13.3
Impact Factor
Theranostics 2019; 9(19):5444-5463. doi:10.7150/thno.29367 This issue Cite
Research Paper
Flow cytometry-based FRET identifies binding intensities in PPARγ1 protein-protein interactions in living cells
1. Institute of Biochemistry I - Pathobiochemistry, Faculty of Medicine, Goethe University Frankfurt, Theodor-Stern-Kai 7, 60590 Frankfurt/Main, Germany
2. Branch for Translational Medicine and Pharmacology, Fraunhofer Institute for Molecular Biology and Applied Ecology IME, Theodor-Stern-Kai 7, 60596 Frankfurt/Main, Germany
3. Department of Physics, Humboldt University Berlin, Newtonstraße 15, 12489 Berlin, Germany
4. Department of Biophysics and Cell Biology, Faculty of Medicine, University of Debrecen, Egyetem tér 1, 4032 Debrecen, Hungary
5. MTA-DE Cell Biology and Signaling Research Group, Faculty of Medicine, University of Debrecen, Egyetem tér 1, 4032 Debrecen, Hungary
6. Institute of Medical Virology and Epidemiology of Viral Diseases, University Hospital Tübingen, Karls University Tübingen, Elfriede-Aulhorn-Str. 6, 72076 Tübingen
* These two authors contributed equally to this work.
#Parts of the work were done in the Institute of Biochemistry I and parts in the Branch for Translational Medicine and Pharmacology TMP, Fraunhofer Institute for Molecular Biology and Applied Ecology IME
Abstract
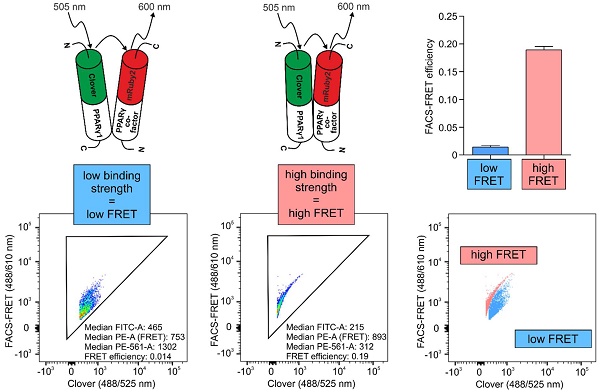
PPARγ is a pharmacological target in inflammatory and metabolic diseases. Upon agonistic treatment or following antagonism, binding of co-factors is altered, which consequently affects PPARγ-dependent transactivation as well as its DNA-independent properties. Therefore, establishing techniques to characterize these interactions is an important issue in living cells.
Methods: Using the FRET pair Clover/mRuby2, we set up a flow cytometry-based FRET assay by analyzing PPARγ1 binding to its heterodimerization partner RXRα. Analyses of PPARγ-reporter and co-localization studies by laser-scanning microscopy validated this system. Refining the system, we created a new readout to distinguish strong from weak interactions, focusing on PPARγ-binding to the co-repressor N-CoR2.
Results: We observed high FRET in cells expressing Clover-PPARγ1 and mRuby2-RXRα, but no FRET when cells express a mRuby2-RXRα deletion mutant, lacking the PPARγ interaction domain. Focusing on the co-repressor N-CoR2, we identified in HEK293T cells the new splice variant N-CoR2-ΔID1-exon. Overexpressing this isoform tagged with mRuby2, revealed no binding to Clover-PPARγ1, nor in murine J774A.1 macrophages. In HEK293T cells, binding was even lower in comparison to N-CoR2 constructs in which domains established to mediate interaction with PPARγ binding are deleted. These data suggest a possible role of N-CoR2-ΔID1-exon as a dominant negative variant. Because binding to N-CoR2-mRuby2 was not altered following activation or antagonism of Clover-PPARγ1, we determined the effect of pharmacological treatment on FRET intensity. Therefore, we calculated flow cytometry-based FRET efficiencies based on our flow cytometry data. As with PPARγ antagonism, PPARγ agonist treatment did not prevent binding of N-CoR2.
Conclusion: Our system allows the close determination of protein-protein interactions with a special focus on binding intensity, allowing this system to characterize the role of protein domains as well as the effect of pharmacological agents on protein-protein interactions.
Keywords: binding affinity and intensity, co-localization analysis, flow cytometry-based FRET assay, FRET, N-CoR2, NHR co-factors protein-protein interactions, PPARγ1, RXRα
 Global reach, higher impact
Global reach, higher impact