13.3
Impact Factor
Theranostics 2022; 12(8):3946-3962. doi:10.7150/thno.73268 This issue Cite
Research Paper
The interplay between lncRNAs, RNA-binding proteins and viral genome during SARS-CoV-2 infection reveals strong connections with regulatory events involved in RNA metabolism and immune response
1. COVID-19 International Research Team (COV-IRT).
2. Instituto de Medicina Molecular João Lobo Antunes, Faculdade de Medicina, Universidade de Lisboa, 1649-028 Lisboa, Portugal.
3. MEtRICs, Department of Sciences and Technology of Biomass, NOVA School of Science and Technology,FCT NOVA, Universidade NOVA de Lisboa, 2829-516 Caparica, Portugal.
4. Department of Radiation Medicine, Georgetown University School of Medicine, Washington, DC 20007, USA.
5. Center for Translational Data Science, Biological Sciences Division, The University of Chicago, Chicago, IL 60615, USA.
6. Clever Research Lab, IL, USA.
7. Department of Biochemistry, Atal Bihari Vajpayee Institute of Medical Sciences & Dr Ram Manohar Lohia Hospital, New Delhi-110001, India.
8. Facultad de Ingeniería, Universidad Nacional de Asunción, San Lorenzo, Central, Paraguay.
9. Department of Biomedical and Health Informatics, The Children's Hospital of Philadelphia, Philadelphia, PA 19104, USA.
10. Department of Pediatrics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
11. Bioinformatics and Computational Biology Program, Center for Metabolic Biology, Department of Genetics, Development and Cell Biology, Iowa State University, Ames, IA 50011, USA.
12. Logyx LLC, Mountain View, CA 94043, USA.
13. Space Biosciences Division, NASA Ames Research Center, Moffett Field, CA 94035, USA.
14. Johns Hopkins School of Medicine, Baltimore, MD 21287, USA.
15. Neuroscience Institute, Department of Neurobiology/ Department of Pharmacology and Toxicology, Morehouse School of Medicine, Atlanta, GA 30310, USA.
16. Department of Agricultural and Biological Engineering, Purdue University, West Lafayette, IN 47907, USA.
17. Center for Mitochondrial and Epigenomic Medicine, Children's Hospital of Philadelphia, Philadelphia, PA 19104, USA.
18. McAllister Heart Institute, Department of Pharmacology, and Department of Pathology and Lab Medicine, The University of North Carolina at Chapel Hill, NC 27599, USA.
19. Department of Physiology, Biophysics and Systems Biology, Weill Cornell Medicine, New York, NY, USA.
20. The HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine, Weill Cornell Medicine, New York, NY, USA.
21. New York Genome Center, New York, NY, USA.
22. The Feil Family Brain and Mind Research Institute, Weill Cornell Medicine, New York, NY, USA.
23. KBR, Space Biosciences Division, NASA Ames Research Center, Moffett Field, CA, 94035, USA.
24. Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, 02142, USA.
Abstract
Rationale: Viral infections are complex processes based on an intricate network of molecular interactions. The infectious agent hijacks components of the cellular machinery for its profit, circumventing the natural defense mechanisms triggered by the infected cell. The successful completion of the replicative viral cycle within a cell depends on the function of viral components versus the cellular defenses. Non-coding RNAs (ncRNAs) are important cellular modulators, either promoting or preventing the progression of viral infections. Among these ncRNAs, the long non-coding RNA (lncRNA) family is especially relevant due to their intrinsic functional properties and ubiquitous biological roles. Specific lncRNAs have been recently characterized as modulators of the cellular response during infection of human host cells by single stranded RNA viruses. However, the role of host lncRNAs in the infection by human RNA coronaviruses such as SARS-CoV-2 remains uncharacterized.
Methods: In the present work, we have performed a transcriptomic study of a cohort of patients with different SARS-CoV-2 viral load and analyzed the involvement of lncRNAs in supporting regulatory networks based on their interaction with RNA-binding proteins (RBPs).
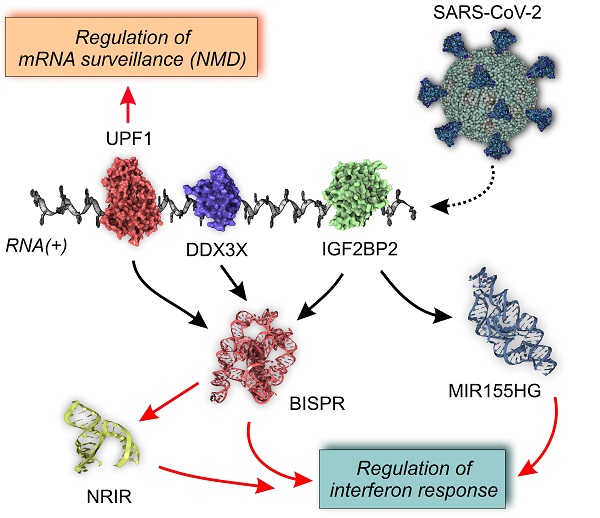
Results: Our results revealed the existence of a SARS-CoV-2 infection-dependent pattern of transcriptional up-regulation in which specific lncRNAs are an integral component. To determine the role of these lncRNAs, we performed a functional correlation analysis complemented with the study of the validated interactions between lncRNAs and RBPs. This combination of in silico functional association studies and experimental evidence allowed us to identify a lncRNA signature composed of six elements - NRIR, BISPR, MIR155HG, FMR1-IT1, USP30-AS1, and U62317.2 - associated with the regulation of SARS-CoV-2 infection.
Conclusions: We propose a competition mechanism between the viral RNA genome and the regulatory lncRNAs in the sequestering of specific RBPs that modulates the interferon response and the regulation of RNA surveillance by nonsense-mediated decay (NMD).
Keywords: SARS-CoV-2, long non-coding RNA, RNA-binding protein, regulatory network
 Global reach, higher impact
Global reach, higher impact