13.3
Impact Factor
Theranostics 2026; 16(1):156-192. doi:10.7150/thno.117085 This issue Cite
Review
Advancements in CRISPR-based therapies for ocular pathologies: from disease mechanisms to intervention strategies
1. State Key Laboratory Cultivation Base, Shandong Provincial Key Laboratory of Ophthalmology, Eye Institute of Shandong First Medical University, Qingdao 266071, China.
2. Eye Hospital of Shandong First Medical University (Shandong Eye Hospital), Jinan 250021, China.
3. School of Ophthalmology, Shandong First Medical University, Jinan 250001, China.
4. State Key Laboratory of Structural Chemistry, Fujian Institute of Research on the Structure of Matter, Chinese Academy of Sciences, Fuzhou 350002, China.
5. Department of Pharmaceutics, Wuya College of Innovation, Shenyang Pharmaceutical University, Shenyang 110016, China.
6. Departments of Diagnostic Radiology, Surgery, Chemical and Biomolecular Engineering, and Biomedical Engineering, Yong Loo Lin School of Medicine and College of Design and Engineering, National University of Singapore, Singapore 119074, Singapore.
# These authors contributed equally to this work.
Received 2025-5-7; Accepted 2025-8-31; Published 2026-1-1
Abstract
Eye diseases caused by genetic mutations affect over 2.2 billion people worldwide. The development of CRISPR technology has opened exciting possibilities for how we diagnose and treat these conditions. However, designing effective CRISPR systems, managing potential risks, and considering the ethical questions around gene therapy in clinical practice are major challenges. To move forward successfully, it's important to evaluate how practical CRISPR-based treatments are for eye diseases from a clinical perspective, while also understanding how CRISPR systems work. In this review, we start by covering the basic principles behind CRISPR technology and explore its different types. Next, we look at various ways CRISPR is being used in eye research and treatments, from early studies to new clinical approaches. Lastly, we address the regulatory environment and ethical issues involved, discussing existing rules, safety concerns, and guidelines for genetic modifications in medical settings. Our goal is to share new insights into innovative treatments for eye diseases and to support the safe use of CRISPR in clinical eye care. This review aims to be a helpful resource for researchers, doctors, and regulators working on CRISPR-based therapies.
Keywords: CRISPR gene therapy, eye diseases, ocular immune privilege, genetic modification, clinical application
Introduction
The human eye is one of the body's most detailed organs, tasked with transmitting visual information to the brain. An estimated about 27% of people worldwide suffer from various eye conditions, with around 2.2 billion individuals experiencing some forms of visual impairment or blindness [1]. Sight-threatening ocular diseases primarily include hereditary diseases such as corneal dystrophy, aniridia, Leber congenital amaurosis (LCA), and retinitis pigmentosa (RP), and multifactorial diseases like corneal scarring, glaucoma, and age-related macular degeneration (AMD) [2]. With in-depth investigation into disease mechanisms, an increasing number of specific gene mutations associated with ocular diseases have been identified, serving either as genetic risk factors or pathogenic variants. These diseases have long lacked effective curative treatments, while gene therapy designed to directly repair or compensate for defective genes holds promise for achieving a one-time cure [3]. The eye's unique immune privilege makes it an ideal target for gene therapy. The clinical evidence for this privileged status is primarily reflected in the high success rate of corneal transplantation and the feasibility of experimental tissue grafting into the eye, indicating a relatively tolerant ocular immune system toward foreign vectors [4]. Anatomically, the corneal and blood-retinal barrier (BRB) confine delivery vectors within the eye, allowing local administration of gene therapy medicines to limit widespread side effects [5]. Plus, the transparent nature of the eye makes it easier to monitor treatment progress directly, which is highly beneficial for clinical applications [6].
Gene therapy has achieved revolutionary breakthroughs in the field of ophthalmology [7, 8]. For instance, gene augmentation therapy represented by Luxturna delivers functional copies of the RPE65 gene to retinal cells via subretinal injection, restoring the visual cycle and improving visual function in patients [9]. However, current gene replacement therapy is limited to autosomal recessive genetic disorders. When facing autosomal dominant mutations (such as autosomal dominant RP, aniridia) and multifactorial diseases (AMD, glaucoma), simply supplementing a normal gene copy cannot overcome the toxic effects of the original mutant protein [10]. Consequently, gene therapy is rapidly evolving towards more complex and precise approaches.
Thanks to advances in genetic engineering-most notably the discovery of Clustered regularly interspaced short palindromic repeats (CRISPR) and its associated proteins (Cas)-there has been a major shift in how ophthalmic conditions are researched and treated [11, 12]. Originally identified as part of bacterial immune defense [13], CRISPR/Cas system has quickly become a groundbreaking tool for genome editing because of its high precision and adaptability [14]. CRISPR/Cas9 is the most renowned and widely utilized version of CRISPR tools. Its core components consist of the Cas9 protein and a single guide RNA (sgRNA), which incorporates both the trans-activating CRISPR RNA (tracrRNA) and CRISPR RNA (crRNA) [15]. The sgRNA directs the Cas9 protein to bind and cleave the target DNA sequence by recognizing a protospacer adjacent motif (PAM) located downstream of the target genomic site, generating double-strand breaks (DSBs) [16]. Then, the cellular intrinsic genome repair mechanisms-non homologous end joining (NHEJ) and homology directed repair (HDR)-are activated to achieve the insertion, deletion, or replacement of the target sequence [16].
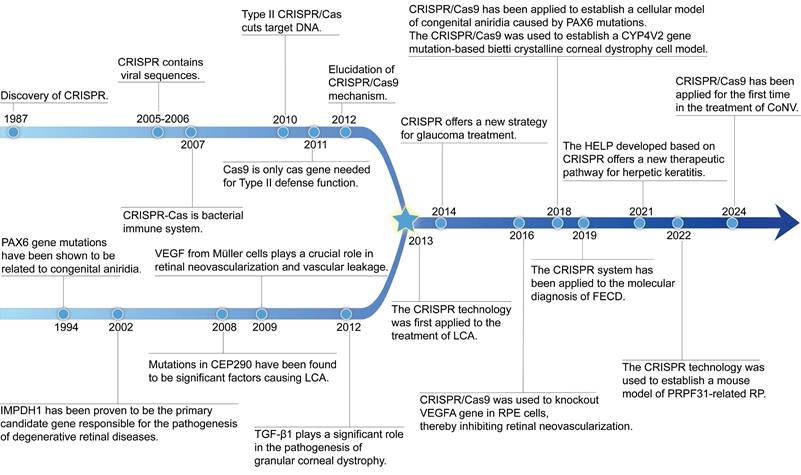
Since Doudna and Charpentier's pioneering work in 2012 demonstrated CRISPR's effectiveness in mammalian cells, this technology has become essential for revealing disease mechanisms, studying genes linked to eye conditions, and creating experimental models [17, 18] (Figure 1). A notable milestone was reached in 2020 when the first in vivo application of CRISPR/Cas9 in LCA10 patients was performed, involving subretinal delivery targeting CEP290 mutations [19]. This demonstrated that gene editing directly within the eye can be a feasible therapeutic strategy. Beyond hereditary diseases, CRISPR also shows promise for multifactorial diseases caused by infections, immune responses, or abnormal vessel growth. For instance, using CRISPR to target herpes simplex virus (HSV) DNA in viral keratitis to interrupt viral replication and lower recurrence risks [20]. CRISPR gene editing technology overcomes the limitation of conventional gene replacement therapy, which only target autosomal recessive disorders, and significantly expanding the scope of gene therapy applications [21]. Compared to gene augmentation therapies relying on transgenic expression, CRISPR/Cas achieves precise in situ edit of target genes, eliminating potential risks of non-physiological expression associated with exogenous therapeutic genes [22].
Despite promising results seen in early studies and laboratory experiments, there are still several major challenges to overcome before CRISPR can be widely used in clinics [23, 24]. Issues such as unintended effects on other parts of the genome, challenges in delivering the editing tools effectively, improving how well the editing works, and ensuring safety over the long term are all critical concerns. This tension between the exciting potential to treat diseases and the need to address safety and ethical concerns emphasizes the necessity for careful oversight and thorough scientific evaluation when applying CRISPR in eye health. Therefore, conducting a detailed and honest review of how CRISPR technology might be used to treat eye diseases is essential to move forward safely and responsibly.
In this review, we take a close look at the latest developments, the healing possibilities, and the challenges involved in using gene editing with CRISPR in eye medicine (Table 1). We also discuss how current rules and future pathways could shape the field. This review is intended as a helpful resource for researchers, ophthalmologists, and regulators involved in advancing and applying CRISPR-based treatments in ophthalmology. By offering this thorough overview, we hope to provide fresh perspectives on treating eye diseases and support the safe development of CRISPR technology in clinical practice.
Classification and Mechanisms of CRISPR Gene Editing Systems
The CRISPR system, first identified in Escherichia coli, is a highly advanced immune mechanism found in bacteria and other prokaryotes [25]. It helps defend against invading genetic materials, such as viruses called bacteriophages or foreign DNA segments [26]. This powerful defense involves two main parts, each with its own structure and function [27]. The first component consists of characteristic DNA elements featuring short palindromic repeat sequences of 20-40 base pairs (bp), namely CRISPR. The second part includes the Cas genes and the proteins they produce.
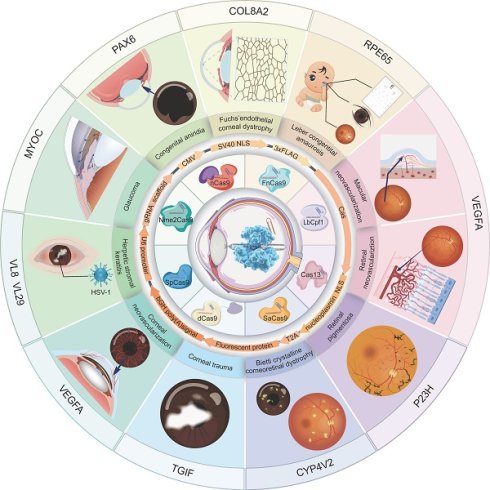
The review of the history of CRISPR system development, the discovery journey of genetic loci for eye diseases, and the advancement of CRISPR treatment in eye diseases.
Applications of CRISPR/Cas systems in ocular diseases.
| Disease | Target | Executor | Vector | Delivery | Model | Strategy | Key findings | Ref |
|---|---|---|---|---|---|---|---|---|
| Anterior eye disease | ||||||||
| Corneal scar and corneal wound healing | Sox2 | dCas9 | Nake Plasmid | Intracameral injection | Sprague-dawley rats | Gene activation | Activating SOX2 using CRISPR/Cas9 technology can achieve functional regeneration of the corneal endothelium. | [75] |
| Aqp5 | Cas9 | Not mentioned | Not mentioned | AQP5 knockout (AQP5-/-) mice | Knockout | AQP5 promotes corneal epithelial wound healing and nerve regeneration by activating the NGF/Akt signaling pathway. | [76] | |
| Tgif1 | Cas9/ dCas9 | Not mentioned | Chemical Transfection | Human corneal stromal fibroblasts | Knockout and activation | CRISPR-mediated editing of the TGIF1 gene activation inhibits the conversion of corneal fibroblasts to myofibroblasts. | [77] | |
| HSK | ICP0 | SpCas9 | Lentiviral particles | lentivirus transduction | L7 cell | Knockout | CRISPR/Cas9 targets and cleaves the ICP0 gene, suppressing ICP0 expression to inhibit viral replication and protect cells from HSV-1 infection. | [85] |
| ICP0 & ICP4 | SpCas9/SaCas9 | AAV1 | Adeno-associated virus transduction | Primary TG neuronal | Knockout | AAV1-mediated SaCas9 gene editing can effectively inhibit HSV-1 replication in trigeminal ganglion neurons by targeting the ICP4 gene. | [86] | |
| UL8 &UL29 | SpCas9 | HELP | Intrastromal injection | HSV-1 infected mouse HSK model | Knockout | HELP effectively suppresses HSV-1 replication in mouse corneas and trigeminal ganglia by targeting the UL8 and UL29 genes. | [20] | |
| NECTIN-1 | Cas9 | Lentiviral particles | Not mentioned | HCECs | knockdown | After knocking down NECTIN-1 in HCECs via the CRISPR/Cas9 system, the HSV-1 DNA content dropped to 30% of the control group's level. | [88] | |
| Corneal dystrophy | TGFBI | Cas9 | Plasmid | Transfection | Human corneal keratocytes derived from a GCD2 patient | HDR | CRISPR/Cas9-mediated HDR successfully corrected the TGFBI R124H mutation. | [94] |
| Col8a2 | SpCas9 | Adenovirus | Anterior chamber injection | Col8a2 mutant mice | knockdown | A single intraocular injection of Ad-Cas9-Col8a2gRNA efficiently prevented endothelial cell loss, rescued corneal endothelium pumping function in adult Col8a2 mutant mice. | [96] | |
| CoNV | Vegfa | SpCas9 | AAV2/9 | Subconjunctival injection | Suture-induced mouse CoNV model | Knockout | AAV2/9-mediated CRISPR/Cas9 knockout of the VEGFA gene significantly inhibits pathologic CoNV. | [99] |
| Midsection eye disease | ||||||||
| Glaucoma | MYOC | Cas9 | Adenovirus | Intravitreal injection | Tg-MYOCY437H mice | Knockout | Using CRISPR/Cas9 to disrupt the mutant MYOC gene reduces endoplasmic reticulum stress, lowers intraocular pressure, and prevents further glaucomatous damage in mouse eyes. | [46] |
| Car2 | SpCas9 | AAV ShH10 | Intravitreal injection | Glaucoma mice model | Knockout | Using CRISPR/Cas9 to disrupt the Car2 gene shows promise for achieving the goal of “one-shot treatment, lifelong benefits” for glaucoma. | [105] | |
| Congenital aniridia | Pax6 | SpCas9 | Not mentioned | Cotransfection | Human limbal epithelial cells | Knock-in | The study successfully modeled aniridia-related keratopathy in vitro using CRISPR/Cas9 gene editing. | [108] |
| Pax6 | SpCas9 | Not mentioned | Electroporation/microinjection | Sey mouse model/Fey mouse model | Germline correction | Germline CRISPR/Cas9-mediated gene editing prevents vision loss in a Fey mouse model of Aniridia | [109] | |
| Pax6 | ABE8e | LNP | Not mentioned | Fey embryonic mouse cortical neurons | Base editing | ABE8e restored 24.8% of Pax6 expression in ex vivo neurons, laying the foundation for translating CRISPR therapy to human aniridia patients. | [110] | |
| Posterior eye disease | ||||||||
| LCA | CEP290 | SpCas9 | AAV5 | Subretinal injection | Wild-type mice | Knockout | AAV5-CRISPR/Cas9 system can delete the homologous CEP290 intronic fragment in wild-type mice retinas via subretinal injection. Also, a self-limiting CRISPR/Cas9 system is developed to restrict the sustained expression of SpCas9. | [115] |
| CEP290 | SaCas9 | AAV5 | Subretinal injection | humanized CEP290 mice | Knockout | Subretinal delivery of EDIT-101 in humanized CEP290 mice showed rapid and sustained CEP290 gene editing. | [170] | |
| Kcnj13 | ABE8e | Silica nanocapsules | Subretinal injection | LCA16 mouse model | Base editing | Using Silica nanocapsules to deliver the ABE8e can efficiently correct the W53X mutation in KCNJ13, preserve vision in the LCA16 mouse model, and restore Kir7.1 channel function in RPE cells. | [118] | |
| Rpe65 | SpCas9 | AAV9 | Subretinal injection | Rd12 mice model | HDR | Dual AAV9-delivered CRISPR/Cas9 can achieve HDR-mediated correction and pathogenic mutation deletion in the rd12 mice model. | [121] | |
| Rpe65 | ABE | Lentiviral particles | Subretinal injection | Rd12 mice model | Base editing | ABE corrects the RPE65 mutation in rd12 mice via subretinal injection, restoring its function and visual function to near-normal levels with minimal off-target and indel mutations. | [123] | |
| RP | Rho | SaCas9 | AAV2/8 | Subretinal injection | RHO humanized mouse model | Knockout | AAV2/8-CRISPR/SaCas9 system specifically target and knockout the T17M mutant allele of the RHO gene, significantly improving retinal function and protecting photoreceptors in RHO humanized mice. | [132] |
| Rho | SaCas9 | AAV2/8 | Subretinal injection | P23H rat model | Knockout | AAV2/8-delivered SaCas9/gRNA selectively ablates the RHO-P23H mutant allele, preserves long-term vision in P23H rat. | [133] | |
| Rho | SpCas9 | Nake Plasmid | Subretinal injection | P23H mutant mouse model | Knockout | CRISPR/Cas9 achieves efficiently edit the mutant RHO gene in P23H mouse retinas. | [134] | |
| Rho | SpCas9 | AAV2/8 | Subretinal injection | Humanized hRHOC110R/hRHOWT mice model | Ablation and replacement | CRISPR-based ablation and replacement strategy via dual AAV2/8 in humanized adRP mice efficiently knocks out mutant RHO and introduces wild-type genes. | [137] | |
| Rho | SaCas9 | AAV9 | Subretinal injection | Rho-associated adRP mouse model | Reduction and replacement | CRISPR/SaCas9-mediated reduction and replacement strategy effectively restores retinal electrophysiological function in adRP mouse model. | [138] | |
| Rho | dCas9 | AAV5 | Subretinal injection | Transgenic Pro23His mutant pigs | CRISPRi | The AAV5-delivered CRISPRi system targets the RHO promoter, reducing mutant RHO protein expression, decreasing endoplasmic reticulum stress and apoptosis markers, and preserving retinal function. | [139] | |
| Prpf31 | SpCas9 | AAV2-7m8 | Subretinal injection | hiPSC-derived RPE cells and retinal organoids | Gene augmentation | PRPF31 mutations lead to structural and functional defects in hiPSC-derived RPE and photoreceptor cells, while AAV-mediated PRPF31 gene augmentation restores cellular morphology, improves electrophysiological responses. | [141] | |
| PHD2 | SpCas9 | AAV8 | Subretinal injection | RP mice model | Knockout | CRISPR editing of PHD2 can enhance retinal aerobic glycolysis, reduce mitochondrial oxidation, maintain photoreceptor survival and function in both autosomal recessive and dominant RP mouse models. | [142] | |
| BCD | Cyp4v2 | SaCas9 | rAAV2/8 | Subretinal injection | Humanized Cyp4v3 mutant mice | HITI | The CRISPR/SaCas9-HITI system delivered by rAAV2/8 achieves precise integration of CYP4V2 in hCyp4v3mut/mut mice, improves retinal morphology and electrophysiological responses. | [50] |
| STGD1 | Abca4 | ABE | AAV9 | Subretinal injection | Female cynomolgus macaques | Base editing | An ABE strategy achieves high-level ABCA4 gene correction in nonhuman primates, with mean editing rates of 75% in cone photoreceptors and 87% in RPE cells. | [147] |
| Retinal neovascularization | Vegfa | Cas13bt3 | AAV2-7m8 | Intravitreal injection | Mouse model of proliferative retinopathy | RNA silencing | AAV2-7m8 delivered CRISPR/Cas13bt3 efficiently silences VEGFA mRNA, reducing retinal neovascular leakage and vessel density. | [155] |
| Vegfr2 | SpCas9 | AAV2/8 | Intravitreal injection | OIR mouse model | Knockout | AAV2/8-mediated CRISPR/SpCas9 editing of the VEGFR2 gene effectively suppressed pathological retinal angiogenesis in OIR mouse model. | [157] | |
| Vegfr2 | PE | dual non-integrating lentivirus | Intravitreal injection | OIR mouse model | Prime editing | The PE6x system efficiently edits the VEGFR2 gene, generating DN-VEGFR2 to inhibit pathological retinal angiogenesis. | [158] | |
| CNV | Vegfa/Hif1α | LbCpf1 | AAV9 | Intravitreal injection | Laser-induced CNV mice model | Knockout | AAV9-delivered LbCpf1 targeting Vegfa or Hif1a significantly reducing the area of laser-induced CNV. | [161] |
| Vegfa/Hif1α/Vegfr2 | Nme2Cas9 | AAV8 | Subretinal injection | Laser-induced CNV mouse model | Knockout | AAV8-delivered Nme2Cas9 efficiently edits target genes, with early intervention targeting Vegfa significantly reducing CNV area by 49.5%. | [162] | |
| Vegfa | SpCas9 | Lentivirus | Subretinal injection | Laser-induced CNV mouse model | Knockout | The RPE-specific CRISPR/pVMD2-Cas9 system efficiently knockout Vegfa, leading to significant regression of laser-induced CNV in mice. | [163] | |
| RB | Rb1/ Rbl1 | SpCas9 | Not mentioned | Microinjection | Xenopus tropicalis | Knockout | CRISPR/Cas9-mediated knockout of rb1 and rbl1 in Xenopus tropicalis tadpoles leads to rapid and highly penetrant RB development, recapitulating human histopathological features, thus providing an efficient preclinical model. | [167] |
| Rb1 | SpCas9 | Not mentioned | Electroporation | Teratoma model mimicking TRb neural tumors | Knockout | CRISPR/Cas9-mediated knockout of RB1 leads to mitochondrial dysfunction, with teratomas recapitulating TRb-like neural expansion, providing a novel model for TRb. | [169] | |
Abbreviations: CRISPR: clustered regularly interspaced short palindromic repeats; Cas: CRISPR associated; NGF: nerve growth factor; TG: trigeminus; AAV: adeno-associated virus; HELP: HSV-1-erasing lentiviral particles; HSK: herpetic stromal keratitis; HSV: herpes simplex virus; HDR: homology-directed repair; hCECs: human corneal endothelial cells; GCD2: granular corneal dystrophy2; Ad: adenovirus; CoNV: corneal neovascularization; VEGF: vascular endothelial growth factor; ABE: adenine base editor; PE: prime editor; LCA: leber congenital amaurosis; RP: retinitis pigmentosa; CRISPRi: CRISPR interference; BCD: bietti crystalline corneoretinal dystrophy; HITI: homology independent targeted integration; RPE: retinal pigment epithelium; CNV: choroidal neovascularization; DN-VEGFR2: dominant-negative VEGF receptor 2; RB: retinoblastoma; TRb: trilateral retinoblastoma; STGD1: stargardt disease.
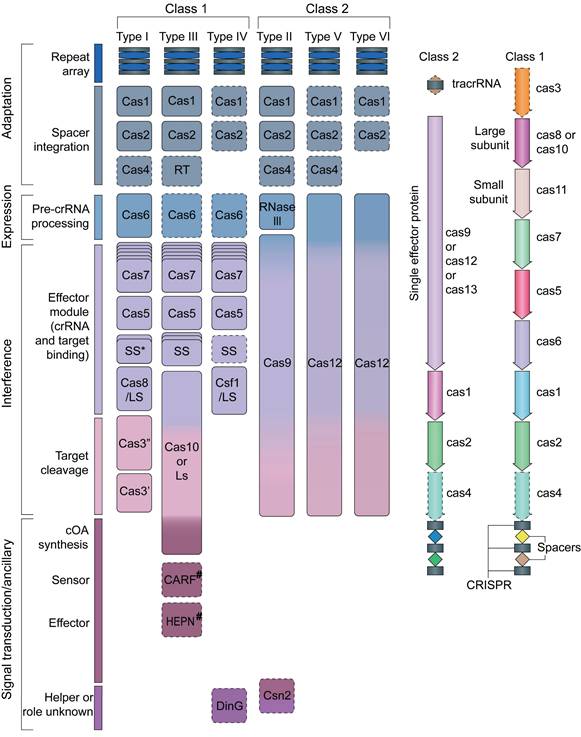
Modern classification divides CRISPR systems into two main types, with six subtypes (Figure 2) [28]. Understanding these differences helps scientists grasp the wide range of functions and uses of CRISPR-Cas systems. This detailed molecular knowledge has changed how we manipulate genes and develop new treatments. Today, CRISPR-Cas tools are central to modern biology and medicine, opening up exciting possibilities for research and therapy.
Class 1 CRISPR-Cas Systems
Class 1 CRISPR-Cas systems (comprising Types I, III, and IV) are characterized by their unique multisubunit effector complexes, which enable precise gene editing through careful interactions between proteins and nucleic acids [29]. However, Class 1 CRISPR-Cas systems, due to their reliance on multi-subunit complexes composed of multiple Cas proteins, have long been considered to face numerous challenges in practical applications:
(1). Inefficient assembly of multi-subunit complexes causes unstable editing efficiency. Type I systems require Cas1-Cas2 mediated spacer acquisition, and the Cascade complex participates in the interference stage [30]. This complexity makes system simplification and application more challenging.
(2). The effector complexes of Class 1 systems typically consist of 4-5 protein subunits, with a total gene size generally exceeding 3.8 kb (e.g., Type I-E reaches 4.4 kb), limiting their application in in vivo gene therapy [31].
(3). Class 1 systems exhibit higher uncontrollability. For instance, Type III-E systems consume ATP and accumulate toxic ITP, which may lead to cellular metabolic disturbances [32].
Due to the above-mentioned reasons, no related literature on the application of Class 1 CRISPR-Cas systems in ophthalmology has been reported to date.
Class 2 CRISPR-Cas Systems
Compared with Class 1 systems, Class 2 systems have significantly greater advantages in ocular pathologies research due to their structural simplicity, operational convenience, and extensive applicability [33]. Class 2 systems only require a single effector protein (e.g., Cas9, Cas12a, Cas13a) to bind with gRNA to function, offering simpler operation and higher editing efficiency [34]. Then, Class 2 effector proteins have a compact size, enabling compatibility with virus vectors for in vivo delivery [35]. Additionally, the simple structure of Class 2 systems facilitates engineering modifications to enhance safety [36].
Class 2 systems include three main types-Type II, Type V, and Type VI-each with its own unique way of working and special features.
The Type II CRISPR-Cas System
The Type II CRISPR-Cas system, often called CRISPR/Cas9, is the most well-known and widely used version of the CRISPR-Cas tools [37] (Figure 3A). As a groundbreaking gene editing technology, CRISPR/Cas9 originates from the natural immune mechanism of prokaryotic organisms (Figure 3B). It mainly involves the Cas9 protein, along with a gRNA integrating tracrRNA with crRNA [15]. Cas9 acts as the main protein responsible for cutting DNA [38]. The crRNA, which is produced from CRISPR sequences, guides Cas9 to the specific DNA target, while tracrRNA binds to crRNA's repeat regions and to Cas9 to help in maturing the RNA and enabling DNA cutting [39]. The PAM sequence is a short DNA motif located just downstream of the target DNA sequence, plays an essential role in helping Cas9 recognize its specific target [40]. When the gRNA pairs with the target DNA, Cas9 creates an R-loop structure that activates its two main nuclease domains: HNH and RuvC [41]. The HNH domain cuts the DNA strand complementary to the guide, while RuvC cuts the opposite strand, resulting in a double-strand break (DSBs). The cell then repairs this break either through nonhomologous end joining (NHEJ) or homology directed repair (HDR) [42] (Figure 3C-D).
Schematic diagram of the classification of CRISPR/Cas gene editing systems. Adapted with permission from [28], copyright 2019, the authors.
The NHEJ repair is currently the primary pathway for gene knockout and has been widely applied in both hereditary ophthalmic diseases and multifactorial diseases [44]. The NHEJ repair pathway directly ligates broken DNA, bypassing the time-consuming template search step required in homologous recombination. This process introduces insertions or deletions (indels) that frequently cause frameshift mutations, thereby deactivating genes with dominant-negative gain-of-function mutations [45]. For instance, CRISPR/Cas9 exploits NHEJ pathway to disrupt the glaucoma-associated MYOC gene and lower intraocular pressure [46]. Unlike NHEJ, HDR requires the provision of an exogenous homologous repair template to guide the cell for precise repair [47]. However, HDR mainly occurs during the cell division phase (S/G2 phase), and its editing efficiency is usually below 20%, which greatly limits its application in in vivo gene editing [47].
Homology-Independent Targeted Integration (HITI) is another breakthrough CRISPR/Cas9-mediated gene editing technology that overcomes the cell cycle dependency of traditional HDR, allowing efficient and precise gene knock in in both dividing and non-dividing cells (such as retinal cells) [48]. Its core principle is to utilize the NHEJ pathway to achieve targeted integration of exogenous DNA. After Cas9 cleaves both the target gene and the donor DNA, the cellular NHEJ machinery integrates the donor DNA into the target locus during DSBs repair [48]. HITI, leveraging its cell cycle independent high efficiency integration capability to directly supplement the corresponding wild-type gene fragment, may become a transformative tool for ophthalmic gene therapy, particularly for inherited retinal diseases (IRDs) [49]. Utilizing HITI technology to repair the CYP4V2 gene mutation in a humanized CYP4V2 mutant mouse model has achieved precise DNA repair and functional protein expression, offering potential for the treatment of Bietti crystalline dystrophy [50].
Moreover, the CRISPR/Cas9 system has also given rise to several gene editing tools:
(1). The Adenine base editor (ABE) is made by fusing nuclease-deficient Cas proteins such as nickase Cas9 (nCas9) with adenine deaminase [51]. Under the guidance of sgRNA, nCas9 binds to and nicks a single strand of the target DNA. Adenine deaminase then converts the target adenine (A) to inosine (I), which pairs like guanine (G), achieving an A-to-G conversion [52]. Like ABE in structure and editing, cytosine base editor (CBE) is formed by fusing nCas9 with cytosine deaminase [51]. It converts target cytosine (C) to uracil (U), which is later changed to thymine (T), thus achieving a C-to-T conversion. Base Editor precisely edits adenine bases at specific genomic sites without causing DSBs and with high specificity and low off-target effects [53].
(2). Prime editor (PE) consists of a reverse transcriptase fused to a Cas9 nickase and a prime editing guide RNA (pegRNA) containing both a gRNA and a template RNA [54]. pegRNA guides the prime editing protein to the target site. The Cas9 nickase then creates a single strand break on the target DNA strand. Subsequently, the reverse transcriptase initiates reverse transcription using the pegRNA as a template and directly polymerizes the DNA product onto the nicked target DNA strand [55]. PE overcomes the limitations of traditional CRISPR/Cas9 and base editors, enabling targeted insertions, deletions, and all 12 types of base substitutions without the need for DSBs or donor DNA templates [56]. Due to these advantages, PE has shown great application potential in the field of ophthalmology, especially in the treatment of IRDs. A recently developed PE system effectively corrected the PDE6B Y347X mutation associated with RP, successfully restored PDE6B protein expression, and significantly improved retinal function [57].
(3). CRISPR/dCas9 is a gene editing tool derived from the CRISPR/Cas9 system [58]. The dCas9 protein is engineered through mutagenesis of key amino acid residues in Cas9, resulting in loss of DNA cleavage activity while retaining DNA-binding capability [59]. By fusing dCas9 to transcriptional activators (e.g., VPR, p65, RTA) or repressors (e.g., KRAB domain), CRISPR/dCas9 enables targeted activation or suppression of gene expression [60]. After dCas9 binds to the gene promoter or enhancers, activators can recruit the transcription machinery, to promote gene transcription (CRISPR activation, CRISPRa); whereas repressors sterically hinder RNA polymerase binding/elongation, suppressing gene transcription (CRISPR interference, CRISPRi) [61]. For instance, dCas9 fused to the VPR showed effective transcriptional activation and long-term expression of cone photoreceptor-specific M-opsin in rhodopsin-deficient mouse models of RP [62]. Diverging from DSB-dependent CRISPR/Cas9 editing, the CRISPR/dCas9 platform introduces no permanent genetic modifications and allows reversible toggling of gene expression states while preserving genomic integrity. Collectively, CRISPR/dCas9 provides a safer and more precise approach to transcriptional regulation.
The mechanism of action and related structures of CRISPR/Cas9: (A) The overall structure of the Cas9-sgRNA-DNA three-dimensional system. Adapted with permission from [41], copyright 2014, Elsevier Inc. (B) DNA interference in CRISPR-Cas9-mediated bacterial adaptive immunity. Adapted with permission from [16], copyright 2014, Elsevier Inc. (C) Schematic diagram of the mechanism of CRISPR/Cas9-mediated target DNA recognition and cleavage. Adapted with permission from [39], copyright 2017 by Annual Reviews. (D) The general mechanism of CRISPR/Cas9 genome editing.
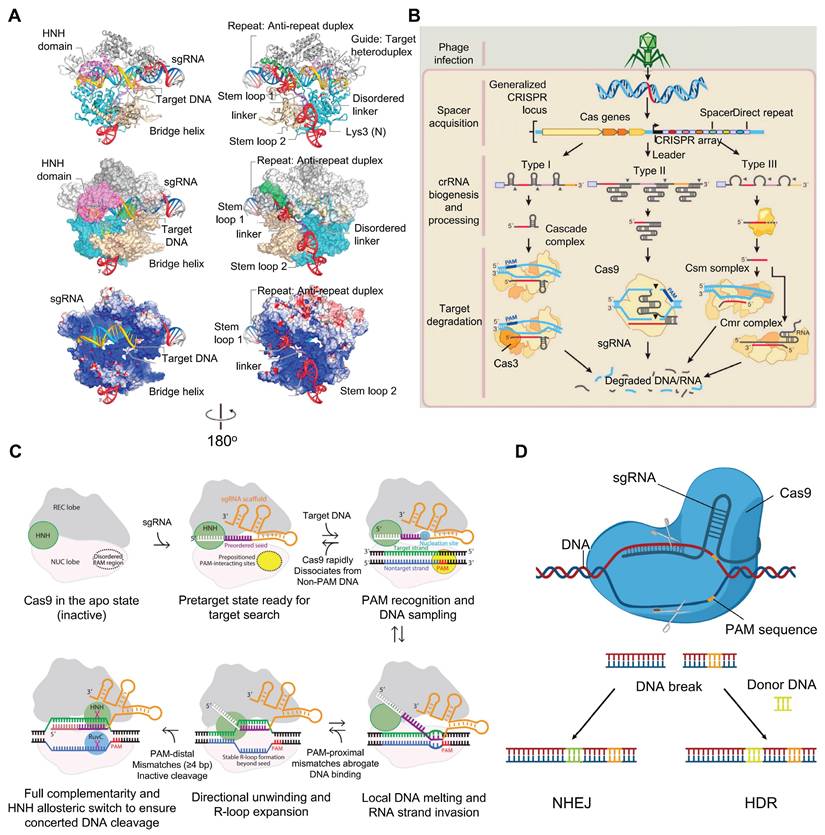
The Type II CRISPR-Cas system has become a widely used tool for gene editing, regulation, and research. Its simplicity, efficiency, and accuracy make it an essential resource for studying and treating eye diseases and developing new gene therapies.
The Type V CRISPR-Cas System
The Type V CRISPR-Cas system relies on proteins from the Cas12 family as its main effectors [63]. These proteins have a distinctive two-part (bilobed) structure: one part, called the REC domain, is responsible for recognizing the crRNA and target DNA, while the other part, known as the NUC domain, contains RuvC nuclease activity that cuts DNA [64-66]. This system is mainly divided into subtypes V-A, which includes Cas12a, and V-B, which includes Cas12b. Cas12a can process its own crRNA, identify specific AT-rich sequences called PAMs, and make staggered cuts in DNA using its RuvC domain [65]. This differs from Cas9, which recognizes NGG PAM sequences and cuts to produce blunt ends. Newer subtypes like V-M (Cas12m2, for example) don't cut double stranded DNA but can still turn off gene expression by strongly binding to DNA helping defend against mobile genetic elements like viruses. The unique structures and functions of Cas12 proteins open up exciting possibilities for gene editing and regulation [67]. Because of these features, Cas12 proteins are becoming increasingly important tools in gene editing research today.
The Type VI CRISPR-Cas System
The type VI CRISPR-Cas system uses Cas13 protein as the core effector protein and is known for its ability to use its HEPN domain for ribonuclease activity to target RNA [68, 69]. The subtypes are further divided into four classes VI-A to VI-D, with the most extensively studied being VI-A, represented by Cas13a. Upon crRNA binding, this activates the HEPN domain of Cas13 traditionally via structural conformations, which allows for sequence-specific recognition of foreign RNA that is independent of PAM sequences [70]. It has two modes: cis cleavage, in which it accurately degrades target RNA, and trans-cleavage, an indiscriminate collateral effect where it breaks down adjacent RNA [71]. Such collateral activity has been harnessed for molecular diagnostics, especially through SHERLOCK technology that reports high-sensitivity detection of pathogens including Zika virus and COVID-19 via fluorescent signal amplification [72, 73]. The system mutually partakes in microbial immunity and biomedicine as functionalities through the unitary mechanism of combinatorial nanoparticle-mediated virus identification, trigger-based defensing and multi-modal response execution into a precise antiviral defense and transportable diagnostic utility.
The contents of the above discussion represent the current classifications of the CRISPR gene editing systems. As science continues its path of exploration, we expect that new types of CRISPR systems will be uncovered. Herein, we will explore the use of the CRISPR gene editing system for ocular diseases.
Molecular Mechanisms and Potential CRISPR-Based Therapies
Anterior Eye Disease
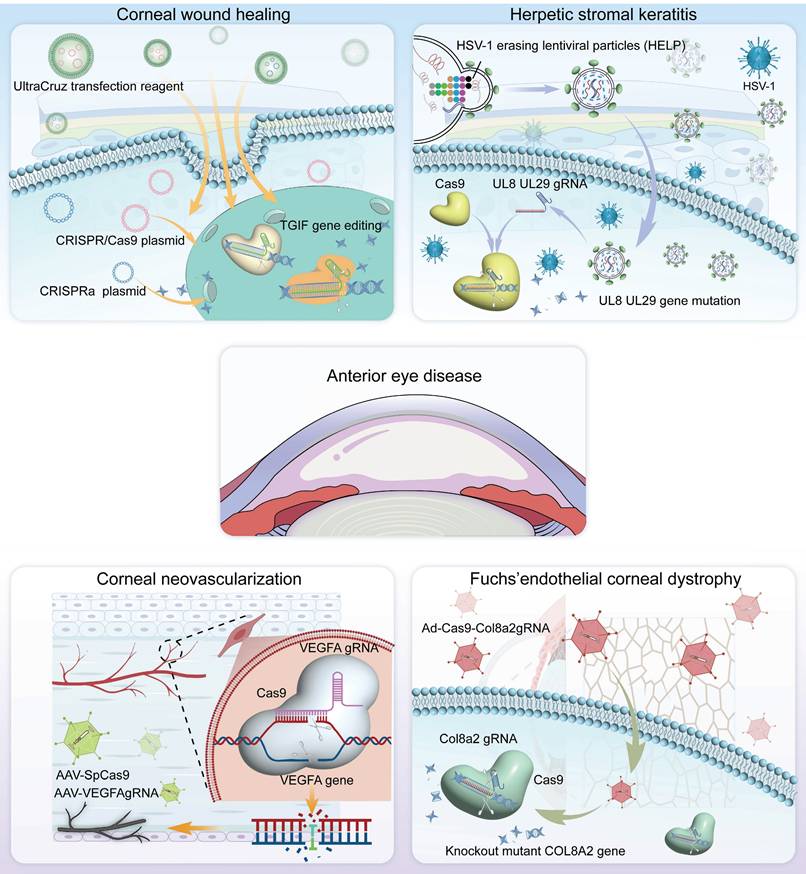
CRISPR technology has immense potential in the treatment of ocular diseases, with a growing focus on its use in corneal-related conditions. In this chapter, we summarize the pathogenesis of these conditions and explore the feasibility of using novel CRISPR-based gene therapies for corneal disease treatment (Figure 4).
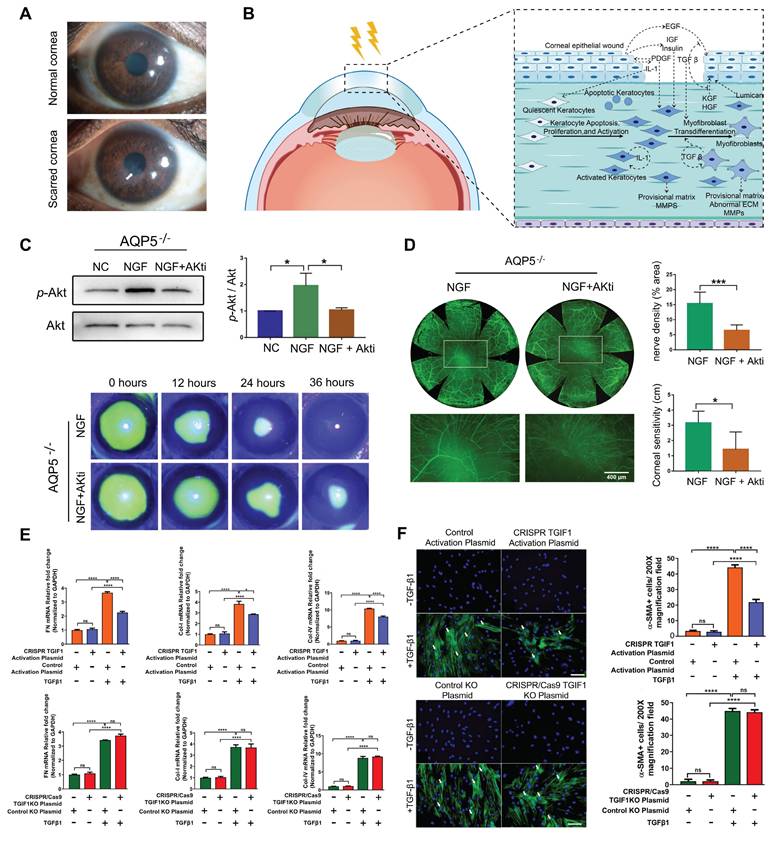
Corneal Scar and Corneal Wound Healing
Corneal scarring is a critical condition that compromises corneal transparency, and severe corneal injuries have been recognized as one of the major factors leading to corneal fibrosis and scar formation [74] (Figure 5A-B). Currently, corneal transplantation is the only option for treating corneal scar. As a result, several studies aim to restore corneal transparency by manipulating the gene expression of certain genes. Utilizing the CRISPR/dCas9 activation system to enhance the expression of Sex Determining Region Y-Box 2 (SOX2) successfully increased the proliferative capacity, and quantity of human corneal endothelial cells (hCECs), thereby holding promise for addressing the challenge of hCECs regeneration [75]. Liu et al. used CRISPR/Cas9 technology to generate an AQP5 gene knockout mouse model to study corneal injury. AQP5 deficiency reduced nerve growth factor (NGF) expression in the corneal epithelium, thereby inhibiting the activation of the Akt signaling pathway and adversely affecting corneal epithelial wound healing and nerve regeneration. Akt inhibitor (Akti) was used to block the reactivation of the Akt signaling pathway, and when applied alongside NGF, it reversed the corneal regeneration effects of NGF in CRISPR/Cas9 mediated AQP5-KO mice [76] (Figure 5C-D).
Recent research in this field has increasingly focused on the TGF-β signaling pathway. CRISPR/dCas9-mediated CRISPRa enables targeted gene expression activation without genomic sequence alteration, serving as a powerful tool for functional genomics research. Tripathi et al. built in vitro models by knocking out (CRISPR/Cas9) or activating (CRISPRa) the TGIF1 gene using CRISPR technology and identified that activation of TGIF1 gene significantly prevented the human corneal stromal fibroblasts (hCSFs) from changing into corneal myofibroblasts (CMFs) and inhibited profibrotic gene expression [77] (Figure 5E-F). The study showed that activation of TGIF1 inhibited corneal fibrosis and promoted tissue repair, suggesting a potential gene therapy strategy for corneal scars.
Herpetic Keratitis
Herpetic keratitis (HK), especially the herpetic stromal keratitis (HSK) type, is very destructive and can result in corneal blindness [78] (Figure 6A). HSK is driven primarily by immune mediated inflammation from interactions of HSV infected resident corneal cells and infiltrating inflammatory cells [79-81] (Figure 6B). ICP0 and ICP4 are two important genes that are needed for the replication and reactivation of HSV-1 [82-84] (Figure 6C). Roehm et al. delivered Cas9 and sgRNA targeting the ICP0 gene into ICP0-complementing L7 cells via plasmid transfection, which successfully introduced InDel mutations at the target site and significantly reduced HSV-1 viral yield [85] (Figure 6D-F). Chen and colleagues employed an AAV1-SaCas9 system to target and edit the ICP4 gene of HSV-1 within trigeminal ganglion neurons, significantly suppressing viral replication and establishing a foundation for developing curative therapies against HSK [86].
Representative applications of CRISPR gene editing system in anterior segment diseases.
Moreover, the CRISPR/Cas9 system has been utilized to regulate other important HSV genes. Yin et al. used a CRISPR/Cas9-based HELP (HSV-1 erasing lentiviral particles) system by incorporating spCas9 mRNA and gRNAs that interrupt the essential HSV-1 genes UL8 and UL29 into mRNA-carrying lentiviral particles, demonstrating potent HSV-1 replication inhibition [20] (Figure 6G). HELP system cleared HSV-1 not only in the cornea but also retrogradely transported to the trigeminal ganglion where latent HSV-1 virus was cleared (Figure 6H). Utilizing Virus-like particles (VLP) delivery technology, BDgene Therapeutics has developed BD111, a CRISPR/Cas9 based gene editing drug that directly targets and cleaves the HSV-1 genome, thereby enabling precise treatment for herpetic keratitis. In investigator-initiated trials (IITs), BD111 has demonstrated favorable safety and tolerability (ClinicalTrials.gov Identifier: NCT0456079). Additionally, construction of HSV-1 mutants with UL7 gene alterations via the CRISPR/Cas9 system demonstrated that the UL7 protein is essential for viral infection and proliferation, in addition to elucidating at the molecular level its mechanism of regulating viral replication and pathogenicity via the transcription of α4 genes [87]. These insights provided an important theoretical basis for the development of novel anti-HSV-1 therapeutics.
Presentation of key data from recent studies on corneal scar and wound healing: (A) Images of corneal scar. Adapted with permission from [74], copyright 2022, the authors. (B) Molecular mechanisms involved in corneal repair, remodeling, and regeneration after injury. (C-D) Injecting NGF in AQP5 knockout mice promotes corneal epithelial wound healing and nerve regeneration via Akt pathway activation. Adapted with permission from [76], copyright 2021, Elsevier Inc. (E) Effects of CRISPR/Cas9 mediated TGIF1 activation or knockout on the expression of pro-fibrotic genes. Adapted with permission from [77], copyright 2022, Elsevier Ltd. (F) CRISPR-activated TGIF1 strongly inhibited α-SMA protein expression. Adapted with permission from [77], copyright 2022, Elsevier Ltd.
Presentation of research results related to the treatment of herpes keratitis based on the CRISPR system: (A) Slit-lamp photos reveal central corneal stromal opacity in a patient with HSK. Adapted with permission from [89], copyright 2024, the authors. (B) HSV-1 infection process. Adapted with permission from [90], copyright 2023, the authors. Licensee MDPI, Basel, Switzerland. (C) ICP4 activates the Rap1b-CDC42-RAC1-Cofilin pathway, promoting HSV-1 invasion. Adapted with permission from [84], copyright 2023, the authors. Licensee MDPI, Basel, Switzerland. (D) Nucleic acid gel electrophoresis analysis of three types of InDel mutations. Adapted with permission from [85], copyright 2016, the authors. (E) Representative confocal images of L7 cells after immunostaining with an antibody against ICP0. Adapted with permission from [85], copyright 2016, the authors. (F) Viral production assay. Adapted with permission from [85], copyright 2016, the authors. (G) The construction of the HELP system. Adapted with permission from [20], copyright 2021, the authors. (H) HELP blocks HSV-1 infection of corneas and trigeminal ganglion. Adapted with permission from [20], copyright 2021, the authors. (I) NECTIN-1 knockout reduces HSV-DNA expression. Adapted with permission from [88]. copyright 2022, the authors.
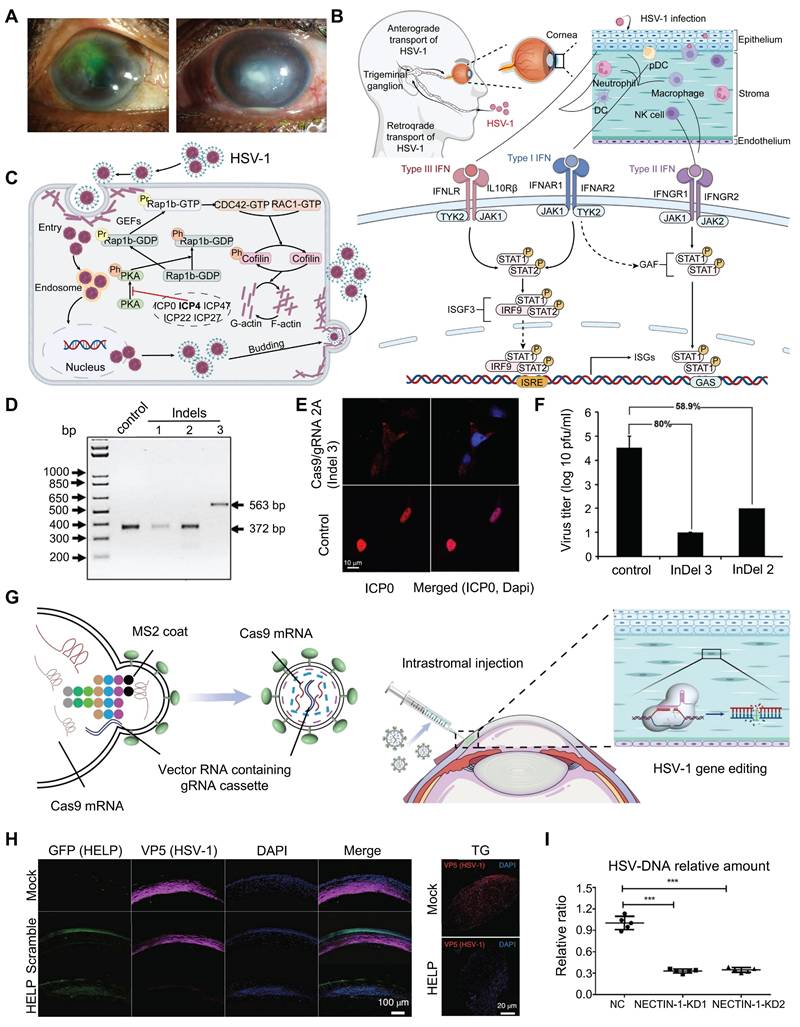
In addition to interfering with the replication of HSV, blocking HSV receptors and their adhesion processes represents a viable therapeutic strategy for herpes simplex keratitis. Using the CRISPR/Cas9 system, sgRNAs targeting the second exon of the nectin cell adhesion molecule 1 (NECTIN-1) gene were designed by Hong's group, and the delivered Cas9 and sgRNAs to hCECs via a lentiviral vector resulting in significant downregulating the expression of NECTIN-1, which significantly reduces HSV-1 infection and improves cell survival [88] (Figure 6I).
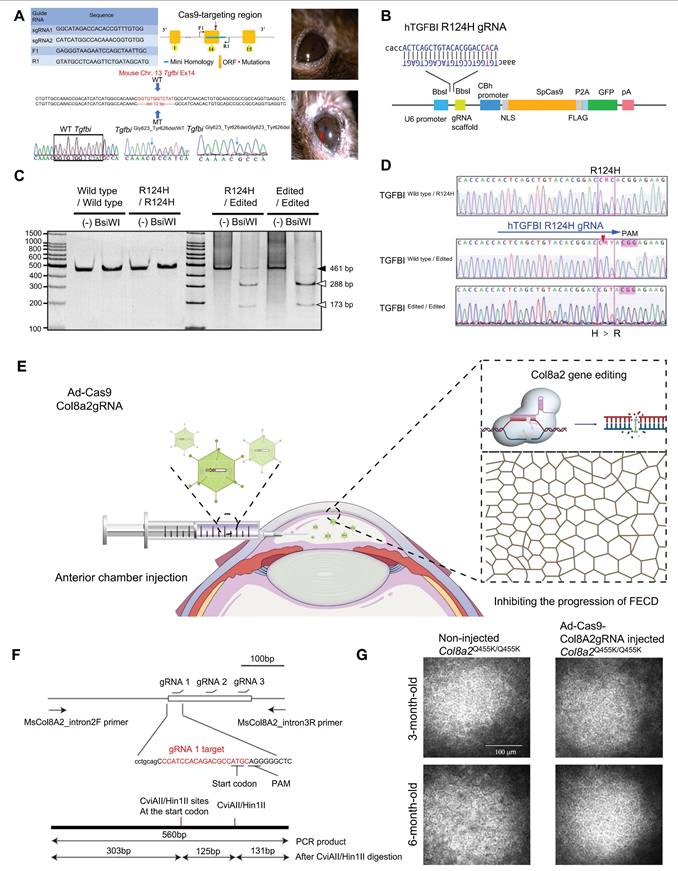
Corneal Dystrophy
Corneal dystrophy is a hereditary progressive disease involving all layers of the cornea, with complex pathogenesis and structural pathological changes [91]. Corneal dystrophies are mainly categorized into four major types: epithelial and subepithelial dystrophies, epithelial-stromal TGF-β1-induced dystrophies, stromal dystrophies, and endothelial dystrophies [92]. In a study of TGF-β1-induced corneal dystrophy, CRISPR/Cas9 technology was utilized to establish a humanized mouse model of TGF-β1-linked Thiel-Behnke corneal dystrophy (TBCD) reproduced the corneal opacity phenotype observed in human patients [93] (Figure 7A). Taketani's team successfully achieved in vitro gene correction of the R124H mutation in the TgfbI gene within primary human corneal keratocytes derived from a granular corneal dystrophy patient, utilizing CRISPR/Cas9 technology combined with a single-stranded oligodeoxynucleotide donor template [94]. Fuchs' endothelial corneal dystrophy (FECD) is the best-characterized of the endothelial dystrophies [95] (Figure 7B-D). Mutations in the Col8a2 gene are known causes of FECD, resulting in alterations to the α2 chain of type VIII collagen encoded by this gene. Col8a2 mutant knock-in mice exhibited corneal endothelial excrescences known as guttae, as well as the endothelial cell loss and disruption of the hexagonal structure, which are hallmarks of human FECD. The research of Uehara indicated that in vivo adenovirus-mediated delivery of SpCas9 and gRNA allowed efficient ablation of the Col8a2 mutants in corneal endothelial cells, preventing cell death and restoring normal corneal endothelial function in a mouse model of early-onset FECD [96] (Figure 7E-G).
Corneal Neovascularization (CoNV)
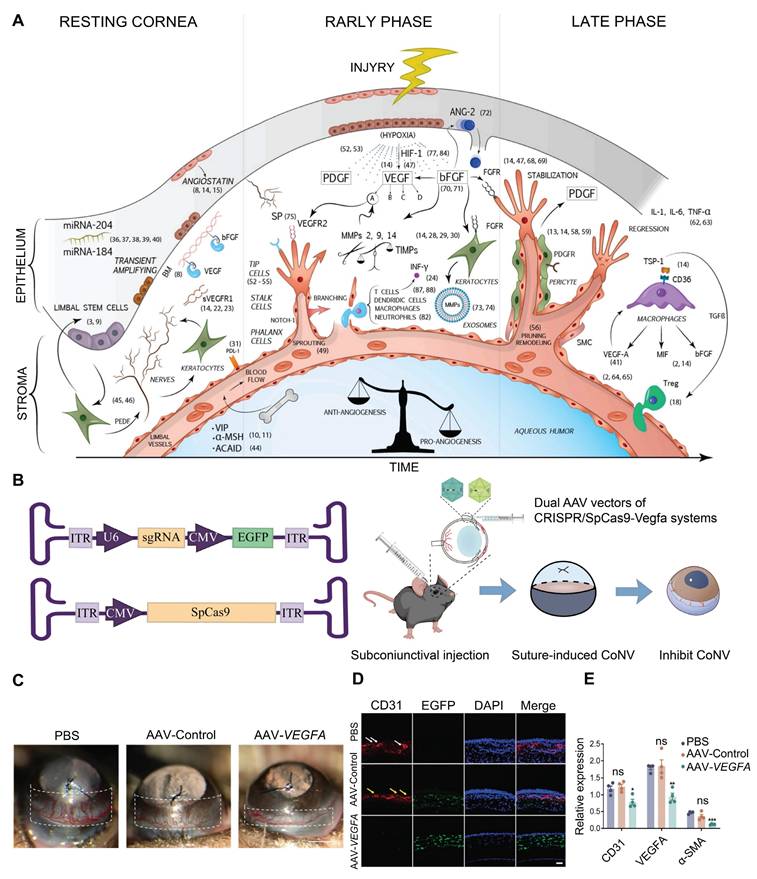
The transparency of a healthy cornea is mainly ensured by maintaining a delicate balance of antiangiogenic and proangiogenic factors in its microenvironment [97]. When the cornea is injured, such as in herpes simplex keratitis, chemical burns, and corneal transplant rejection, the secretion of angiogenic factors increases, inducing angiogenesis. VEGFA is a crucial angiogenic factor, and its upregulation is a primary contributor to corneal pathologies [98] (Figure 8A). Building on this mechanism, Zeng et al. designed and constructed a CRISPR/Cas9 system targeting the Vegfa gene and delivered it to mouse corneas via subconjunctival injection (Figure 8B). This system significantly suppressed suture-induced CoNV in mice and downregulated protein expression levels of VEGFA, CD31 (endothelial cell marker), and α-SMA (myofibroblast/smooth muscle cell marker) [99] (Figure 8C-E). The core advantage of the highly precise CRISPR/Cas9 gene editing system lies in its ability to specifically recognize and cleave target gene sequences. In the treatment of CoNV, CRISPR/Cas9 technology offers significant improvement over traditional pharmacological approaches, such as anti-VEGFA antibodies and small-molecule inhibitors, by directly modifying the genome to produce sustained biological effects.
Midsection Eye Disease
The ocular midsegment, which includes the iris, lens, and ciliary body, plays a crucial role in light modulation and the maintenance of visual function. Among the diseases of the ocular midsegment, glaucoma is the most prevalent and impactful, significantly reducing patients' quality of life, whereas traditional therapies often fail to address the underlying issues effectively. Leveraging the precise gene repair capabilities of CRISPR, we anticipate the potential to revolutionize the treatment of these conditions, achieving lasting and effective therapeutic outcomes through genetic intervention. In the following sections, we review the genetic mechanisms underlying these diseases and explore the potential applications of CRISPR technology in ocular midsegment disorders, along with prospects for future development (Figure 9).
Glaucoma
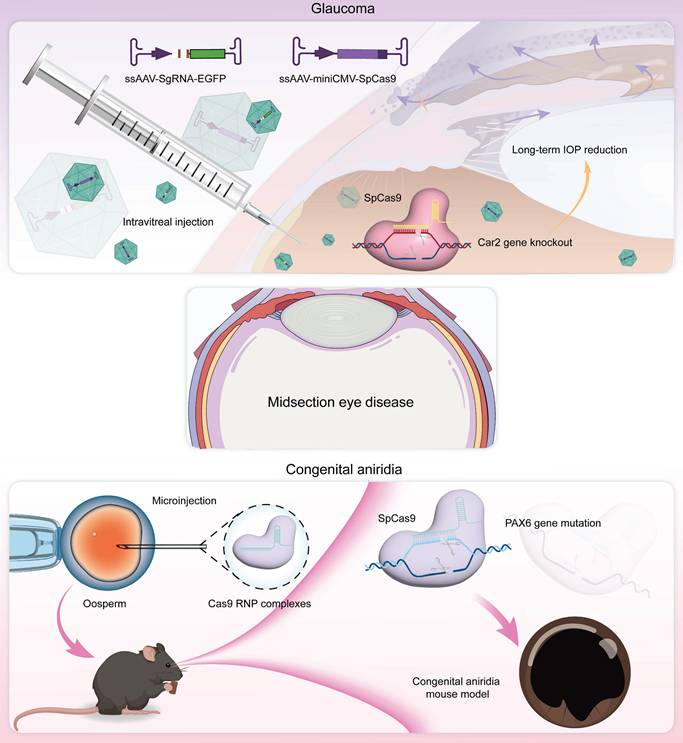
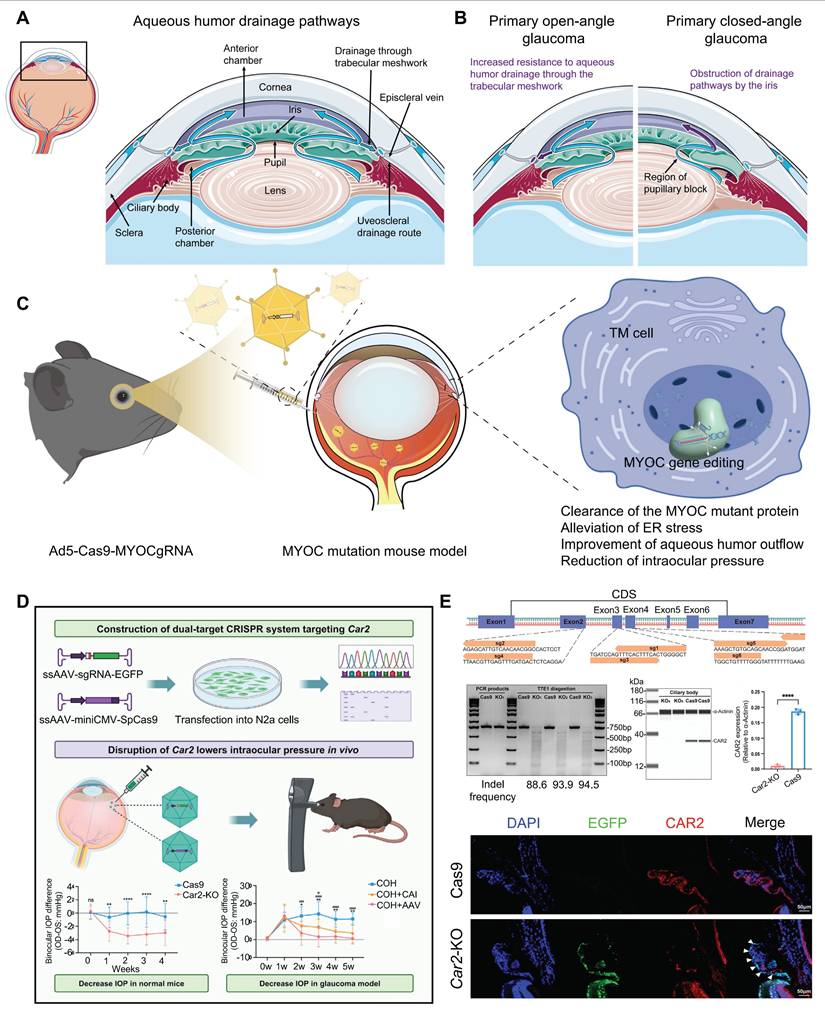
Glaucoma is a progressive neuropathy primarily characterized by the degeneration of retinal ganglion cells (RGCs) and changes in the appearance of the optic disc, with elevated intraocular pressure (IOP) being the most notable clinical feature [101] (Figure 10A-B). Glaucoma exhibits a complex pathogenesis, with MYOC gene mutations representing significant genetic risk factors for this condition [102]. Targeting this gene, Ankur Jain et al. delivered the CRISPR-Cas9 system via an adenoviral vector into the eyes of the MYOC mutant mouse model, which successfully knocked out the MYOC gene in vivo. This intervention alleviated endoplasmic reticulum stress within cells, significantly reduced IOP, and prevented further glaucoma damage in the mice's eyes [46] (Figure 10C). Meanwhile, BDgene Therapeutics has developed BD113, a therapy utilizing engineered lentiviral vectors to deliver gRNA/Cas9 ribonucleoprotein (RNP) complexes that target mutant MYOC gene for knockout. Currently, an ongoing single-dose, two-arm clinical study (ClinicalTrials.gov Identifier: NCT06465537), initiated by Beijing Tongren Hospital, aims to evaluate the efficacy of BD113 in treating primary open-angle glaucoma. Two BD113-treated patients exhibited no treatment-related serious adverse events, achieving normalized IOP and discontinuation of pressure-lowering medications.
Recent key findings in the study of corneal dystrophy: (A) TGFBI-related murine TBCD model was made by CRISPR/Cas9. Adapted with permission from [93], copyright 2021, the authors. (B) Linear structure of the plasmid for CRISPR/Cas9-mediated HDR of a TgfbI mutation. Adapted with permission from [94], copyright 2017, the authors. (C-D) Correction of the mutation in TgfbI mutant keratocytes using CRISPR-mediated HDR. Adapted with permission from [94], copyright 2017, the authors. (E) Construction and mechanism of Ad-Cas9-Col8a2 gRNA. (F) Schematic diagram of Col8a2 gRNA design and PCR product restriction endonuclease detection. Adapted with permission from [96], copyright 2021, Uehara et al. (G) Ad-Cas9-Col8a2 gRNA significantly reduced corneal endothelial cell loss. Adapted with permission from [96], copyright 2021, Uehara et al.
Presentation of key data from representative studies on CoNV: (A) Summary Schematic Diagram of the Molecular Pathways in CoNV. Adapted with permission from [100], copyright 2021, Elsevier Ltd. (B) CRISPR/SpCas9-Vegfa dual AAV vector system. Adapted with permission from [99], copyright 2024, the authors. (C) CRISPR/SpCas9-Vegfa system reduces suture-induced CoNV. Adapted with permission from [99], copyright 2024, the authors. (D-E) CD31, VEGFA, and α-SMA protein expression in corneal tissue. Adapted with permission from [99], copyright 2024, the authors.
Essentially, IOP is controlled from a structural abundance of aqueous humor production and outflow [103]. Carbonic anhydrase, which regulates aqueous humor formation, has long been targeted for the treatment of glaucoma, yet local therapies face uveoscleral penetration/low bioavailability and systemic side effects [104]. To address this issue, Zhang and colleagues recently proposed a CRISPR/Cas9 approach that directly targets the Car2 gene in the ciliary body, using the serotype AAV vector ShH10, achieving a mean reduction of ~ 18% IOP in normal mice and ~ 40% IOP in a model of glaucoma [105] (Figure 10D-E). Importantly, CRISPR/Cas9 mediated Car2-KO remarkably alleviated RGC death and optic nerve fiber degeneration in a chronic high IOP model.
Representative applications of CRISPR system in midsection eye diseases.
Congenital Aniridia
Congenital aniridia is a rare genetic eye disorder characterized by varying degrees of iris and foveal hypoplasia [106] (Figure 11A-B). This condition is inherited in an autosomal dominant pattern; mutations in the Pax6 gene on the short arm of chromosome 11 (11p13) are responsible for roughly 90% of cases [107]. Based on these mechanisms, using the CRISPR/Cas9 system, Roux and colleagues generated a Pax6 gene mutation in human limbal stem cells as a cell model of aniridia-related corneal disease [108]. The small eye (Sey) mouse carries a Pax6 nonsense variant (c.580G>T, p.G194X), presenting with microphthalmia, aniridia, and other ocular phenotypes like human aniridia, making it a critical model for studying aniridia pathophysiology and therapies. Elizabeth et al. delivered an in vitro-optimized CRISPR/Cas9 system into Sey mutant zygotes via electroporation, successfully inserting a 3×FLAG tag into the Sey allele. This generated a novel mouse strain termed Fey, which retains the mutant characteristics of the Sey allele. The 3×FLAG tag enables researchers to distinguish wild type from edited Pax6 proteins, allowing precise evaluation of editing efficacy. Subsequently, the optimized CRISPR/Cas9 system was microinjected into Fey mouse zygotes, correcting the Sey mutation at 25% efficiency and restoring normal Pax6 expression. Genetically rescued mice exhibited wild-type ocular phenotypes [109].
Display of key results from recent glaucoma research: (A) The aqueous humor circulation pathway. (B) The pathogenesis of glaucoma. (C) Using CRISPR/Cas9 technology to interfere with the MYOC gene in vivo. (D-E) AAV-mediated CRISPR/Cas9 Car2 gene knockout for glaucoma treatment. Adapted with permission from [105], copyright 2024, Elsevier Inc.
Displaying the key findings of recent studies on congenital aniridia: (A) Ocular manifestations of aniridia syndrome. Adapted with permission from [106], copyright 2023, Elsevier Inc. (B) Keratoprosthesis surgery in advanced aniridia. Adapted with permission from [106], copyright 2023, Elsevier Inc. (C) ABE8e strategy. (D) ABE8e strategy restores Pax6 expression. Adapted with permission from [110], copyright 2023, the authors.
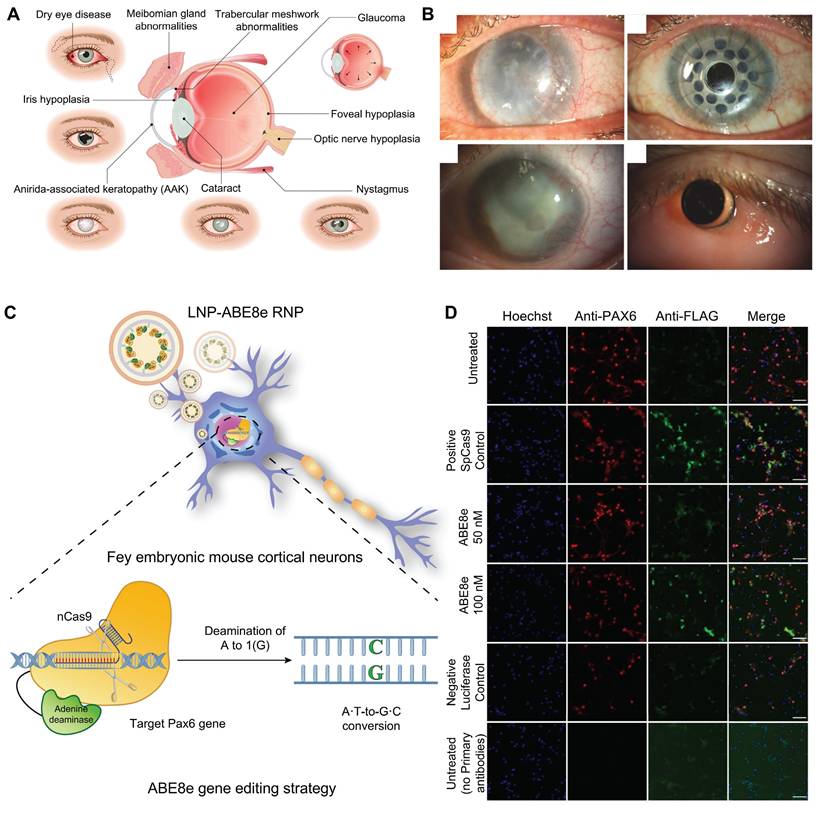
ABE8e allows for the direct conversion from adenine (A) to guanine (G) in the genome and can obtain single-base editing without introducing DSBs. This strategy is milder than CRISPR/Cas9 technologies and minimizes nonspecific effects and potential off-target effects, and it is particularly appropriate for correcting specific point mutations responsible for the development of aniridia. Adair et al. encapsulated optimized ABE8e-RNP into lipid nanoparticle (LNPs) and delivered to Fey embryonic mouse cortical neurons, resulting in c.580G>T mutation correction and restoration of Pax6 protein levels (24.8%) based on this principle [110] (Figure 11C-D).
Representative applications of CRISPR system in posterior segment diseases.
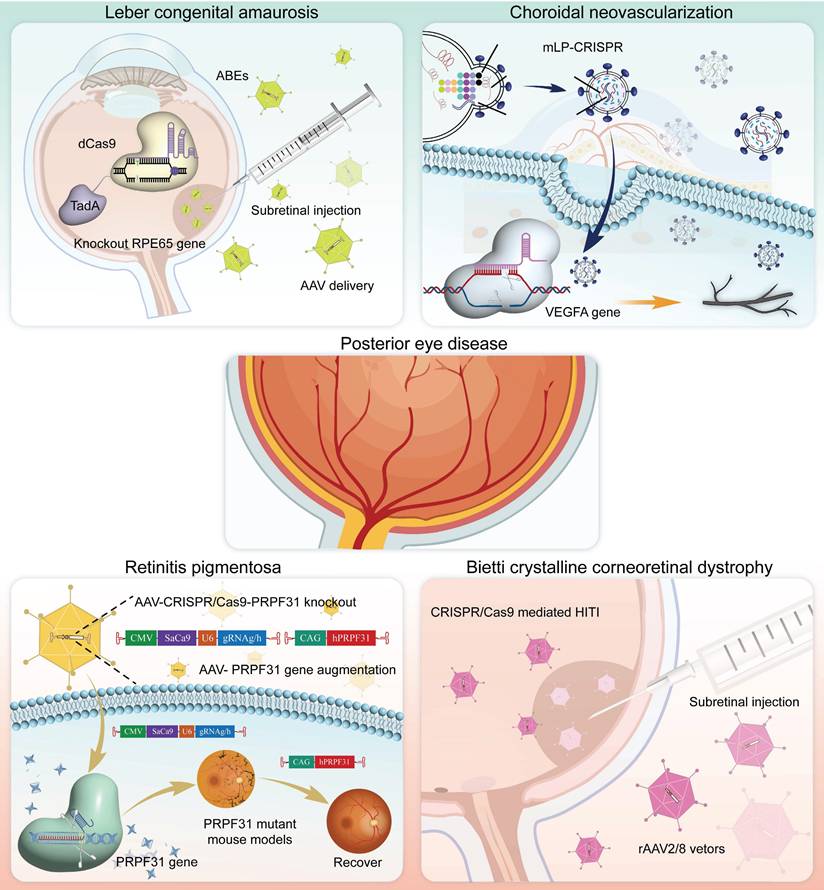
Posterior Eye Disease
The posterior segment of the eye, which is composed of intricate structures such as the retina, choroid, and optic nerve, is the core area for the transformation and transmission of visual signals. CRISPR technology, with its precise gene-editing capabilities, offers a revolutionary solution for correcting genetic defects, protecting neural cells, and promoting functional recovery. Below, we review the currently employed applications and advancements of CRISPR gene editing technology in the treatment of posterior segment diseases (Figure 12).
Leber's Congenital Amaurosis (LCA)
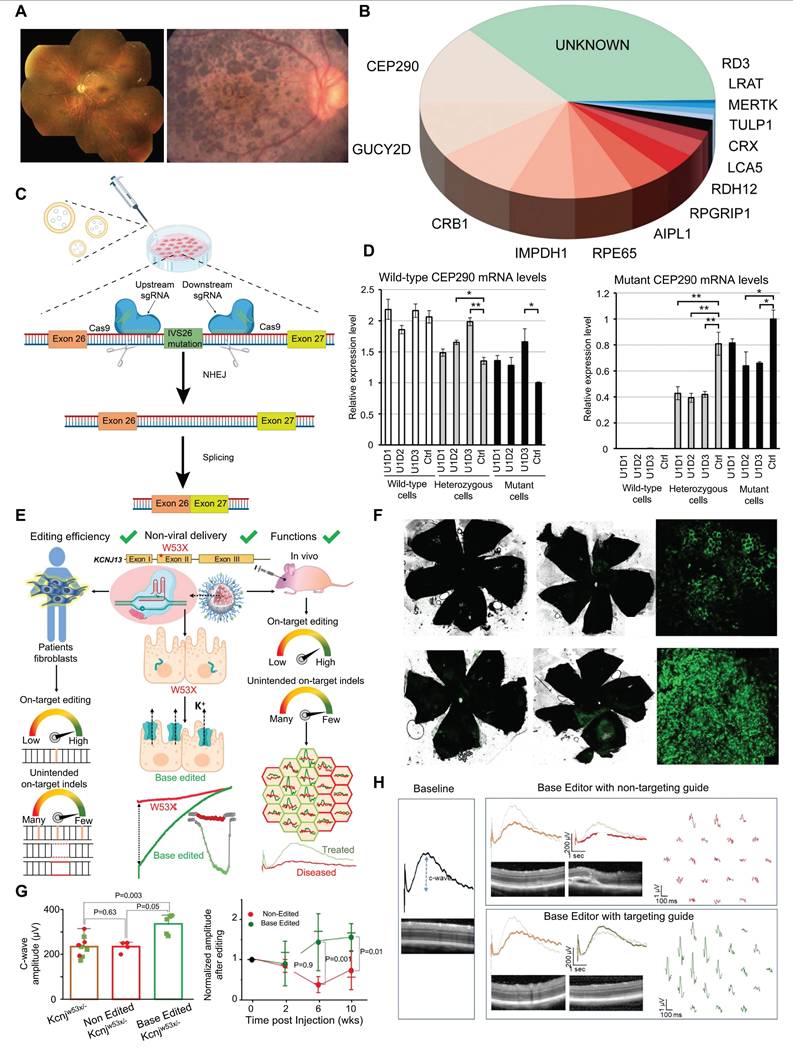
LCA is a severe retinal dystrophy, usually manifesting at birth or in infancy, with progressive loss of vision. Clinical features of LCA can include nystagmus, sluggish pupillary response, night blindness, and severely reduced or absent ERG signals [111] (Figure 13A). Most LCA cases follow an autosomal recessive inheritance pattern and exhibit significant genetic and allelic heterogeneity [112]. Now, more than 20 genes associated with LCA have been identified through genetic studies, including CEP290, GUCY2D, CRB1, KCNJ13, and RPE65 (Figure 13B) [113]. LCA10 is the most common subtype of LCA, caused by a deep intronic point mutation in intron 26 of the CEP290 gene (IVS26 mutation) [114]. Using the CRISPR/Cas9 system, two gRNAs were designed-one upstream and one downstream of the IVS26 mutation site-to direct the Cas9 nuclease in precisely excising the intronic fragment that contains the IVS26 mutation. Following deletion of the mutant region, the CEP290 gene can undergo normal RNA splicing and produce full-length, functional CEP290 protein (Figure 13C-D) [115]. Tailored for CEP290-associated IRDs such as LCA10, EDIT-101 is a CRISPR/Cas9-based gene editing therapy developed by Editas Medicine to excise the aberrant splice donor generated by the IVS26 mutation and thereby restore normal CEP290 expression. Subsequently, phase 1-2, open-label, single-ascending-dose study (ClinicalTrials.gov ID, NCT03872479) results demonstrated that 9 out of 14 patients treated with EDIT-101 showed meaningful improvements from baseline in the best corrected visual acuity. No treatment-related serious adverse events or dose-limiting toxicities were observed [116].
KCNJ13 is critical for retinal pigment epithelium (RPE) function and encodes a potassium channel protein that modulates the electrophysiological response of the RPE to light-induced alterations in subretinal potassium concentration [117]. Kabra and colleagues confirmed that ABE8e had improved KCNJ13 W53X mutation gene editing efficiency and reduced off-target effects in HEK293T cells and patient-derived-of fibroblasts. To further study in vivo editing effects, researchers delivered ABE8e mRNA to mouse eyes via silica nanocapsules. Results showed a significant increase in the C-wave amplitude of edited mice's retinal response, indicating restored Kir7.1 channel function and maintained retinal structure [118] (Figure 13E-H). Additionally, Yuki Kanzaki et al. generated KCNJ13 KO human induced pluripotent stem cells (hiPSCs) via the CRISPR/Cas9 system to successfully establish an LCA16 cell model [119].
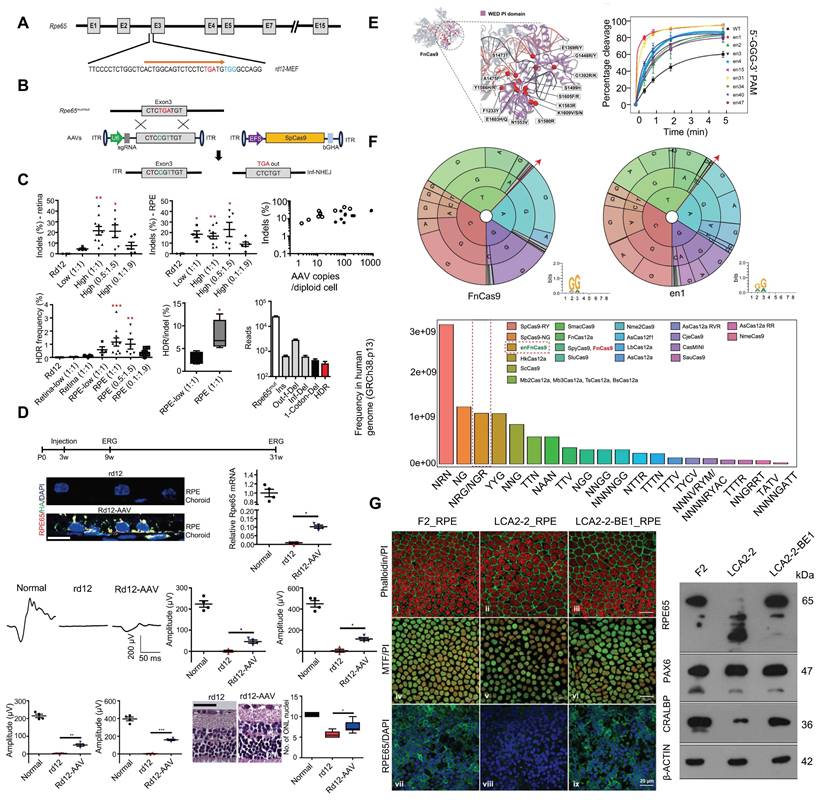
RPE65 encodes a key isomerase that participates in generating the chromophore of cone and rod visual pigments. Loss of translation due to mutations in RPE65 leads to premature termination of the translation process, which is one of the major causes of LCA. Jo and colleagues used a dual AAV vector encoding CRISPR/Cas9, and an RPE65 donor sequence to target exon 3 of RPE65 leading to the removal of the premature stop codon that caused LCA and restored retinal function in rd12 mouse model [121] (Figure 14A-D). In addition, Acharya's team altered FnCas9 to make three enFnCas9 variants (en1, en15 and en31). These possess increased editing efficiency and wider PAM specificity (Figure 14E-F). They offer potential for gene editing and disease treatment, including the correction of RPE65 mutations and restoration of pigment expression in RPE cells, which appear to be possible strategies for LCA treatment [122] (Figure 14G). Furthermore, Retinal injections were performed in mice using a lentivirus expressing ABE and sgRNA targeting the de novo nonsense mutation in the RPE65 gene by Susie Suh and colleagues. Results showed a restored expression of RPE65 and vitamin A isomerase activity, as well as retinal and visual function approaching the normal range [123]. Jang et al. delivered PE via AAV vectors to the subretinal space of rd12 mouse models, successfully corrected the nonsense mutation in the RPE65 gene, effectively ameliorated disease phenotypes in the mice, and detected no off target editing [124].
Biallelic mutations in the NMNAT1 gene are also an important cause of LCA [125]. To investigate the mechanism by which NMNAT1 mutations lead to retinal degeneration, during the differentiation of NMNAT1-knockout iPSCs created used CRISPR/Cas9 technology into retinal organoids, the formation of the retinal primordial structure failed, confirming the important role of NMNAT1 in early retinal development [126]. Since the NMNAT1 gene mutation is recessive, supplementing the retina with a normal copy of NMNAT1 has become a means to protect vulnerable cells from the effects of disease progression. Scott and colleagues performed gene augmentation therapy by subretinal injection of AAV carrying a normal human copy of NMNAT1 in a mouse model harboring the NMNAT1-p.Val9Met mutation, rescuing retinal structure and function [127]. To determine which cell types need to express NMNAT1 for therapeutic effects, they treated NMNAT1 mutant mice with AAV driven by specific cell type promoters and found that therapy promoting NMNAT1 expression in photoreceptors could protect retinal morphology, suggesting that gene therapy for NMNAT1 related diseases should target photoreceptors [128].
Presentation of important data from recent studies on LCA, Part I: (A) Fundus photography images of LCA patients. Adapted with permission from [120], copyright 2015, the authors. (B) The mutation genes associated with LCA. (C) The CRISPR/Cas9 strategy used to remove the IVS26 mutation. Adapted with permission from [115], copyright 2016, The American Society of Gene and Cell Therapy. (D) Rescue of the expression of WT CEP290. Adapted with permission from [115], copyright 2016, The American Society of Gene and Cell Therapy. (E) Using silica nanocapsules to deliver ABE8e to RPE cells for the treatment of inherited retinal diseases caused by KCNJ13 gene mutations. Adapted with permission from [118], copyright 2023, Kabra et al. (F-H) ABE8e can effectively restore RPE cell function and improve retinal structure and function in vivo. Adapted with permission from [118], copyright 2023, Kabra et al.
Presentation of important data from recent studies on LCA, Part II: (A) The structure of the RPE65 gene is shown. Adapted with permission from [121], copyright 2019, the authors. (B) The positions of the premature stop codon. Adapted with permission from [121], copyright 2019, the authors. (C) RPE65 gene correction in rd12 mice. Adapted with permission from [121], copyright 2019, the authors. (D) Therapeutic effects of HDR-mediated correction of the RPE65 gene in rd12 mice. Adapted with permission from [121], copyright 2019, the authors. (E) FnCas9 crystal structure and in vitro cleavage assay. Adapted with permission from [122], copyright 2024, the authors. (F) PAM wheels and sequence logos for FnCas9 and enFnCas9, with bar plots showing the genome-wide availability of PAMs for each Cas effector in the human genome. Adapted with permission from [122], copyright 2024, the authors. (G) Therapeutic base editing by en31-ABEmax8.17d in a LCA patient specific iPSC line. Adapted with permission from [122], copyright 2024, the authors.
Retinitis Pigmentosa (RP)
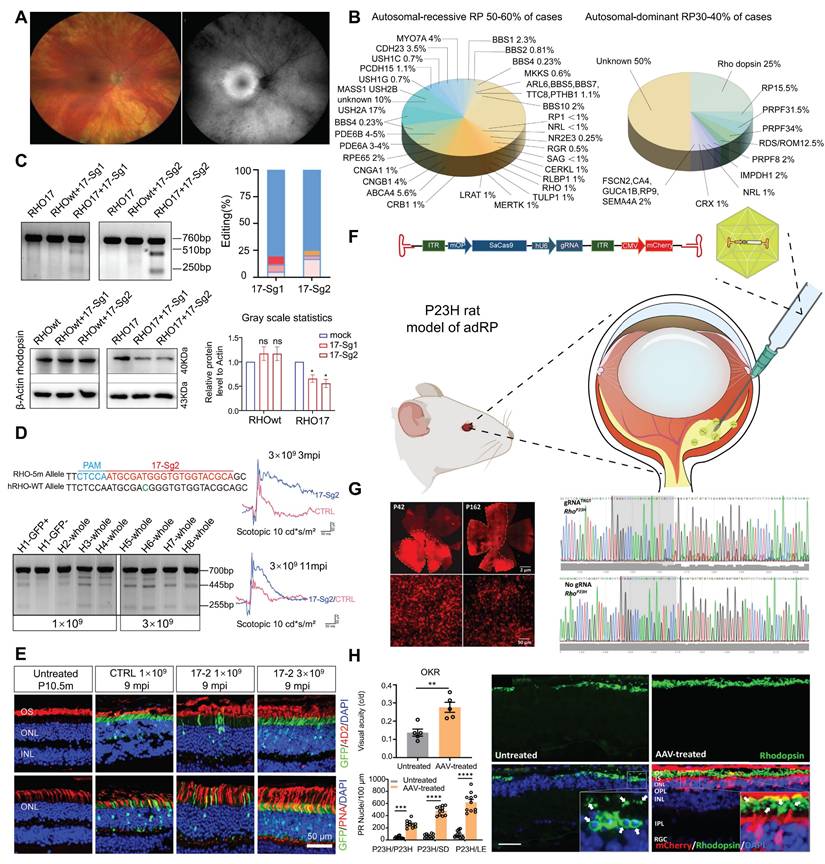
RP refers to a class of inherited retinal dystrophies in which the rod photoreceptors and RPE undergo progressive degeneration, resulting in photoreceptor cell death and vision loss [129] (Figure 15A). RP can be inherited through autosomal recessive (50-60 %), autosomal dominant (30-40 %), and X-linked recessive (5-15 %) patterns (Figure 15B) [130]. RHO gene have been recognized as a leading cause of autosomal dominant RP (adRP) [131]. Liu et al. developed an allele-specific CRISPR/Cas9 gene editing therapy capable of selectively targeting the mutant RHO-T17M allele while preserving wild-type function (Figure 15C-D). Through subretinal injection delivery into a humanized mouse model, it effectively delayed retinal degeneration and restored visual function [132] (Figure 15E). Saba Shahin and coworkers developed an AAV2-SaCas9/gRNA system effectively knocked out the RHO S334ter site, suppressing the P23H mutation. Through subretinal injection into P23H mutant rat models, this system delayed apoptosis of photoreceptors, and maintained long term vision and retinal function [133] (Figure 15F-H). Latella et al. delivered CRISPR/Cas9 plasmids designed to edit the human mutant RHO gene into mouse retinas via subretinal electroporation, achieving targeted gene editing and a significant reduction in mutant RHO protein levels. Notably, the engineered sgRNA exhibited high specificity for the human RHO gene without affecting the endogenous mouse RHO gene [134]. Li and coworkers developed the engineered Cas9 variant and truncated sgRNA to achieve efficient discrimination of a single-nucleotide mutation in RHO-P23H mice. They achieved approximately 45% editing of the mutant P23H allele at the DNA level, while the editing rate of the wild-type allele was only 1.3% and the thickness of the photoreceptor cell layer in the treated retinal regions significantly increased. This study accomplished allele-specific editing of single-nucleotide mutations, expanding the application scope of CRISPR/Cas9 in the treatment of dominant genetic diseases [135].
Although CRISPR therapies targeting the specific P23H mutation have achieved numerous breakthroughs, developing separate treatments for over 150 distinct RHO mutations is not only costly but also clinically impractical. Wu et al. developed a CRISPR-based, mutation-independent gene ablation and replacement (AR) compound therapy, providing a universal solution for RHO-related adRP. This work overcomes the challenge of genetic heterogeneity in treating autosomal dominant disorders and holds direct promise for clinical transformation [137]. Additionally, Du and colleagues utilized CRISPR/SaCas9-mediated delivery of dual AAV vectors for a “reduction and replacement” system to specifically silence the mutant RHO gene, while driving expression from the normal RHO allele [138].
CRISPRi inhibits dominant pathogenic gene expressions for avoiding the aggregate of toxic proteins, nudging cell longevity. Burnight et al. constructed a CRISPRi system that targets to RHO mutation gene pro23his. After subretinal transduction of human retinal explants and transgenic Pro23his mutant pigs, this system effectively suppressed Rho protein expression, maintenance of outer nuclear layer thickness and slowed retinal degeneration [139].
ZVS203e is a novel CRISPR/Cas9 based gene editing product designed to target and silence the mutated RHO gene. The drug was granted orphan drug designation by the U.S. Food and Drug Administration (FDA) in July 2022. To date, a single-arm, open-label Phase I/II clinical trial is currently underway to evaluate the safety and efficacy of ZVS203e in treating RP caused by RHO gene mutations (ClinicalTrials.gov ID, NCT05805007).
In addition to RHO gene, PRPF31 mutations are also a common cause of adRP. Xi et al. employed an AAV-mediated CRISPR/Cas9 KO system to successfully replicate the morphological and functional damage associated with RP in mice, demonstrating the successful establishment of an RP animal model. Additionally, AAV-mediated PRPF31 gene enhancement restored retinal structure and function in an RP mouse model [140]. This study not only creates a novel system for studying the pathogenesis of PRPF31-associated RP but also provides valuable experimental evidence for the PRPF31 gene therapy. Amélie Rodrigues et al. also used an AAV-mediated PRPF31 gene enhancement technology repaired PRPF31 gene mutations and improved the survival rate of photoreceptor cells [141].
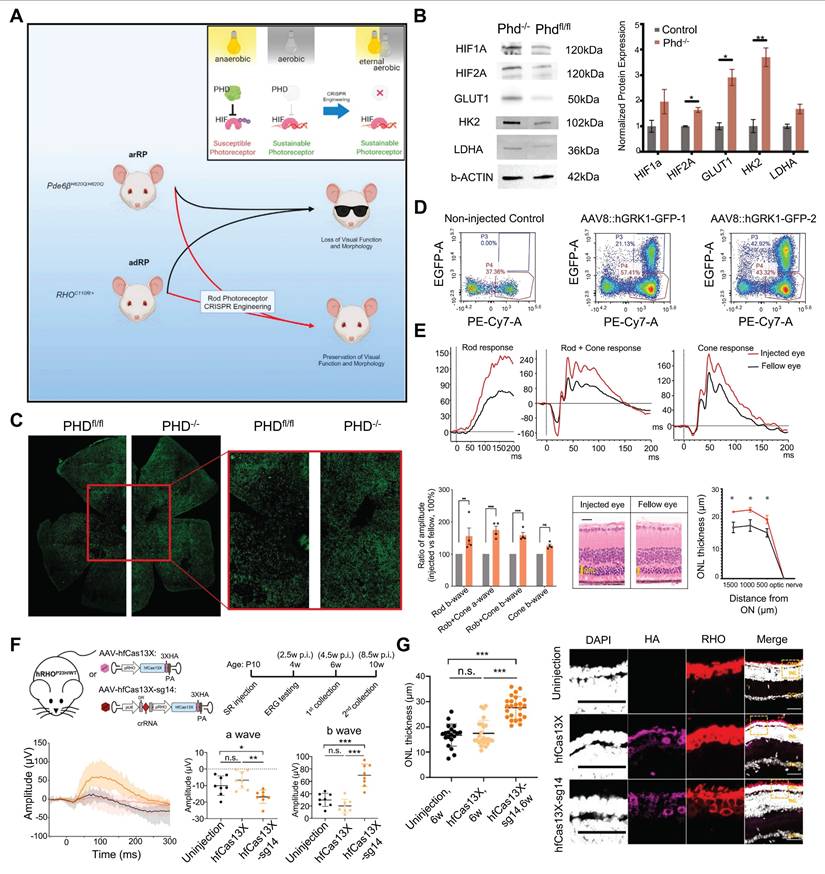
Recently, Nolan and colleagues developed a new dual gRNA-guided CRISPR/Cas9 system targeting exon 1 of the prolyl hydroxylase 2 (PHD2) gene based on the role of the PHD-von hippel lindau (VHL)-hypoxia inducible factor (HIF) axis in glycolysis (Figure 16A). This dual gRNA-guided CRISPR/Cas9 system successfully knocked out the PHD2 gene, promotes aerobic glycolysis and contribute to the survival of photoreceptor cells and improvements in retinal function [142] (Figure 16B-E). This research represents a successful attempt to promote photoreceptor glycolysis through metabolic reprogramming via the CRISPR system, providing a novel perspective for treating this disease from a metabolic standpoint.
Cas13 editing of RNA offers a therapeutic approach that avoids the risks associated with permanent genomic changes. Cas13X degrades target RNA with high specificity and minimizes off-target effects, ensuring therapeutic efficacy while preserving nontarget RNA. Yan et al. designed a compact and high-fidelity Cas13X (hfCas13X) sgRNA CRISPR system targeting the human mutant ribozyme transcript RHO-P23H (Figure 16F). This system was delivered by AAV injected into the vitreous cavity of RP mouse models, leading to retinal function improvement and inhibition of photoreceptor apoptosis and retinal thinning [143] (Figure 16G).
Key data presentation of the latest achievements in RP, Part I: (A) Fundus photography images of RP patients. Adapted with permission from [136], copyright 2023, the authors. (B) The currently known genes associated with RP and their relative contributions. (C) SaCas9/17-Sg1 and -Sg2 can specifically cut the mutant RHO-T17M sequence. Adapted with permission from [132], copyright 2023, Liu et al. (D) SaCas9/17-sg2 improves retinal function by editing mutant RHO gene. Adapted with permission from [132], copyright 2023, Liu et al. (E) The photoreceptor cells of RHO mutation mice treated with SaCas9/17-sg2 were preserved. Adapted with permission from [132], copyright 2023, Liu et al. (F) AAV-CRISPR/SaCas9 gene editing in the P23H rat model of adRP. (G) The CRISPR/Cas9 knocks out m-RhoP23H. Adapted with permission from [133], copyright 2022, the authors. (H) CRISPR/SaCas9-mediated ablation of m-RhoP23H protects photoreceptor cells. Adapted with permission from [133], copyright 2022, the authors.
Bietti Crystalline Corneal Retinal Dystrophy (BCD)
BCD is an autosomal recessive progressive degenerative disease of the chorioretina, which is characterized by abundant yellow-white crystalline deposits in the retina, particularly around the posterior pole [144]. Currently, CYP4V2 is the only gene identified to be associated with BCD [144]. Given that there are no approved treatments for BCD, researchers are increasingly exploring CRISPR-based therapies for this genetic condition. Reports indicate that approximately 80% of BCD patients have mutations in exons 7-11 of the CYP4V2 gene [144]. HITI is a precise gene editing technique that enables the insertion of DNA sequences at specific loci without relying on homologous recombination [48]. Meng et al. created a sgRNA to direct the Cas9 nuclease to make DSBs in intron 6 of the CYP4V2 gene and used the HITI method to successfully insert the sequence carrying exons 7-11 at the intron 6 site of the CYP4V2 gene. In patient-derived iPSCs, the HITI method revitalized the viability and functionality of RPE cells. In a humanized Cyp4v3 mouse model, this strategy successful increased numbers of RPE cells, metabolic activity and significant improved retinal function postediting [50]. CRISPR/Cas9-mediated HITI has a promising application to those IRDs genes with mutation hotspots and can serve as an effective complement to gene replacement therapy.
Key data presentation of the latest achievements in RP research, Part II: (A) CRISPR-mediated editing of the PHD gene for RP therapy. Adapted with permission from [142], copyright 2024, Elsevier Inc. (B) PHD deficiency up-regulates key regulators of glycolytic metabolism in photoreceptors. Adapted with permission from [142], copyright 2024, Elsevier Inc. (C) PHD knockout protects cone photoreceptors. Adapted with permission from [142], copyright 2024, Elsevier Inc. (D) AAV8 delivers hGRK1-GFP to mouse retinas. Adapted with permission from [142], copyright 2024, Elsevier Inc. (E) Detection of photosensitive function in RP mice. Adapted with permission from [142], copyright 2024, Elsevier Inc. (F) Strategy for treatment of RP mice with hfCas13X. Adapted with permission from [143], copyright 2023, the authors. (G) The protective role of AAV-hfCas13X for photoreceptor degeneration. Adapted with permission from [143], copyright 2023, the authors.
Stargardt Disease (STGD1)
STGD1 is currently an untreatable autosomal recessive macular dystrophy primarily caused by biallelic mutations in the ABCA4 gene [145]. The most prevalent mutation associated with STGD1 is a G-to-A point mutation in ABCA4 (c.5882G>A, p. Gly1961Glu), affecting approximately 15% of patients [146]. An ABE strategy utilizing AAV vectors was developed to correct the ABCA4 c.5882G>A mutation. High-level gene correction was achieved in mutation-carrying mice and nonhuman primates, with mean editing rates of 75% in cone photoreceptors and 87% in RPE cells. However, functional improvements following editing could not be validated due to the absence of disease phenotype in animal models [147].
Moreover, several deep intronic variants (DIVs) in ABCA4 have been identified as pathogenic for STGD1, such as c.5197-557G>T, which causes aberrant splicing, resulting in the generation of a premature termination codon [148]. Angeli et al. designed three CRISPR/Cas9 approaches (sgRNA/SpCas9, dual gRNAs/SpCas9, and dual gRNAs/SpCas9 nickase) to address this splicing defect. In patient-derived cone photoreceptor precursor cells carrying the c.5197-557G>T variant, these strategies were able to restore correct splicing by up to 83% and increase the total amount of correctly spliced transcripts by 1.8-fold [149]. This study demonstrates that CRISPR/Cas9 technology can effectively repair splicing defects caused by ABCA4 DIVs, providing a novel therapeutic strategy for STGD1.
Retinal Neovascularization
The retinal structure is morphologically divided into ten layers, with the inner five layers receiving blood from the central retinal artery and its branches [150]. Hypoxia and the accumulation of metabolic byproducts in the retina often leads to the development of retinal neovascularization [151]. This condition is not classified as a distinct disease; rather, it is closely associated with various systemic diseases and retinal disorders, such as diabetic retinopathy and retinopathy of prematurity [152, 153] (Figure 17A). Clinically, treatment for retinal neovascularization mainly involves anti-VEGF antibodies (such as ranibizumab) or antagonists of the VEGFR2 (such as aflibercept) [154]. However, both approaches require repeated long-term administration, imposing significant economic and lifestyle burdens on patients and presenting substantial challenges. CRISPR technology may offer a promising new solution to these issues.
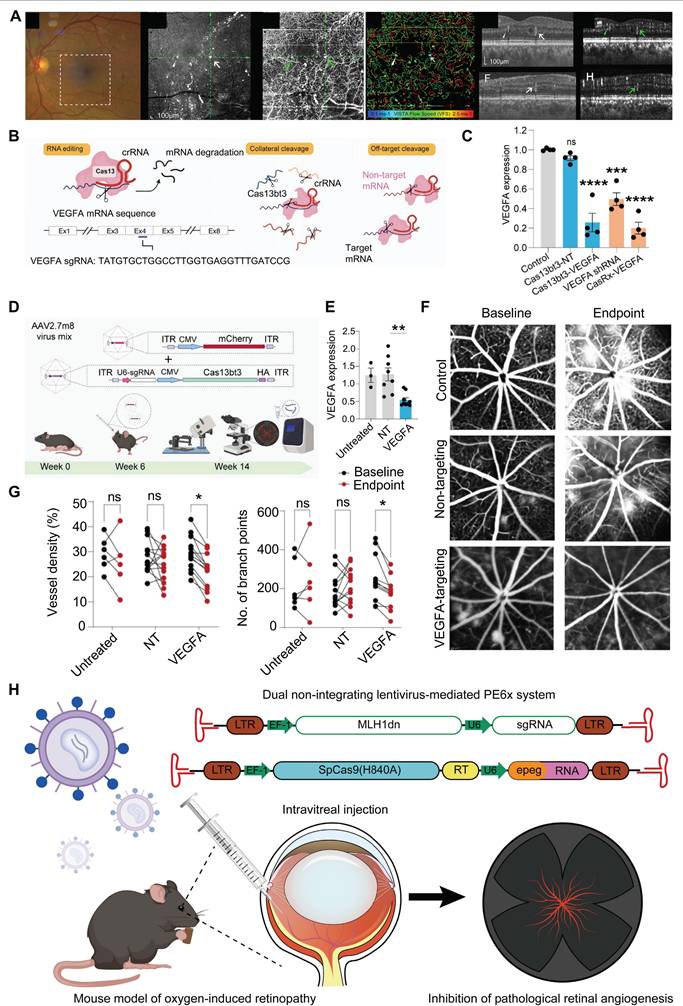
CRISPR/Cas13-based Cas13bt3 gene therapy can effectively silences VEGFA mRNA in both human retinal organoids and humanized VEGF transgenic mouse models (Figure 17B-C). Intravitreal delivery of Cas13bt3 by AAV2.7m8 significantly inhibits retinal neovascularization in the mouse [155] (Figure 17D-G). Cas13bt3 is smaller (∼775 amino acids) compared to traditional CRISPR/Cas13 systems, which allows it to be delivered via AAV vectors successfully [68]. With further preclinical research and optimization, Cas13bt3 is expected to become an effective gene therapy tool for treating retinal neovascular diseases.
VEGFR2 mediates almost all known VEGF-induced effects, including its effects on increasing microvascular permeability and angiogenesis [156]. Addressing this mechanism, Wu et al. developed an AAV-mediated CRISPR/SpCas9 system targeting VEGFR2. This system was delivered into retinal vascular endothelial cells in a mouse model of oxygen-induced retinopathy via intravitreal injection, enabling VEGFR2 genome editing and suppressing pathological retinal neovascularization [157]. Additionally, Huang et al. established a PE6x system within two lentiviral vectors, with one carrying an enhanced PE gRNA and Cas9 nickase fused with an optimized reversal transcriptase, and the other conveying a nicking gRNA and a dominant negative DNA mismatch repair protein (DN-MLH1) to improve PE efficiency [158]. The PE6x system was employed to introduce a T17967A mutation at the VEGFR2 gene locus, which resulted in the generation of an earlier stop codon (TAG, K796stop) from the original AAG (K796). Because of the PE6x induced stop codon, a dominant negative VEGFR2 (DN-VEGFR2) with 795 amino acids was produced during translation. This DN-VEGFR2 effectively blocked VEGF induced VEGFR2 phosphorylation and inhibited the activation of the VEGF/VEGFR2 signaling, thereby impeding pathological retinal neovascularization [158] (Figure 17H). Unlike traditional gene editing methods, PE6x allows for the various gene modifications, such as base substitutions and small insertions/deletions, to be performed without generating a DSB. This greatly minimizes off-target effects and potential genomic instability.
Choroidal Neovascularization (CNV)
CNV is defined as new blood vessels that proliferate from the choroid and protrude through fenestrations in Bruch's membrane into the subretinal pigment epithelium or subretinal space [159]. So far, studies on CRISPR gene editing of CNV are strongly biased towards traditional pro-angiogenic elements such as VEGFA [160]. Utilizing the AAV9 vector to deliver the crRNA-guided endonuclease LbCpf1 derived from Lachnospiraceae bacteria, Taeyoung Koo and colleagues targeted the retinas of mice to knockout the angiogenesis-related genes Vegfa and Hif1α [161] (Figure 18A). The coding sequence of LbCpf1 (~3.7 kbp) is less than that of SpCas9, enabling its packaging into AAV vectors for delivery. Importantly, targeting Vegfa or Hif1α with LbCpf1 significantly reduced the area of laser-induced CNV [161] (Figure 18B-C).
Recent key research achievements in retinal neovascularization: (A) Multimodal imaging of a patient with diabetic retinopathy imaged by highspeed, swept-source OCT. Adapted with permission from [152], copyright 2025, the authors. (B) Schematic of single and multiplex targeting with Cas13bt3. Adapted with permission from [155], copyright 2024, the authors. (C) VEGFA mRNA knockdown from Cas13bt3. Adapted with permission from [155], copyright 2024, the authors. (D) Schematic of experimental procedure for intravitreal treatment of Kimba mice. Adapted with permission from [155], copyright 2024, the authors. (E) Expression of human VEGFA mRNA in Kimba retinas. Adapted with permission from [155], copyright 2024, the authors. (F) Central fluorescein fundus angiography images after Cas13bt3 treatment. Adapted with permission from [155], copyright 2024, the authors. (G) Evaluate the situation of angiogenesis. Adapted with permission from [155], copyright 2024, the authors. (H) Schematic of PE6x system-mediated suppression of angiogenesis in an oxygen-induced retinopathy mouse model.
Summary of recent representative research results on CNV: (A) LbCpf1 crRNA targets Vegfa and Hif1a genes. Adapted with permission from [161], copyright 2018, the authors. (B-C) LbCpf1 gene editing can reduce the area of laser-induced CNV. Adapted with permission from [161], copyright 2018, the authors. (D) Nme2Cas9 gene editing can significantly alleviate CNV. Adapted with permission from [162], copyright 2023, John Wiley & Sons Australia, Ltd. (E) Nme2Cas9 sgRNA targets Hif1α, Vegfa, Vefgr2. Adapted with permission from [162], copyright 2023, John Wiley & Sons Australia, Ltd. (F-H) AAV-delivered Nme2Cas9 knocks out Vegfa, inhibiting laser-induced CNV. Adapted with permission from [162], copyright 2023, John Wiley & Sons Australia, Ltd.
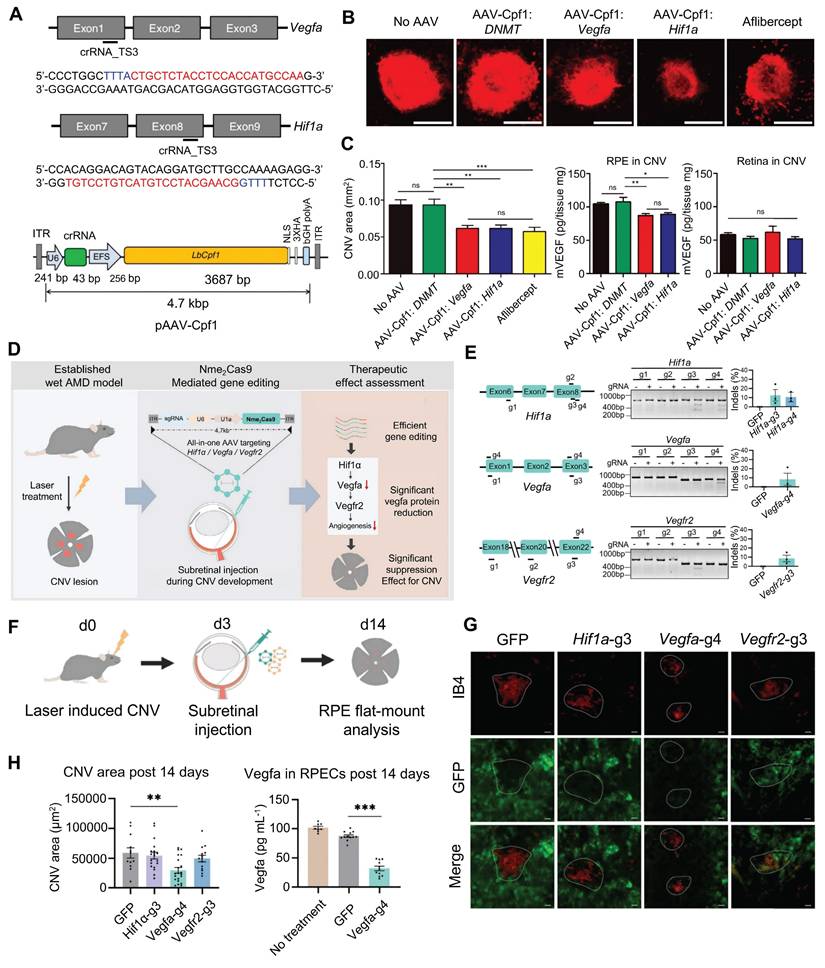
Nme2Cas9 is a Neisseria meningitidis-derived Cas9 protein. Compared with SpCas9, Nme2Cas9 is smaller in size and can be packaged into a single AAV vector. Hu et al. designed sgRNAs targeting the Hif1α, Vegfa, and Vegfr2 genes and delivered the AAV-mediated Nme2Cas9/sgRNA system to mouse RPE cells via subretinal injection (Figure 18D-F). The results demonstrated that only early intervention with the Vegfa gene significantly reduced the area of CNV lesions (by 49.5%), whereas intervention with Hif1α and Vegfr2 did not yield significant therapeutic effects [162] (Figure 18G-H).
Moreover, Park and colleagues previously substituted the CMV promoter with an RPE-specific one (VMD2) in a lentiviral vector expressing Cas9, resulting in specific Cas9 expression and high editing efficiency in RPE cells [163]. The VMD2 driven gVEGFA/Cas9 system knockout effectively inhibited laser-induced CNV, with little adverse effects on the neural retina, providing a new strategy for the safer and effective treatment of CNV [163]. Ling and colleagues synthesized an integrated CRISPR lentiviral vector (mLP-CRISPR) carrying both Cas9 and gRNA.
This mLP-CRISPR system can effectively inhibit the formation of CNV in a laser-induced mouse model [164]. Notably, the mLP-CRISPR system does not elicit potent innate or adaptive immune responses during in vivo applications, with reduced potential safety risks. HG202 developed by HuidaGene Therapeutics is a CRISPR/Cas13-based RNA editing candidate for the treatment of nAMD, and it works by locally reducing the expression of VEGFA mRNA within the retina. Currently, HG202 is undergoing clinical trials to evaluate its safety, tolerability, and efficacy (ClinicalTrials.gov ID, NCT06031727).
Retinoblastoma
Retinoblastoma (RB), the predominant pediatric retinal malignancy, arises from biallelic inactivation of the chromosome 13-located tumor suppressor gene Rb1 in germline or somatic lineages [165]. Although recent advances have improved RB cure rates, current treatments still lack effective molecular targeted drugs, partly because of the preclinical research is limited by existing animal models characterized by long tumor latency and low penetrance [166]. Pioneering this frontier, Naert et al. engineered CRISPR/Cas9 mediated co-knockout of Rb1 and retinoblastoma-like 1 (Rbl1) in Xenopus tropicalis, establishing the non-mammalian genetic RB model exhibiting rapid-onset tumors (minimum 36 days) and high penetrance (73%) [167]. This paradigm provides an ethically and economically optimized platform for deconvoluting RB pathogenesis and accelerating drug discovery. Notably, 6% of heritable RB cases progress to trilateral retinoblastoma (TRB), characterized by synchronous intracranial neuroectodermal malignancies [168]. Leveraging human embryonic stem cells (hESCs) as a developmental model, Avior et al. established Rb1-null hESCs via CRISPR/Cas9 editing. These cells generated teratomas exhibiting neural overproliferation and mitochondrial dysfunction-key features of TRB pathology [169]. This model elucidates both the developmental roles of Rb1 and its tumorigenic effects, providing a robust platform for drug discovery.
CRISPR Delivery Systems
The development of gene editing technologies, particularly that of the CRISPR/Cas9 system, has opened new avenues for the treatment of various genetic disorders [171, 172]. However, the effective delivery of these systems to target cells in vivo necessitates the development of suitable delivery strategies. In recent years, substantial advancements have been made in delivery methods for CRISPR systems, evolving from early viral vectors and physical delivery to the current extensively researched LNPs and virus-like particles (VLPs) [173] (Figure 19).
The physical delivery methods of the CRISPR system involve directly penetrating the cell membrane or cell wall through physical means, such as electroporation and microinjection, to deliver CRISPR components into cells [174]. Viral vector delivery systems, including AAVs, lentivirus (LVs) and adenovirus (AVs), have been widely studied because of their high transduction efficiency. AVs leverage high transduction efficiency and large cargo capacity to mediate transient gene expression, making them suitable for vaccine development and large-fragment gene repair [175]. AAVs enable safe and targeted delivery due to their low immunogenicity, sustained expression, and serotype-dependent tissue tropism, establishing AAVs as the most validated and widely deployed platform for in vivo gene delivery [176]. LVs enable long-term stable expression through genomic integration capability, infect both dividing and non-dividing cells, and are suitable for establishing stable cell lines and long-term in vivo editing [177]. However, the limited payload capacity of viral vectors and potential for eliciting immune responses limit their application.
These disadvantages of AAV vectors have led to the exploration of non-viral delivery systems, which include mainly cell-penetrating peptide (CPPs), gold nanoparticles (AuNPs), DNA nanoclew, LNPs and VLPs [178]. Non-viral delivery methods have many important benefits such as their ability to avoid the limitations of viral packaging sizes and a greatly increased ability to tightly control the dosage, duration, and specificity of action [179]. Of these, LNPs are the most well representative non-viral delivery vehicles to date. Typically, negatively charged nucleic acids form complexes with positively charged lipids through electrostatic interactions, creating LNPs that protect the nucleic acids from nuclease degradation and facilitate cellular uptake via endocytosis. By optimizing lipid formulations, LNPs achieve efficient and low-toxicity targeted delivery to specific tissue cells [180]. VLPs are self-assembled empty shells composed of viral structural proteins, capable of efficiently delivering mRNA and RNP to specific cells [181]. Combining viral transduction efficiency with the biosafety of non-viral vectors, VLPs have emerged as a promising delivery platform for CRISPR-based therapy. Additionally, CPPs enable carrier-independent delivery of CRISPR systems via short peptide mediated efficient transmembrane transport [182]; AuNPs possess dual capabilities for multiplexed cargo loading and photothermal/photo-triggered release [183]; DNA nanoclew utilizes rolling circle replication technology to self-assemble into high density DNA nanostructures, achieving efficient CRISPR RNP delivery [184].
In summary, the development and refinement of CRISPR/Cas9 systems are progressing rapidly, with an increasing number of studies focusing on optimizing their composition and delivery strategies to enable broader clinical applications (Table 2).
Schematic diagram of CRISPR delivery vectors.
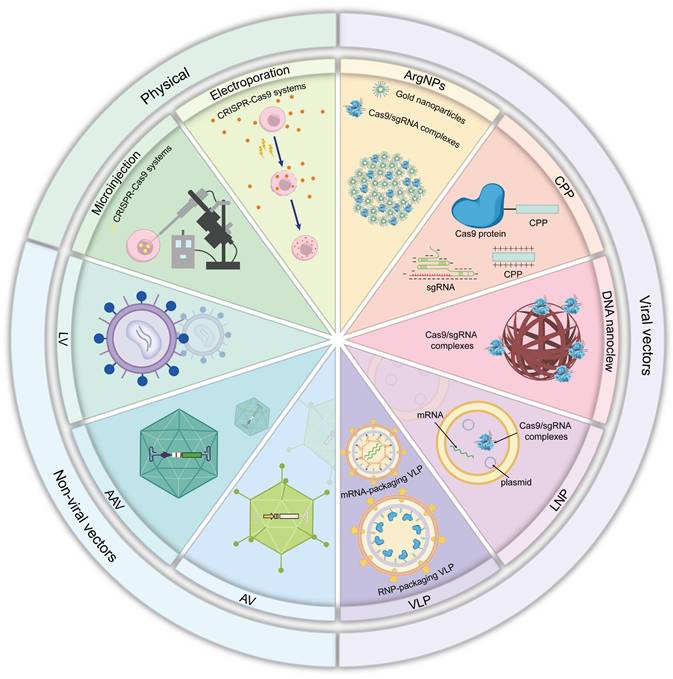
The advantages and disadvantages of CRISPR delivery systems and their applications in ocular diseases.
| Category | Applications | Cargo options | Advantages | Limitations |
|---|---|---|---|---|
| Physical | ||||
| Microinjection | Congenital aniridia [109]/RB [167] | DNA/mRNA/ proteins | High-precision single-cell delivery/Low off-target risk/Suitability for hard-to-transfect cells. | Labor-intensive operation with substantial damage/Low throughput/Stage-dependency |
| Electroporation | Corneal scar and corneal wound healing [75]/ Congenital aniridia [108]/ LCA [119]/RP [134]/STGD1 [149]/Retinal neovascularization [157] | DNA/mRNA/ proteins | Efficient batch processing/Low mechanical damage/Enhanced HDR efficiency | Potential cytotoxicity/Slightly higher off-target risk |
| Viral Delivery | ||||
| AAVs | HSK [86]/CoNV [99]/ Glaucoma [105]/ LCA [115]/RP [133]/ BCD [50]/STGD1 [147]/ Retinal neovascularization [155]/CNV [161] | DNA | High biosafety/Low immunogenicity/Tissue tropism/Stable episomal expression/Clinical validation | Small packaging capacity/Preexisting antibody restriction/Potential integration risk |
| AVs | Corneal dystrophy [96]/ Glaucoma [46] | DNA | Large packaging capacity/Efficient transduction/Episomal expression/Scalable production | High immunogenicity/Short expression duration |
| LVs | LCA [123]/ RP [132]/Retinal neovascularization [158] /CNV [163] | RNA | Large-capacity integration/Efficient in vitro screening | Random integration risk/Long-term off-target effects/ High immunogenicity |
| Non-Viral Delivery | ||||
| CPPs | Not mentioned | DNA/mRNA/ Proteins | Rapid action/High stability/Low cytotoxicity | Prone to proteolytic degradation/Endosomal escape barrier/Insufficient targeting |
| LNPs | Congenital aniridia [110] | DNA/mRNA/ proteins | Efficient endosomal escape/Compatible with multiple delivery forms/Low immunogenicity | Targeting depends on ligands/Particle size limitation/Difficult production standardization |
| Polymers | Not mentioned | RNPs | Flexible modification/High loading capacity/Biocompatibility | Cytotoxicity/Poor serum stability/Low nuclear transport efficiency |
| DNA nanoclew | Not mentioned | RNPs | Sequence programmability/Precise design/Stimuli-responsive release | Exhibits certain immunogenicity/Poor stability/Low in vivo delivery efficiency |
| AuNPs | Not mentioned | RNPs | Strong nucleic acid binding and protection/Cell penetrability/Low immunogenicity | Potential cytotoxicity/Insufficient targeting |
| VLPs | HSK [20] | RNPs/RNA | No integration risk/Transient expression safety/High loading capacity | In vivo stability challenge/Tissue penetration limitation |
Abbreviations: AAV: adeno-associated virus; AVs: adenovirus; LVs: lentivirus; CPPs: cell-penetrating peptide; LNPs: lipid nanoparticles; AuNPs: gold nanoparticles; VLPs: virus-like particles; CoNV: corneal neovascularization; HSK: herpetic stromal keratitis; LCA: leber congenital amaurosis; RP: retinitis pigmentosa; CNV: choroidal neovascularization; RB: retinoblastoma; STGD1: stargardt disease.
Safety and Ethical Regulations of CRISPR/Cas Gene Editing Systems
From 2025 onwards, there are going to be 10 to 20 gene therapies approved annually by the FDA, which indicative of the rapid development and increasing maturity of gene therapy [185]. Luxturna, a gene augmentation therapy delivering functional RPE65 to RPE cells for treating LCA, marking a milestone in ophthalmic gene therapy [186]. CRISPR gene editing technology overcomes the limitation of conventional gene replacement therapy, which only target autosomal recessive disorders, and significantly expanding the scope of gene therapy applications. Notably, multiple candidate drugs demonstrate significant clinical promise, such as the EDIT-101, HG202, ZVS203e, BD111 and BD113. These collective advances establish a robust foundation for future regulatory approvals of CRISPR-based ocular therapeutics.
Despite significant advancements in the precision of CRISPR technology, safety concerns such as off-target effects, immune responses, and the long-term stability of gene editing systems remain critical issues [187]. Therefore, comprehensive preclinical studies and clinical trials to evaluate the safety and potential long-term impacts of these technologies are essential. Ophthalmic safety assessments should include the following:
(1) Behavioral tests such as localization tasks, alongside electrophysiological tests such as visual evoked potential, to evaluate changes in vision resulting from CRISPR gene editing.
(2) Optical coherence tomography (OCT) for obtaining high-resolution images of retinal structure, aiding in the assessment of retinal changes post-CRISPR treatment, including changes in thickness, layering, and structural integrity.
(3) Fundus fluorescein angiography (FFA) to observe the health of retinal blood vessels and identify leakage or pathological changes.
(4) Histological analyses to examine cellular morphology, structural changes, and expression of damage markers in ocular structures.
(5) Measurement of biomarkers of ocular inflammation (e.g., cytokines and chemokines) to evaluate the immune response elicited by gene editing technology.
By employing these methodologies, researchers can comprehensively assess the extent of damage and potential side effects of CRISPR gene editing technologies on the eye, thus providing crucial experimental data to support clinical applications.
The application of CRISPR technology has raised many ethical concerns. Therapeutic editing aims to repair or replace defective genes to cure diseases, whereas enhancement editing seeks to improve an individual's traits or capabilities. The ambiguity surrounding these boundaries can lead to ethical debates, such as whether gene editing should be permitted for healthy individuals to enhance vision or other characteristics. Furthermore, patient autonomy and informed consent are paramount in gene editing, particularly when it involves minors or individuals with limited cognitive ability; ensuring that their interests are adequately represented is a complex challenge.
To ensure the safe and responsible application of CRISPR technology, effective regulation is essential. Ethical review mechanisms concerning CRISPR applications need to be strengthened to promote transparency and compliance in research, including training for researchers and the formation of independent review boards. Regulatory agencies must establish clear guidelines and regulations to ensure both the safety and ethical integrity of the technology. As gene editing technologies such as CRISPR continue to evolve, the scientific community, ethicists, and policymakers must work together to formulate ethical guidelines that align with societal values.
Ocular Diseases Remain Unaddressed by CRISPR/Cas Gene Editing Systems
CRISPR technology was initially applied primarily to treat rare blinding hereditary eye diseases such as LCA and RP. As the technology matures, CRISPR has begun to demonstrate promise in common ophthalmic conditions like AMD and viral keratitis. Nevertheless, numerous vision-threatening diseases still lack effective treatments. CRISPR offers new hope for managing these conditions.
(1) Keratoconus (KC): KC is a common progressive corneal disorder characterized by corneal thinning and asymmetric conical protrusion, which can cause severe visual impairment [188]. The pathogenesis of KC is highly complex, involving interactions among genetic, environmental, and mechanical factors [189]. Polygenic loci such as ZNF469, VSX1, and SOD1 have been identified [190]. CRISPR-assisted high throughput genetic screening can help uncover disease associated genetic variants, facilitating the discovery of novel genetic markers. The cornea's biophysical properties make it an ideal candidate for CRISPR therapy. For instance, CRISPRi technology could knockout matrix metalloproteinase-related genes (such as Timp3) to inhibit corneal stromal degradation, halting disease progression. Additionally, developing keratoconus animal models using CRISPR technology represents a promising avenue for research.
(2) High myopia: High myopia can lead to complications such as macular degeneration, retinal detachment, and glaucoma, representing a major cause of irreversible blindness in young adults [191]. It is currently understood as a multifactorial disorder involving both genetic and environmental factors, with over 30 associated loci reported (including LRRC46, SCO2, CCDC111, SLC39A5, CPSF1) demonstrating a complex polygenic inheritance pattern [192]. CRISPR technology shows significant research potential for high myopia. Specifically, using a CRISPR activation system to upregulate COL1A1 expression, delivered via AAV vectors targeted to scleral cells through posterior scleral injection, holds promise for enhancing scleral collagen cross-linking and slowing myopia progression.
(3) Dry eye disease (DED): DED is a multifactorial ocular surface disorder that significantly impairs patients' quality of life and severe cases may lead to corneal ulcers and perforation [193]. Chronic inflammation represents a core mechanism in DED pathogenesis [194]. CRISPR technology enables targeted modulation of key genes involved in ocular surface inflammation and immune responses [195]. For instance, CRISPR/Cas9-mediated gene knockout can specifically reduce expression of pro-inflammatory cytokines (e.g., TNF-α, IL-1β) in ocular surface cells, thereby mitigating inflammatory cascades. For aqueous-deficient DED caused by lacrimal gland dysfunction, CRISPR-based overexpression of regenerative growth factors may promote repair and functional restoration of damaged lacrimal tissue.
(4) Congenital stationary night blindness (CSNB): CSNB is a group of inherited retinal disorders caused by dysfunctional signal transduction within the retina [196]. To date, over 20 CSNB-associated pathogenic genes have been identified [196]. While animal studies have successfully restored visual function in CSNB models using AAV-mediated gene augmentation therapy, CRISPR technology remains unexplored for this condition [197]. Unlike conventional gene replacement approaches, CRISPR enables precise genomic level modification of disease-causing genes. For instance, CRISPR/Cas9-mediated HITI technology holds promise for precisely repairing CSNB-related mutations, potentially achieving lifelong therapeutic benefits from a single treatment.
Conclusion and Outlook
This review provides an in-depth exploration of the innovative applications of CRISPR technology in the treatment of ocular diseases. The precise gene-editing capabilities of CRISPR have demonstrated significant potential across a range of major ocular conditions, from those that affect the anterior segment to those that affect the posterior segment. This technology not only aids in elucidating pathological mechanisms but also promotes the advancement of personalized therapies. However, the clinical application of CRISPR is not without challenges, particularly concerning off-target effects and ethical considerations, which necessitate further safety validation and regulatory measures. Nevertheless, CRISPR is poised to lead the future of precision medicine in ophthalmology, resulting in revolutionary changes in the field.
The application of CRISPR technology in ophthalmic diseases undoubtedly marks a new chapter in precision medicine. Precision medicine emphasizes tailoring treatment plans based on the unique genomic characteristics of individuals, enabling the formulation of specific therapeutic strategies aligned with patients' genetic profiles. CRISPR not only facilitates the editing of known pathogenic genes but can also be combined with genomic sequencing technologies to enable a comprehensive analysis of a patient's genomic data, thereby identifying potential pathogenic mutations. Additionally, with the rapid development of artificial intelligence (AI), breakthroughs in CRISPR applications within ophthalmology are expected to accelerate further. AI can assist in the design and optimization of gRNAs, significantly enhancing the editing efficiency and specificity of CRISPR systems. AI and machine learning techniques can be employed to identify new gene targets, predict the outcomes of gene editing, and monitor potential off-target effects, thereby improving the comprehensive application of CRISPR gene editing.
Notably, as the clinical applications of CRISPR have expanded in ophthalmology, it is imperative to consider the ethical and legal challenges that accompany these advancements to ensure the safety and affordability of the technology. Establishing robust international standards and regulatory frameworks is crucial to mitigate potential ethical disputes and protect patient rights. Only under a stringent ethical framework and legal guidance can CRISPR technology be safely and responsibly applied in clinical practice, ultimately providing tangible health benefits to a wide range of patients.
The rapid advancement of scientific technology is remarkable, with concepts such as silicon-based life, brain-machine interfaces, and AI robots becoming a reality. CRISPR technology has the potential to revolutionize the field of ophthalmology, transitioning from science fiction to reality. It promises to bring about multifaceted, innovative changes that transcend current treatment limitations and create a new medical paradigm.
Abbreviations
Cas: CRISPR associated; gRNA: guide RNA; RP: retinitis pigmentosa; LCA: leber congenital amaurosis; HSV: herpes simplex virus; CoNV: corneal neovascularization; VEGF: vascular endothelial growth factor; CRISPR: clustered regularly interspaced short palindromic repeats; crRNA: CRISPR RNA; PAM: protospacer adjacent motif; NHEJ: non-homologous end joining; HDR: homology-directed repair; ABE: adenine base editor; CBE: cytosine base editor; PE: prime editor; hCECs: human corneal endothelial cells; SOX2: sex-determining region Y-box 2; AQP5: aquaporin 5; NGF: nerve growth factor; hCSFs: human corneal stromal fibroblasts; CMFs: corneal myofibroblasts; HK: herpetic keratitis; HSK: herpetic stromal keratitis; HELP: HSV-1 erasing lentiviral particles; mLPs: mRNA-carrying lentiviral particles; NECTIN-1: nectin cell adhesion molecule 1; TBCD: thiel-behnke corneal dystrophy; GCD: granular corneal dystrophy; FECD: fuchs' endothelial corneal dystrophy; hUVECs: human umbilical vein endothelial cells; RGCs: retinal ganglion cells; TM: trabecular meshwork; LSCs: limbal stem cells; LNPs: lipid nanoparticles; RPE: retinal pigment epithelium; hiPSCs: human induced pluripotent stem cells; Rho: rhodopsin; CRISPRi: CRISPR interference; PHD: prolyl hydroxylase; VHL: von hippel-lindau; HIF: hypoxia-inducible factor; hfCas13X: high-fidelity Cas13X; BCD: bietti crystalline corneoretinal dystrophy; HITI: homology independent targeted integration; VECs: vascular endothelial cells; DN-VEGFR2: dominant-negative VEGF receptor 2; NILVs: non-integrative lentiviral vectors; RB: retinoblastoma; TRb: trilateral retinoblastoma; STGD1: stargardt disease; AAVs: adeno-associated virus; AVs: adenovirus; LVs: lentivirus; VLPs: virus-like particles; FDA: Food and Drug Administration; VEP: visual evoked potentials; AI: artificial intelligence.
Acknowledgements
This study was supported by National Natural Science Foundation of China (82571179, 82401219, 82371032), Natural Science Foundation of Shandong Province of China (ZR2024QH257), and Shandong Province College Youth Innovation Team Project (2024KJJ014), Major Basic Research Project of Natural Science Foundation of Shandong Province (ZR2023ZD60), Taishan Scholar Program (20231255), Academic Promotion Program of Shandong First Medical University (2019RC009).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Flaxman SR, Bourne RRA, Resnikoff S, Ackland P, Braithwaite T, Cicinelli MV. et al. Global causes of blindness and distance vision impairment 1990-2020: a systematic review and meta-analysis. Lancet Glob Health. 2017;5:e1221-e34
2. Yu W, Wu Z. Ocular delivery of CRISPR/Cas genome editing components for treatment of eye diseases. Adv Drug Deliv Rev. 2021;168:181-95
3. Choi EH, Suh S, Sears AE, Hołubowicz R, Kedhar SR, Browne AW. et al. Genome editing in the treatment of ocular diseases. Exp Mol Med. 2023;55:1678-90
4. Peng Z, Nagarajan V, Horai R, Jittayasothorn Y, Mattapallil MJ, Caspi RR. Ocular immune privilege in action: The living eye imposes unique regulatory and anergic gene signatures on uveitogenic T cells. Cell Rep. 2025;44:115780
5. Katamay R, Nussenblatt RB. Blood-retinal barrier, immune privilege, and autoimmunity. Retina. 2012: 579.
6. Kantor A, McClements ME, Peddle CF, Fry LE, Salman A, Cehajic-Kapetanovic J. et al. CRISPR genome engineering for retinal diseases. Prog Mol Biol Transl Sci. 2021;182:29-79
7. Al-Saikhan FI. The gene therapy revolution in ophthalmology. Saudi Journal of Ophthalmology. 2013;27:107-11
8. Askou AL, Jakobsen TS, Corydon TJ. Retinal gene therapy: an eye-opener of the 21st century. Gene therapy. 2021;28:209-16
9. Darrow JJ. Luxturna: FDA documents reveal the value of a costly gene therapy. Drug Discov Today. 2019;24:949-54
10. Liu MM, Tuo J, Chan CC. Gene therapy for ocular diseases. Br J Ophthalmol. 2011;95:604-12
11. Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096
12. Villiger L, Joung J, Koblan L, Weissman J, Abudayyeh OO, Gootenberg JS. CRISPR technologies for genome, epigenome and transcriptome editing. Nat Rev Mol Cell Biol. 2024;25:464-87
13. Simeonov DR, Marson A. CRISPR-based tools in immunity. Annu Rev Immunol. 2019;37:571-97
14. Koch L. CRISPR editing within microbial communities. Nat Rev Genet. 2022;23:72
15. Jiang F, Doudna JA. CRISPR-Cas9 structures and mechanisms. Annual review of biophysics. 2017;46:505-29
16. Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262-78
17. Manghwar H, Lindsey K, Zhang X, Jin S. CRISPR/Cas system: recent advances and future prospects for genome editing. Trends Plant Sci. 2019;24:1102-25
18. Sharma G, Sharma AR, Bhattacharya M, Lee SS, Chakraborty C. CRISPR-Cas9: a preclinical and clinical perspective for the treatment of human diseases. Mol Ther. 2021;29:571-86
19. Ledford H. CRISPR treatment inserted directly into the body for first time. Nature. 2020;579:185
20. Yin D, Ling S, Wang D, Dai Y, Jiang H, Zhou X. et al. Targeting herpes simplex virus with CRISPR-Cas9 cures herpetic stromal keratitis in mice. Nat Biotechnol. 2021;39:567-77
21. Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096
22. Li T, Yang Y, Qi H, Cui W, Zhang L, Fu X. et al. CRISPR/Cas9 therapeutics: progress and prospects. Signal Transduct Target Ther. 2023;8:36
23. Guo C, Ma X, Gao F, Guo Y. Off-target effects in CRISPR/Cas9 gene editing. Front Bioeng Biotechnol. 2023;11:1143157
24. Ma Y, Zhang L, Huang X. Genome modification by CRISPR/Cas9. Febs j. 2014;281:5186-93
25. Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429-33
26. Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167-70
27. Liu L, Zhao D, Ye L, Zhan T, Xiong B, Hu M. et al. A programmable CRISPR/Cas9-based phage defense system for Escherichia coli BL21(DE3). Microb Cell Fact. 2020;19:136
28. Koonin EV, Makarova KS. Origins and evolution of CRISPR-Cas systems. Philos Trans R Soc Lond B Biol Sci. 2019;13:374
29. Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759-71
30. Hidalgo-Cantabrana C, Barrangou R. Characterization and applications of Type I CRISPR-Cas systems. Biochem Soc Trans. 2020;48:15-23
31. Ramos JN, Baio PVP, Veras JFC, Vieira É MD, Mattos-Guaraldi AL, Vieira VV. Novel configurations of type I-E CRISPR-Cas system in Corynebacterium striatum clinical isolates. Braz J Microbiol. 2023;54:69-80
32. Li X, Han J, Yang J, Zhang H. The structural biology of type III CRISPR-Cas systems. J Struct Biol. 2024;216:108070
33. Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E. et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol cell. 2015;60:385-97
34. Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W. et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15:169-82
35. Wang Y, Jiang H, Li M, Xu Z, Xu H, Chen Y. et al. Delivery of CRISPR/Cas9 system by AAV as vectors for gene therapy. Gene. 2024;927:148733
36. Shivram H, Cress BF, Knott GJ, Doudna JA. Controlling and enhancing CRISPR systems. Nat Chem Biol. 2021;17:10-9
37. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816-21
38. Gupta D, Bhattacharjee O, Mandal D, Sen MK, Dey D, Dasgupta A. et al. CRISPR-Cas9 system: A new-fangled dawn in gene editing. Life Sci. 2019;232:116636
39. Jiang F, Doudna JA. CRISPR-Cas9 Structures and Mechanisms. Annu Rev Biophys. 2017;46:505-29
40. Amitai G, Sorek R. CRISPR-Cas adaptation: insights into the mechanism of action. Nat Rev Microbiol. 2016;14:67-76
41. Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N. et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935-49
42. Tyumentseva M, Tyumentsev A, Akimkin V. CRISPR/Cas9 landscape: current state and future perspectives. Int J Mol Sci. 2023 24
43. Savić N, Schwank G. Advances in therapeutic CRISPR/Cas9 genome editing. Transl Res. 2016;168:15-21
44. Chang HHY, Pannunzio NR, Adachi N, Lieber MR. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol. 2017;18:495-506
45. Scully R, Panday A, Elango R, Willis NA. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol. 2019;20:698-714
46. Jain A, Zode G, Kasetti RB, Ran FA, Yan W, Sharma TP. et al. CRISPR-Cas9-based treatment of myocilin-associated glaucoma. Proc Natl Acad Sci U S A. 2017;114:11199-204
47. Liao H, Wu J, VanDusen NJ, Li Y, Zheng Y. CRISPR-Cas9-mediated homology-directed repair for precise gene editing. Mol Ther Nucleic Acids. 2024;35:102344
48. Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ. et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144-9
49. Tornabene P, Ferla R, Llado-Santaeularia M, Centrulo M, Dell'Anno M, Esposito F. et al. Therapeutic homology-independent targeted integration in retina and liver. Nat Commun. 2022;13:1963
50. Meng X, Jia R, Zhao X, Zhang F, Chen S, Yu S. et al. In vivo genome editing via CRISPR/Cas9-mediated homology-independent targeted integration for Bietti crystalline corneoretinal dystrophy treatment. Nat Commun. 2024;15:3773
51. Lapinaite A, Knott GJ, Palumbo CM, Lin-Shiao E, Richter MF, Zhao KT. et al. DNA capture by a CRISPR-Cas9-guided adenine base editor. Science. 2020;369:566-71
52. Richter MF, Zhao KT, Eton E, Lapinaite A, Newby GA, Thuronyi BW. et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat Biotechnol. 2020;38:883-91
53. Chen L, Zhu B, Ru G, Meng H, Yan Y, Hong M. et al. Re-engineering the adenine deaminase TadA-8e for efficient and specific CRISPR-based cytosine base editing. Nat Biotechnol. 2023;41:663-72
54. Chen PJ, Hussmann JA, Yan J, Knipping F, Ravisankar P, Chen PF. et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell. 2021;184:5635-52.e29
55. Nelson JW, Randolph PB, Shen SP, Everette KA, Chen PJ, Anzalone AV. et al. Engineered pegRNAs improve prime editing efficiency. Nat Biotechnol. 2022;40:402-10
56. Zhao Z, Shang P, Mohanraju P, Geijsen N. Prime editing: advances and therapeutic applications. Trends Biotechnol. 2023;41:1000-12
57. Fu Y, He X, Ma L, Gao XD, Liu P, Shi H. et al. In vivo prime editing rescues photoreceptor degeneration in nonsense mutant retinitis pigmentosa. Nat Commun. 2025;16:2394
58. Cai R, Lv R, Shi X, Yang G, Jin J. CRISPR/dCas9 tools: epigenetic mechanism and application in genetranscriptional regulation. Int J Mol Sci. 2023 24
59. Radzisheuskaya A, Shlyueva D, Müller I, Helin K. Optimizing sgRNA position markedly improves the efficiency of CRISPR/dCas9-mediated transcriptional repression. Nucleic Acids Res. 2016;44:e141-e
60. Zhou H, Liu J, Zhou C, Gao N, Rao Z, Li H. et al. In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR-dCas9-activator transgenic mice. Nat Neurosci. 2018;21:440-6
61. Tong X, Zhang K, Han Y, Li T, Duan M, Ji R. et al. Fast and sensitive CRISPR detection by minimized interference of target amplification. Nat Chem Biol. 2024;20:885-93
62. Böhm S, Splith V, Riedmayr LM, Rötzer RD, Gasparoni G, Nordström KJ. et al. A gene therapy for inherited blindness using dCas9-VPR-mediated transcriptional activation. Sci Adv. 2020;6:eaba5614
63. Paul B, Montoya G. CRISPR-Cas12a: Functional overview and applications. Biomed J. 2020;43:8-17
64. Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E. et al. Discovery and functionalcharacterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60:385-97
65. Chen W, Ma J, Wu Z, Wang Z, Zhang H, Fu W. et al. Cas12n nucleases, early evolutionary intermediates of type V CRISPR, comprise a distinct family of miniature genome editors. Mol Cell. 2023;83:2768-80.e6
66. Senthilnathan R, Ilangovan I, Kunale M, Easwaran N, Ramamoorthy S, Veeramuthu A. et al. An update on CRISPR-Cas12 as a versatile tool in genome editing. Mol Biol Rep. 2023;50:2865-81
67. Yan F, Wang W, Zhang J. CRISPR-Cas12 and Cas13: the lesser known siblings of CRISPR-Cas9. Cell Biol Toxicol. 2019;35:489-92
68. Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J. et al. RNA editing with CRISPR-Cas13. Science. 2017;358:1019-27
69. Yang H, Patel DJ. Structures, mechanisms and applications of RNA-centric CRISPR-Cas13. Nat Chem Biol. 2024;20:673-88
70. Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ. et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280-4
71. Yoon PH, Zhang Z, Loi KJ, Adler BA, Lahiri A, Vohra K. et al. Structure-guided discovery of ancestral CRISPR-Cas13 ribonucleases. Science. 2024;385:538-43
72. Lotfi M, Rezaei N. CRISPR/Cas13: A potential therapeutic option of COVID-19. Biomed Pharmacother. 2020;131:110738
73. Zhou Q, Chen Y, Wang R, Jia F, He F, Yuan F. Advances of CRISPR-Cas13 system in COVID-19 diagnosis and treatment. Genes Dis. 2022;10:2414-24
74. Ghosh A, Singh VK, Singh V, Basu S, Pati F. Recent advancements in molecular therapeutics for corneal scar treatment. Cells. 2022 11
75. Chang YK, Hwang JS, Chung TY, Shin YJ. SOX2 activation using CRISPR/dCas9 promotes wound healing in corneal endothelial cells. Stem Cells. 2018;36:1851-62
76. Liu Y, Di G, Wang Y, Chong D, Cao X, Chen P. Aquaporin 5 facilitates corneal epithelial wound healing and nerve regeneration by reactivating akt signaling pathway. Am J Pathol. 2021;191:1974-85
77. Tripathi R, Sinha NR, Kempuraj D, Balne PK, Landreneau JR, Juneja A. et al. Evaluation of CRISPR/Cas9 mediated TGIF gene editing to inhibit corneal fibrosis in vitro. Exp Eye Res. 2022;220:109113
78. Rowe AM, St Leger AJ, Jeon S, Dhaliwal DK, Knickelbein JE, Hendricks RL. Herpes keratitis. Prog Retin Eye Res. 2013;32:88-101
79. Chang EJ, Dreyer EB. Herpesvirus infections of the anterior segment. Int Ophthalmol Clin. 1996;36:17-28
80. Green LK, Pavan-Langston D. Herpes simplex ocular inflammatory disease. Int Ophthalmol Clin. 2006;46:27-37
81. Hendricks RL, Tumpey TM, Finnegan A. IFN-gamma and IL-2 are protective in the skin but pathologic in the corneas of HSV-1-infected mice. J Immunol. 1992;149:3023-8
82. Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780-3
83. Harrell TL, Davido DJ, Bertke AS. Herpes simplex virus 1 (HSV-1) infected cell protein 0 (ICP0) targets of ubiquitination during productive infection of primary adult sensory neurons. Int J Mol Sci. 2023 24
84. Zhang B, Ding J, Ma Z. ICP4-associated activation of rap1b facilitates herpes simplex virus type I (HSV-1) infection in human corneal epithelial cells. Viruses. 2023 15
85. Roehm PC, Shekarabi M, Wollebo HS, Bellizzi A, He L, Salkind J. et al. Inhibition of HSV-1 replication by gene editing strategy. Sci Rep. 2016;6:23146
86. Chen Y, Zhi S, Liang P, Zheng Q, Liu M, Zhao Q. et al. Single AAV-mediated CRISPR-SaCas9 inhibits HSV-1 replication by editing ICP4 in trigeminal ganglion neurons. Mol Ther Methods Clin Dev. 2020;18:33-43
87. Xu X, Fan S, Zhou J, Zhang Y, Che Y, Cai H. et al. The mutated tegument protein UL7 attenuates the virulence of herpes simplex virus 1 by reducing the modulation of α-4 gene transcription. Virol J. 2016;13:152
88. Li Y, Wei Y, Li G, Huang S, Xu J, Ding Q. et al. Targeting NECTIN-1 based on CRISPR/Cas9 system attenuated the herpes simplex virus infection in human corneal epithelial cells in vitro. Transl Vis Sci Technol. 2022;11:8
89. Gagan S, Khapuinamai A, Kapoor D, Sharma P, Yadavalli T, Joseph J. et al. Exploring heparanase levels in tears: insights from herpes simplex virus-1 keratitis patients and animal studies. Invest Ophthalmol Vis Sci. 2024;65:7
90. Ren J, Antony F, Rouse BT, Suryawanshi A. Role of innate interferon responses at the ocular surface in herpes simplex virus-1-induced herpetic stromal keratitis. Pathogens. 2023 12
91. Soh YQ, Kocaba V, Weiss JS, Jurkunas UV, Kinoshita S, Aldave AJ. et al. Corneal dystrophies. Nat Rev Dis Primers. 2020;6:46
92. Weiss JS, Møller HU, Aldave AJ, Seitz B, Bredrup C, Kivelä T. et al. IC3D classification of corneal dystrophies-edition 2. Cornea. 2015;34:117-59
93. Wang L, Zhao C, Zheng T, Zhang Y, Liu H, Wang X. et al. Torin 1 alleviates impairment of TFEB-mediated lysosomal biogenesis and autophagy in TGFBI (p.G623_H626del)-linked thiel-behnke corneal dystrophy. Autophagy. 2022;18:765-82
94. Taketani Y, Kitamoto K, Sakisaka T, Kimakura M, Toyono T, Yamagami S. et al. Repair of the TGFBI gene in human corneal keratocytes derived from a granular corneal dystrophy patient via CRISPR/Cas9-induced homology-directed repair. Sci Rep. 2017;7:16713
95. Hu J, Rong Z, Gong X, Zhou Z, Sharma VK, Xing C. et al. Oligonucleotides targeting TCF4 triplet repeat expansion inhibit RNA foci and mis-splicing in Fuchs' dystrophy. Hum Mol Genet. 2018;27:1015-26
96. Uehara H, Zhang X, Pereira F, Narendran S, Choi S, Bhuvanagiri S. et al. Start codon disruption with CRISPR/Cas9 prevents murine Fuchs' endothelial corneal dystrophy. Elife. 2021 10
97. Mohan RR, Kempuraj D, D'Souza S, Ghosh A. Corneal stromal repair and regeneration. Prog Retin Eye Res. 2022;91:101090
98. Stevenson W, Cheng SF, Dastjerdi MH, Ferrari G, Dana R. Corneal neovascularization and the utility of topical VEGF inhibition: ranibizumab (Lucentis) vs bevacizumab (Avastin). Ocul Surf. 2012;10:67-83
99. Zeng Z, Li S, Ye X, Wang Y, Wang Q, Chen Z. et al. Genome editing VEGFAprevents corneal neovascularization in vivo. Adv Sci (Weinh). 2024;11:e2401710
100. Di Zazzo A, Gaudenzi D, Yin J, Coassin M, Fernandes M, Dana R. et al. Corneal angiogenic privilege and its failure. Exp Eye Res. 2021;204:108457
101. Weinreb RN, Aung T, Medeiros FA. The pathophysiology and treatment of glaucoma: a review. Jama. 2014;311:1901-11
102. Kwon YH, Fingert JH, Kuehn MH, Alward WL. Primary open-angle glaucoma. N Engl J Med. 2009;360:1113-24
103. Chowdhury UR, Hann CR, Stamer WD, Fautsch MP. Aqueous humor outflow: dynamics and disease. Invest Ophthalmol Vis Sci. 2015;56:2993-3003
104. Scozzafava A, Supuran CT. Glaucoma and the applications of carbonic anhydrase inhibitors. Subcell Biochem. 2014;75:349-59
105. Jiang J, Kong K, Fang X, Wang D, Zhang Y, Wang P. et al. CRISPR-Cas9-mediated deletion of carbonic anhydrase 2 in the ciliary body to treat glaucoma. Cell Rep Med. 2024;5:101524
106. van Velthoven AJH, Utheim TP, Notara M, Bremond-Gignac D, Figueiredo FC, Skottman H. et al. Future directions in managing aniridia-associated keratopathy. Surv Ophthalmol. 2023;68:940-56
107. Hingorani M, Hanson I, van Heyningen V. Aniridia. Eur J Hum Genet. 2012;20:1011-7
108. Roux LN, Petit I, Domart R, Concordet JP, Qu J, Zhou H. et al. Modeling of aniridia-related keratopathy by CRISPR/Cas9 genome editing of human limbal epithelial cells and rescue by recombinant PAX6 protein. Stem Cells. 2018;36:1421-9
109. Mirjalili Mohanna SZ, Hickmott JW, Lam SL, Chiu NY, Lengyell TC, Tam BM. et al. Germline CRISPR/Cas9-mediated gene editing prevents vision loss in a novel mouse model of aniridia. Mol Ther Methods Clin Dev. 2020;17:478-90
110. Adair BA, Korecki AJ, Djaksigulova D, Wagner PK, Chiu NY, Lam SL. et al. ABE8e corrects pax6-aniridic variant in humanized mouse ESCs and via LNPs in ex vivo cortical neurons. Ophthalmol Ther. 2023;12:2049-68
111. Kumaran N, Moore AT, Weleber RG, Michaelides M. Leber congenital amaurosis/early-onset severe retinal dystrophy: clinical features, molecular genetics and therapeutic interventions. Br J Ophthalmol. 2017;101:1147-54
112. Tsang SH, Sharma T. Leber congenital amaurosis. Adv Exp Med Biol. 2018;1085:131-7
113. den Hollander AI, Roepman R, Koenekoop RK, Cremers FP. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog Retin Eye Res. 2008;27:391-419
114. Perrault I, Delphin N, Hanein S, Gerber S, Dufier JL, Roche O. et al. Spectrum of NPHP6/CEP290 mutations in Leber congenital amaurosis and delineation of the associated phenotype. Hum Mutat. 2007;28:416
115. Ruan GX, Barry E, Yu D, Lukason M, Cheng SH, Scaria A. CRISPR/Cas9-mediated genome editing as a therapeutic approach for leber congenital amaurosis 10. Mol Ther. 2017;25:331-41
116. Pierce EA, Aleman TS, Jayasundera KT, Ashimatey BS, Kim K, Rashid A. et al. Gene editing for CEP290-associated retinal degeneration. N Engl J Med. 2024;390:1972-84
117. Sergouniotis PI, Davidson AE, Mackay DS, Li Z, Yang X, Plagnol V. et al. Recessive mutations in KCNJ13, encoding an inwardly rectifying potassium channel subunit, cause leber congenital amaurosis. Am J Hum Genet. 2011;89:183-90
118. Kabra M, Shahi PK, Wang Y, Sinha D, Spillane A, Newby GA. et al. Nonviral base editing of KCNJ13 mutation preserves vision in a model of inherited retinal channelopathy. J Clin Invest. 2023 133
119. Kanzaki Y, Fujita H, Sato K, Hosokawa M, Matsumae H, Shiraga F. et al. KCNJ13 gene deletion impairs cell alignment and phagocytosis in retinal pigment epithelium derived from human-induced pluripotent stem cells. Invest Ophthalmol Vis Sci. 2020;61:38
120. Chacon-Camacho OF, Zenteno JC. Review and update on the molecular basis of Leber congenital amaurosis. World J Clin Cases. 2015;3:112-24
121. Jo DH, Song DW, Cho CS, Kim UG, Lee KJ, Lee K. et al. CRISPR-Cas9-mediated therapeutic editing of Rpe65 ameliorates the disease phenotypes in a mouse model of Leber congenital amaurosis. Sci Adv. 2019;5:eaax1210
122. Acharya S, Ansari AH, Kumar Das P, Hirano S, Aich M, Rauthan R. et al. PAM-flexible Engineered FnCas9 variants for robust and ultra-precise genome editing and diagnostics. Nat Commun. 2024;15:5471
123. Suh S, Choi EH, Leinonen H, Foik AT, Newby GA, Yeh WH. et al. Restoration of visual function in adult mice with an inherited retinal disease via adenine base editing. Nat Biomed Eng. 2021;5:169-78
124. Jang H, Jo DH, Cho CS, Shin JH, Seo JH, Yu G. et al. Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nat Biomed Eng. 2022;6:181-94
125. Koenekoop RK, Wang H, Majewski J, Wang X, Lopez I, Ren H. et al. Mutations in NMNAT1 cause leber congenital amaurosis and identify a new disease pathway for retinal degeneration. Nat Genet. 2012;44:1035-9
126. Kuribayashi H, Iwagawa T, Murakami A, Kawamura T, Suzuki Y, Watanabe S. NMNAT1 is essential for human iPS cell differentiation to the retinal lineage. Invest Ophthalmol Vis Sci. 2024;65:37 -
127. Greenwald SH, Brown EE, Scandura MJ, Hennessey E, Farmer R, Pawlyk BS. et al. Gene therapy preserves retinal structure and function in a mouse model of NMNAT1-associated retinal degeneration. Mol Ther Methods Clin Dev. 2020;18:582-94
128. Brown EE, Scandura MJ, Pierce EA. Expression of NMNAT1 in the photoreceptors is sufficient to prevent NMNAT1-associated retinal degeneration. Mol Ther Methods Clin Dev. 2023;29:319-28
129. Verbakel SK, van Huet RAC, Boon CJF, den Hollander AI, Collin RWJ, Klaver CCW. et al. Non-syndromic retinitis pigmentosa. Prog Retin Eye Res. 2018;66:157-86
130. Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795-809
131. Koyanagi Y, Akiyama M, Nishiguchi KM, Momozawa Y, Kamatani Y, Takata S. et al. Genetic characteristics of retinitis pigmentosa in 1204 Japanese patients. J Med Genet. 2019;56:662-70
132. Liu X, Qiao J, Jia R, Zhang F, Meng X, Li Y. et al. Allele-specific gene-editing approach for vision loss restoration in RHO-associated retinitis pigmentosa. Elife. 2023 12
133. Shahin S, Xu H, Lu B, Mercado A, Jones MK, Bakondi B. et al. AAV-CRISPR/Cas9 gene editing preserves long-term vision in the P23H rat model of autosomal dominant retinitis pigmentosa. Pharmaceutics. 2022 14
134. Latella MC, Di Salvo MT, Cocchiarella F, Benati D, Grisendi G, Comitato A. et al. In vivo editing of the human mutant rhodopsin gene by electroporation of plasmid-based CRISPR/Cas9 in the mouse retina. Mol Ther Nucleic Acids. 2016;5:e389
135. Li P, Kleinstiver BP, Leon MY, Prew MS, Navarro-Gomez D, Greenwald SH. et al. Allele-specific CRISPR-Cas9 genome editing of the single-base P23H mutation for rhodopsin-associated dominant retinitis pigmentosa. Crispr j. 2018;1:55-64
136. Thenappan A, Nanda A, Lee CS, Lee SY. Retinitis pigmentosa masquerades: case series and review of the literature. J Clin Med. 2023 12
137. Wu WH, Tsai YT, Huang IW, Cheng CH, Hsu CW, Cui X. et al. CRISPR genome surgery in a novel humanized model for autosomal dominant retinitis pigmentosa. Mol Ther. 2022;30:1407-20
138. Du W, Li J, Tang X, Yu W, Zhao M. CRISPR/SaCas9-based gene editing rescues photoreceptor degeneration throughout a rhodopsin-associated autosomal dominant retinitis pigmentosa mouse model. Exp Biol Med (Maywood). 2023;248:1818-28
139. Burnight ER, Wiley LA, Mullin NK, Adur MK, Lang MJ, Cranston CM. et al. CRISPRi-mediated treatment of dominant rhodopsin-associated retinitis pigmentosa. Crispr j. 2023;6:502-13
140. Xi Z, Vats A, Sahel JA, Chen Y, Byrne LC. Gene augmentation prevents retinal degeneration in a CRISPR/Cas9-based mouse model of PRPF31 retinitis pigmentosa. Nat Commun. 2022;13:7695
141. Rodrigues A, Slembrouck-Brec A, Nanteau C, Terray A, Tymoshenko Y, Zagar Y. et al. Modeling PRPF31 retinitis pigmentosa using retinal pigment epithelium and organoids combined with gene augmentation rescue. NPJ Regen Med. 2022;7:39
142. Nolan ND, Cui X, Robbings BM, Demirkol A, Pandey K, Wu WH. et al. CRISPR editing of anti-anemia drug target rescues independent preclinical models of retinitis pigmentosa. Cell Rep Med. 2024;5:101459
143. Yan Z, Yao Y, Li L, Cai L, Zhang H, Zhang S. et al. Treatment of autosomal dominant retinitis pigmentosa caused by RHO-P23H mutation with high-fidelity Cas13X in mice. Mol Ther Nucleic Acids. 2023;33:750-61
144. Halford S, Liew G, Mackay DS, Sergouniotis PI, Holt R, Broadgate S. et al. Detailed phenotypic and genotypic characterization of bietti crystalline dystrophy. Ophthalmology. 2014;121:1174-84
145. Hanany M, Rivolta C, Sharon D. Worldwide carrier frequency and genetic prevalence of autosomal recessive inherited retinal diseases. Proc Natl Acad Sci U S A. 2020;117:2710-6
146. Fujinami K, Strauss RW, Chiang JP, Audo IS, Bernstein PS, Birch DG. et al. Detailed genetic characteristics of an international large cohort of patients with Stargardt disease: ProgStar study report 8. Br J Ophthalmol. 2019;103:390-7
147. Muller A, Sullivan J, Schwarzer W, Wang M, Park-Windhol C, Hasler PW. et al. High-efficiency base editing in the retina in primates and human tissues. Nat Med. 2025;31:490-501
148. Sangermano R, Khan M, Cornelis SS, Richelle V, Albert S, Garanto A. et al. ABCA4 midigenes reveal the full splice spectrum of all reported noncanonical splice site variants in stargardt disease. Genome Res. 2018;28:100-10
149. De Angeli P, Reuter P, Hauser S, Schöls L, Stingl K, Wissinger B. et al. Effective splicing restoration of a deep-intronic ABCA4 variant in cone photoreceptor precursor cells by CRISPR/SpCas9 approaches. Mol Ther Nucleic Acids. 2022;29:511-24
150. Grossniklaus HE, Geisert EE, Nickerson JM. Introduction to the retina. Prog Mol Biol Transl Sci. 2015;134:383-96
151. Grossniklaus HE, Kang SJ, Berglin L. Animal models of choroidal and retinal neovascularization. Prog Retin Eye Res. 2010;29:500-19
152. Takahashi H, Hwang Y, Won J, Jamil MU, Yaghy A, Liang MC. et al. Evaluating blood flow speed in retinal microaneurysms secondary to diabetic retinopathy using variable interscan time analysis OCTA. Transl Vis Sci Technol. 2025;14:27
153. Huang Q, Wang C, Yu H, Liu Y, Zhu H, Peng J. et al. Incomplete retinal vascularization with retinopathy of prematurity-like ridges in healthy full-term newborns. Int Ophthalmol. 2023;43:3263-8
154. Jorge R, Oliveira RS, Messias A, Almeida FP, Strambe ML, Costa RA. et al. Ranibizumab for retinal neovascularization. Ophthalmology. 2011;118:1004-e1
155. Kumar S, Hsiao YW, Wong VHY, Aubin D, Wang JH, Lisowski L. et al. Characterization of RNA editing and gene therapy with a compact CRISPR-Cas13 in the retina. Proc Natl Acad Sci U S A. 2024;121:e2408345121
156. Melincovici CS, Boşca AB, Şuşman S, Mărginean M, Mihu C, Istrate M. et al. Vascular endothelial growth factor (VEGF) - key factor in normal and pathological angiogenesis. Rom J Morphol Embryol. 2018;59:455-67
157. Wu W, Lei H. Genome editing inhibits retinal angiogenesis in a mouse model of oxygen-induced retinopathy. Methods Mol Biol. 2023;2678:207-17
158. Huang X, Wu W, Qi H, Yan X, Dong L, Yang Y. et al. Exploitation of enhanced prime editing for blocking aberrant angiogenesis. J Adv Res. 2024
159. Green WR, Wilson DJ. Choroidal neovascularization. Ophthalmology. 1986;93:1169-76
160. Cao D, Zhu J, Guo Y, Zhou Y, Zeng J, Tu Y. et al. Dynamically covalent lipid nanoparticles mediate CRISPR-Cas9 genome editing against choroidal neovascularization in mice. Sci Adv. 2025;11:eadj0006
161. Koo T, Park SW, Jo DH, Kim D, Kim JH, Cho HY. et al. CRISPR-LbCpf1 prevents choroidal neovascularization in a mouse model of age-related macular degeneration. Nat Commun. 2018;9:1855
162. Hu S, Chen Y, Xie D, Xu K, Fu Y, Chi W. et al. Nme(2) Cas9-mediated therapeutic editing in inhibiting angiogenesis after wet age-related macular degeneration onset. Clin Transl Med. 2023;13:e1383
163. Park J, Cui G, Lee H, Jeong H, Kwak JJ, Lee J. et al. CRISPR/Cas9 mediated specific ablation of vegfa in retinal pigment epithelium efficiently regresses choroidal neovascularization. Sci Rep. 2023;13:3715
164. Ling S, Yang S, Hu X, Yin D, Dai Y, Qian X. et al. Lentiviral delivery of co-packaged Cas9 mRNA and a vegfa-targeting guide RNA prevents wet age-related macular degeneration in mice. Nat Biomed Eng. 2021;5:144-56
165. Akdeniz Odemis D, Kebudi R, Bayramova J, Kilic Erciyas S, Kuru Turkcan G, Tuncer SB. et al. RB1 gene mutations and genetic spectrum in retinoblastoma cases. Medicine (Baltimore). 2023;102:e35068
166. Peeler CE, Gonzalez E. Retinoblastoma. N Engl J Med. 2022;386:2412
167. Naert T, Colpaert R, Van Nieuwenhuysen T, Dimitrakopoulou D, Leoen J, Haustraete J. et al. CRISPR/Cas9 mediated knockout of rb1 and rbl1 leads to rapid and penetrant retinoblastoma development in xenopus tropicalis. Sci Rep. 2016;6:35264
168. Dimaras H, Kimani K, Dimba EA, Gronsdahl P, White A, Chan HS. et al. Retinoblastoma. Lancet. 2012;379:1436-46
169. Avior Y, Lezmi E, Yanuka D, Benvenisty N. Modeling developmental and tumorigenic aspects of trilateral retinoblastoma via human embryonic stem cells. Stem Cell Reports. 2017;8:1354-65
170. Maeder ML, Stefanidakis M, Wilson CJ, Baral R, Barrera LA, Bounoutas GS. et al. Development of a gene-editing approach to restore vision loss in leber congenital amaurosis type 10. Nat Med. 2019;25:229-33
171. Nie W, Wu G, Zhang J, Huang LL, Ding J, Jiang A. et al. Responsive exosome nano-bioconjugates for synergistic cancer therapy. Angew Chem Int Ed Engl. 2020;59:2018-22
172. Jia Y, Wang X, Li L, Li F, Zhang J, Liang XJ. Lipid nanoparticles optimized for targeting and release of nucleic acid. Adv Mater. 2024;36:e2305300
173. Madigan V, Zhang F, Dahlman JE. Drug delivery systems for CRISPR-based genome editors. Nat Rev Drug Discov. 2023;22:875-94
174. Wu Z, Asokan A, Samulski RJ. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol Ther. 2006;14:316-27
175. Boucher P, Cui X, Curiel DT. Adenoviral vectors for in vivo delivery of CRISPR-Cas gene editors. J Control Release. 2020;327:788-800
176. Kim DY, Lee JM, Moon SB, Chin HJ, Park S, Lim Y. et al. Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat Biotechnol. 2022;40:94-102
177. Maes ME, Colombo G, Schulz R, Siegert S. Targeting microglia with lentivirus and AAV: recent advances and remaining challenges. Neurosci Lett. 2019;707:134310
178. Moazzam M, Zhang M, Hussain A, Yu X, Huang J, Huang Y. The landscape of nanoparticle-based siRNA delivery and therapeutic development. Mol Ther. 2024;32:284-312
179. Wang X, Zhang M, Zhu T, Wei Q, Liu G, Ding J. Flourishing antibacterial strategies for osteomyelitis therapy. Adv Sci (Weinh). 2023;10:e2206154
180. Rosenblum D, Gutkin A, Kedmi R, Ramishetti S, Veiga N, Jacobi AM. et al. CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci Adv. 2020 6
181. Geilenkeuser J, Armbrust N, Steinmaßl E, Du SW, Schmidt S, Binder EMH. et al. Engineered nucleocytosolic vehicles for loading of programmable editors. Cell. 2025;188:2637-55.e31
182. Ramakrishna S, Kwaku Dad AB, Beloor J, Gopalappa R, Lee SK, Kim H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014;24:1020-7
183. Huang X, El-Sayed MA. Gold nanoparticles: Optical properties and implementations in cancer diagnosis and photothermal therapy. J Adv Res. 2010;1:13-28
184. Sun W, Ji W, Hall JM, Hu Q, Wang C, Beisel CL. et al. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew Chem Int Ed Engl. 2015;54:12029-33
185. Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361:866-9
186. Apte RS. Gene therapy for retinal degeneration. Cell. 2018;173:5
187. Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids. 2015;4:e264
188. Dou S, Wang Q, Zhang B, Wei C, Wang H, Liu T. et al. Single-cell atlas of keratoconus corneas revealed aberrant transcriptional signatures and implicated mechanical stretch as a trigger for keratoconus pathogenesis. Cell Discov. 2022;8:66
189. Davidson AE, Hayes S, Hardcastle AJ, Tuft SJ. The pathogenesis of keratoconus. Eye (Lond). 2014;28:189-95
190. Lopes AG, de Almeida GC Jr, Miola MP, Teixeira RM, Pires F, Miani RA. et al. Absence of significant genetic alterations in the VSX1, SOD1, TIMP3, and LOX genes in brazilian patients with keratoconus. Ophthalmic Genet. 2022;43:73-9
191. Morgan IG, Ohno-Matsui K, Saw SM. Myopia. Lancet. 2012;379:1739-48
192. Jiang D, Li J, Xiao X, Li S, Jia X, Sun W. et al. Detection of mutations in LRPAP1, CTSH, LEPREL1, ZNF644, SLC39A5, and SCO2 in 298 families with early-onset high myopia by exome sequencing. Invest Ophthalmol Vis Sci. 2014;56:339-45
193. Pflugfelder SC, de Paiva CS. The pathophysiology of dry eye disease: what we know and future directions for research. Ophthalmology. 2017;124:S4-S13
194. Yamaguchi T. Inflammatory response in dry eye. Invest Ophthalmol Vis Sci. 2018;59:DES192-DES9
195. Periman LM, Perez VL, Saban DR, Lin MC, Neri P. The immunological basis of dry eye disease and current topical treatment options. J Ocul Pharmacol Ther. 2020;36:137-46
196. Zeitz C, Robson AG, Audo I. Congenital stationary night blindness: an analysis and update of genotype-phenotype correlations and pathogenic mechanisms. Prog Retin Eye Res. 2015;45:58-110
197. Miyadera K, Santana E, Roszak K, Iffrig S, Visel M, Iwabe S. et al. Targeting ON-bipolar cells by AAV gene therapy stably reverses LRIT3-congenital stationary night blindness. Proc Natl Acad Sci U S A. 2022;119:e2117038119
Author contact
![]() Corresponding authors: Wenlong Li (Email: wlliedu.cn) and Hua Gao (Email: hgaoedu.cn).
Corresponding authors: Wenlong Li (Email: wlliedu.cn) and Hua Gao (Email: hgaoedu.cn).
 Global reach, higher impact
Global reach, higher impact