Gao Y, Huang L, Gong R, Wang W. et al. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. 2021;31:157-77
37. Hoshino A, Kim HS, Bojmar L, Gyan KE, Cioffi M, Hernandez J. et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell. 2020;182:1044-61 e18
38. Mead B, Tomarev S. Bone Marrow-Derived Mesenchymal Stem Cells-Derived Exosomes Promote Survival of Retinal Ganglion Cells Through miRNA-Dependent Mechanisms. Stem Cells Transl Med. 2017;6:1273-85
39. Mead B, Ahmed Z, Tomarev S. Mesenchymal Stem Cell-Derived Small Extracellular Vesicles Promote Neuroprotection in a Genetic DBA/2J Mouse Model of Glaucoma. Invest Ophthalmol Vis Sci. 2018;59:5473-80
40. Toma K, Zhao M, Zhang S, Wang F, Graham HK, Zou J. et al. Perivascular neurons instruct 3D vascular lattice formation via neurovascular contact. Cell. 2024;187:2767-84 e23
41. Xu H, Chen M, Manivannan A, Lois N, Forrester JV. Age-dependent accumulation of lipofuscin in perivascular and subretinal microglia in experimental mice. Aging cell. 2008;7:58-68
42. Luhmann UF, Robbie S, Munro PM, Barker SE, Duran Y, Luong V. et al. The drusenlike phenotype in aging Ccl2-knockout mice is caused by an accelerated accumulation of swollen autofluorescent subretinal macrophages. Invest Ophthalmol Vis Sci. 2009;50:5934-43
43. Zhao Z, Xu P, Jie Z, Zuo Y, Yu B, Soong L. et al. gammadelta T cells as a major source of IL-17 production during age-dependent RPE degeneration. Invest Ophthalmol Vis Sci. 2014;55:6580-9
44. Nussenblatt RB, Lee RW, Chew E, Wei L, Liu B, Sen HN. et al. Immune responses in age-related macular degeneration and a possible long-term therapeutic strategy for prevention. Am J Ophthalmol. 2014;158:5-11 e2
45. Taylor AW. Ocular immune privilege. Eye (Lond). 2009;23:1885-9
46. Batlle E, Massague J. Transforming Growth Factor-beta Signaling in Immunity and Cancer. Immunity. 2019;50:924-40
47. Silverman SM, Wong WT. Microglia in the Retina: Roles in Development, Maturity, and Disease. Annu Rev Vis Sci. 2018;4:45-77
48. Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156-72
49. Bose D, Banerjee S, Chatterjee N, Das S, Saha M, Saha KD. Inhibition of TGF-beta induced lipid droplets switches M2 macrophages to M1 phenotype. Toxicol In Vitro. 2019;58:207-14
50. Trempolec N, Degavre C, Doix B, Brusa D, Corbet C, Feron O. Acidosis-Induced TGF-beta2 Production Promotes Lipid Droplet Formation in Dendritic Cells and Alters Their Potential to Support Anti-Mesothelioma T Cell Response. Cancers (Basel). 2020;12:1284
51. Fortmann SD, Frey BF, Rosencrans RF, Adu-Rutledge Y, Ready VE, Kilchrist KV. et al. Prenatally derived macrophages support choroidal health and decline in age-related macular degeneration. J Exp Med. 2025;222:e20242007
52. Hajrasouliha AR, Jiang G, Lu Q, Lu H, Kaplan HJ, Zhang HG. et al. Exosomes from retinal astrocytes contain antiangiogenic components that inhibit laser-induced choroidal neovascularization. J Biol Chem. 2013;288:28058-67
53. Guilliams M, Bruhns P, Saeys Y, Hammad H, Lambrecht BN. The function of Fcgamma receptors in dendritic cells and macrophages. Nat Rev Immunol. 2014;14:94-108
54. Huang C, Fisher KP, Hammer SS, Navitskaya S, Blanchard GJ, Busik JV. Plasma Exosomes Contribute to Microvascular Damage in Diabetic Retinopathy by Activating the Classical Complement Pathway. Diabetes. 2018;67:1639-49
55. Couto N, Elzanowska J, Maia J, Batista S, Pereira CE, Beck HC. et al. IgG(+) Extracellular Vesicles Measure Therapeutic Response in Advanced Pancreatic Cancer. Cells. 2022;11:2800
56. Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58-63
57. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597-610
58. Sass S, Dietmann S, Burk UC, Brabletz S, Lutter D, Kowarsch A. et al. MicroRNAs coordinately regulate protein complexes. BMC Syst Biol. 2011;5:136
Author contact
![]() Corresponding author: Jiyang Cai, Department of Biochemistry & Physiology, University of Oklahoma Health Sciences Center, 940 Stanton L Young Boulevard, BMSB 955, Oklahoma City, OK 73104. Tel.: (405) 271-2226, Ext. 31896; Email: jiyang-caiedu.
Corresponding author: Jiyang Cai, Department of Biochemistry & Physiology, University of Oklahoma Health Sciences Center, 940 Stanton L Young Boulevard, BMSB 955, Oklahoma City, OK 73104. Tel.: (405) 271-2226, Ext. 31896; Email: jiyang-caiedu.
 Global reach, higher impact
Global reach, higher impact