13.3
Impact Factor
Theranostics 2025; 15(16):8031-8048. doi:10.7150/thno.115131 This issue Cite
Review
On the dilemma of using single EV analysis for liquid biopsy: the challenge of low abundance of tumor EVs in blood
Department of Bioengineering, McGill University, Montreal, QC, H3A 0E9, Canada.
Received 2025-4-5; Accepted 2025-6-24; Published 2025-7-24
Abstract
Single extracellular vesicle (EV) analysis holds great promise for non-invasive cancer diagnostics, offering insights into tumor-specific biomarkers and enabling personalized treatment strategies. However, a significant challenge in the path towards clinical applications is the low abundance of tumor-derived EVs (tEVs) in biofluids, which reduces the sensitivity, specificity, and accuracy of detection. This review emphasizes the importance of analyzing a large number of single EVs to overcome this limitation. We estimate that less than 0.1% of total EVs could be from cancer cells in a mixed sample. Additionally, the development of more efficient tEVs isolation methods and targeted enrichment strategies, as well as high-throughput analysis techniques are crucial for improving diagnostic accuracy and advancing liquid biopsy applications in cancer care.
Keywords: extracellular vesicles, liquid biopsy, single EV analysis, low abundance of tumor EVs, non-invasive cancer diagnostics
1. Introduction
Cancer remains one of the leading causes of morbidity and mortality worldwide, underscoring the critical need for early and accurate diagnostic methods. Current technologies for detecting cancer, particularly in its early stages, have made significant progress, but there are still challenges to overcome. Early-stage cancer detection is crucial for improving survival rates, as tumors are often more treatable when detected early. Imaging techniques are well-established for tumor detection, but their ability to detect early-stage cancers is limited by tumor size, location, and metabolic characteristics. They are often more useful for staging and monitoring rather than for initial detection in asymptomatic patients. Biomarker-based immunoassays show promise for early detection, particularly in high-risk individuals, but they are not universally applicable across all cancer types. Their current role is more about monitoring and screening rather than providing definitive early detection. Traditional biopsy techniques can be invasive and may not always provide a comprehensive view of tumor heterogeneity [1].
Liquid biopsy, on the other hand, encompasses a range of minimally invasive techniques for analyzing tumor-derived materials in body fluids, including circulating tumor DNA (ctDNA), circulating tumor cells (CTCs), and cell-free RNA (cfRNA) (Figure 1) [2, 3].
Each of these biomarkers offers distinct advantages and limitations: ctDNA enables mutation profiling but is often present in low concentrations, especially in early-stage disease [5]; CTCs provide rich phenotypic and genomic information, but are rare and difficult to isolate [7]. In contrast, EVs are abundantly released by most cell types, are stable in circulation, and carry a diverse cargo of proteins, lipids, and nucleic acids that reflect their cell of origin [8]. These properties make EVs an attractive and versatile source of biomarkers for disease diagnosis, prognosis, and monitoring, warranting further investigation in the context of liquid biopsy [9].
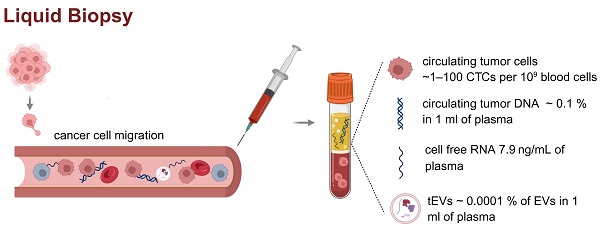
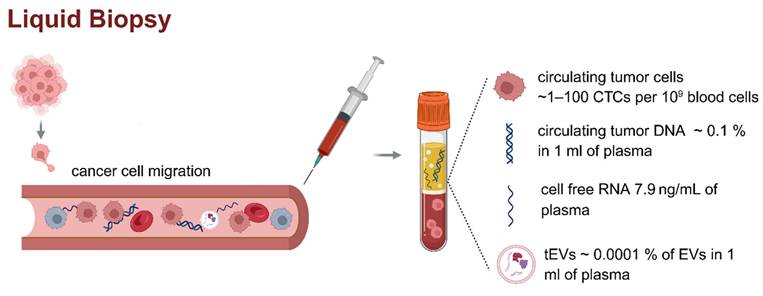
Liquid biopsy and circulating biomarkers in plasma. Liquid biopsy is a minimally invasive method for analyzing tumor-derived components such as ctDNA, cfDNA, cfRNA, and tEVs in peripheral blood. These biomarkers are typically present at very low concentrations (CTCs found in plasma ~ 1-100 CTCs per 109 blood cells, ctDNA ~ 0.1 % in 1 ml of plasma, cfRNA concentration 7.9 ng/ml of plasma, concentration of tEVs ~ 0.0001 % of EVs in 1 ml of plasma), posing significant analytical challenges [4-6]. The figure presents the literature values of the concentrations of individual circulating biomarkers in patient plasma samples. The low abundance of these analytes highlights the need for highly sensitive and specific technologies for clinical application in cancer diagnostics and monitoring. Created in https://BioRender.com.
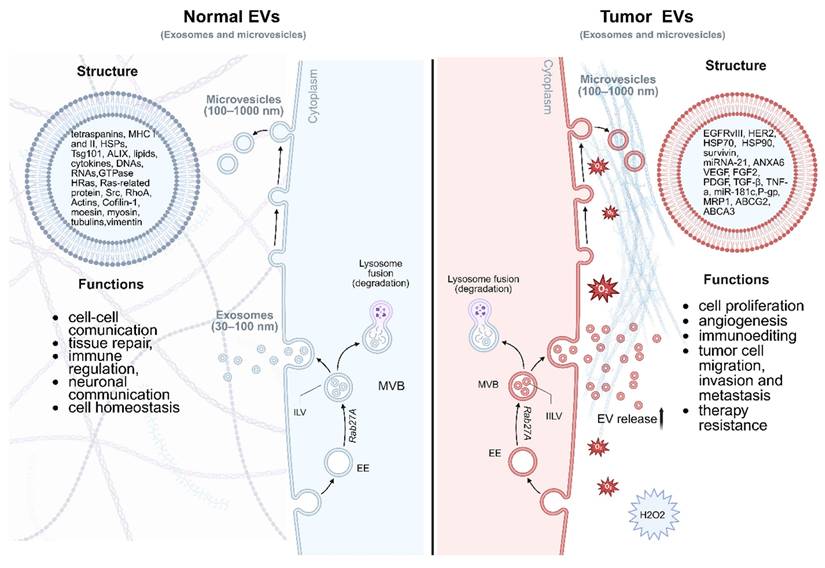
Nearly all cell types release these nanoscale lipid bilayer-enclosed particles that carry a wealth of information, including proteins and lipids. EVs are categorized mainly into three types: exosomes, microvesicles, and apoptotic bodies [8]. They vary in size and origin and serve as crucial mediators of cellular and tissue functions [10]. EVs are involved in numerous biological processes, including immune response [11], cellular signaling [12], and tissue repair [13]. Importantly, the cargo within EVs reflects the physiological state of their parent cells, making them valuable biomarkers for disease diagnosis. tEVs exhibit distinct biological behaviors compared to EVs released by normal cells (nEVs), reflecting the altered molecular and functional state of malignant cells (Figure 2) [14]. One of the most prominent distinctions lies in the upregulated release of tEVs, which is frequently driven by oncogenic signaling pathways such as RAS, EGFR, and MYC, as well as environmental stressors including hypoxia, low pH, and nutrient deprivation within the tumor microenvironment [15, 16]. These factors collectively enhance EV biogenesis through modulation of the endosomal sorting complex required for transport (ESCRT) machinery and other vesicle formation pathways, resulting in increased vesicle production and altered cargo composition. In terms of uptake, tEVs demonstrate preferential interactions with specific recipient cells, including various immune cell subsets. This selective uptake is mediated by surface molecules such as integrins, tetraspanins, and phosphatidylserine, which facilitate vesicle internalization and determine cellular tropism [17].
Notably, tEVs have been shown to exert significant immunomodulatory effects. They are capable of delivering immunosuppressive cargo, including programmed death-ligand 1 (PD-L1), transforming growth factor-β (TGF-β), interleukin-10 (IL-10), and tumor-associated microRNAs such as miR-21, miR-23a, to immune cells [18]. These factors collectively contribute to T cell exhaustion, inhibition of antigen presentation, expansion of regulatory T cells, and polarization of macrophages toward an M2-like immunosuppressive phenotype. Through these mechanisms, tEVs effectively facilitate immune evasion and promote tumor progression [19]. Furthermore, the capacity of tEVs to modulate immune responses may also influence their pharmacokinetics. While nEVs are generally subject to rapid clearance by the mononuclear phagocyte system, particularly in the liver and spleen, tEVs may exhibit prolonged circulation times due to their ability to inhibit immune recognition and phagocytic uptake [20, 21]. This extended half-life enhances their potential utility as circulating biomarkers in liquid biopsy applications.
The molecular and functional divergence between tEVs and nEVs also provides opportunities for the development of selective enrichment and targeting strategies. Surface proteins uniquely or preferentially expressed on tEVs, such as epithelial cell adhesion molecule (EpCAM), human epidermal growth factor receptor 2 (HER2), or mutant forms of epidermal growth factor receptor (EGFR), may serve as effective markers for immunoaffinity-based isolation. In addition, differential glycosylation patterns, lipid compositions, and nucleic acid profiles may offer alternative approaches for tEV-specific separation, including lectin affinity chromatography, aptamer-based capture, or size- and density-based fractionation. Exploiting these differences could significantly improve the sensitivity and specificity of EV-based diagnostics and enable the design of novel therapeutic strategies, including the targeted delivery of drugs or immune modulators via engineered vesicles.
Currently, research is focused on characterization of the size, shape, relative number, and cargo of EVs and detection of specific tumor-associated markers [22-24]. While EVs from cancer and normal cell lines have been thoroughly evaluated, translating cell culture measurements into liquid biopsies has been challenging. Approaches such as EV isolation and enrichment (separation based on size, density, or surface markers) are commonly performed to separate EVs of a certain provenance, with label-free characterization being now explored in many reports, as detailed in the review article by Imanbekova et al [25]. More recently, single EV analysis has gained interest due to the possibility of revealing the heterogeneity of EVs, thus providing deeper insights into disease mechanisms and potential therapeutic targets [26]. In one of the first such demonstrations, Z. Smith et al demonstrated that Raman spectroscopy of optically-trapped single exosomes can reveal heterogeneity within EV populations that is often masked in bulk measurements [27]. Single EV analysis can also detect low-abundance biomarkers that may be overlooked in traditional assays.
Comparison of EVs derived from tumor cells and normal cells. Both types of EVs share a common bilayer membrane structure and carry proteins, lipids, and nucleic acids; however, their content and functions differ significantly. Tumor EVs are typically enriched in oncogenic proteins such as EGFR and HER2, tumor-promoting miRNAs including miR-21, immunosuppressive molecules (PD-L1), supporting cancer progression, angiogenesis, immune evasion, and metastasis. In contrast, EVs from healthy cells contain homeostatic markers (CD9, CD63, CD81, TSG101), functional mRNAs, and regulatory miRNAs, and they play roles in maintaining tissue equilibrium, intercellular communication, and immune surveillance. Tumor cells exhibit an increased rate of EV compared to normal cells, thereby amplifying their paracrine and systemic effects within the tumor microenvironment (TME). The TME is frequently characterized by hypoxic conditions, elevated levels of reactive oxygen species such as hydrogen peroxide, and metabolic stress, all of which influence the biogenesis, cargo composition, and functional impact of tumor-derived EVs. Importantly, tumor-derived EVs also interact dynamically with the extracellular matrix (ECM), contributing to its biochemical and biomechanical remodeling. The ECM often undergoes pathological stiffening due to aberrant deposition and cross-linking of matrix components such as collagen and fibronectin. Tumor EVs can exacerbate this process by delivering matrix-modifying enzymes and fibrogenic mediators, thereby facilitating ECM remodeling that supports tumor invasion, angiogenesis, and metastasis. Early endosome - EE, multivesicular body - MVB, intraluminal vesicles - ILV, major histocompatibility complex class I - MHCI, HSP - heat shock protein, Annexin A6+ - ANXA6, vascular endothelial growth factor - VEGF, platelet-derived growth factor - PDGF, fibroblast growth factor 2 - FGF2, tumor necrosis factor alpha - TNF-α, P-glycoprotein - P-gp, multidrug resistance protein 1 - MRP1, Tumor Susceptibility Gene 101 - TSG10, ALG-2-interacting protein X - ALIX. Created in https://BioRender.com.
Cancer is one of the most promising areas for applications of single EV analysis. By focusing on single EVs, researchers can capture detailed information about the molecular characteristics of tumors, providing insights into the cancer's presence, type, and potential treatment pathways. For instance, in cancer diagnostics, single EVs can carry tumor-specific proteins or genetic material that signify the presence of malignancy [28]. EVs from cancer cells often carry tumor-specific mutations, oncogenic proteins, and microRNAs, which can be used for early detection and monitoring of disease progression. For example, studies have shown that single EV analysis can detect KRAS mutations in pancreatic cancer and HER2 protein levels in breast cancer with high sensitivity [29, 30]. This capability allows for not only early diagnosis but also the potential for personalized treatment strategies based on the molecular profile of the EVs. Single EV analysis can uncover this heterogeneity by allowing characterization of the molecular cargo of individual EVs. This can reveal different tumor subpopulations and their specific markers, which is crucial for tailoring personalized treatment strategies. For example, analyzing EVs from a heterogeneous tumor may identify distinct subclones that respond differently to therapies. The dynamics of EV release can also provide insights into tumor progression and treatment efficacy. As cancer evolves, the cargo of EVs may change, reflecting alterations in the tumor's biological behavior [28]. By performing serial analysis of single EVs over time, clinicians could monitor how the tumor responds to treatment or whether it is developing resistance, potentially allowing for timely adjustments to therapy. One of the most compelling advantages of using single EV analysis for cancer diagnostics is the ability to obtain samples non-invasively. EVs can be isolated from readily accessible body fluids such as blood, urine, or saliva [31]. This not only reduces patient discomfort but also allows repeated over-time sampling, which is essential for effective disease monitoring.
Advancements in technologies such as nanopore sensing, microfluidics, and high-resolution imaging techniques have significantly improved the ability to capture and analyze individual EVs [32]. These innovations increase the likelihood of detecting rare and disease-specific biomarkers that could play a pivotal role in early diagnosis and personalized treatment. Despite the promise of these technologies, the number of EVs available for analysis remains a critical factor in the validity and reproducibility of results. For instance, the success of a single EV analysis depends not only on the ability to isolate and analyze EVs but also on the availability of an adequate number of tEVs in the sample to ensure statistical significance and robust findings.
Although numerous studies have reported promising results using single EV analysis, a consistent challenge is the limited quantity of tEVs obtained from biofluids, which can hinder the accuracy of biomarker detection, especially when trying to detect low-abundance markers [26]. This issue is compounded by the inherent heterogeneity of EVs, where variations in size, content, and origin can make it difficult to draw definitive conclusions without large, well-characterized sample sizes. Furthermore, data interpretation remains complex, with the need to differentiate between EVs originating from the tumor versus those derived from other sources, such as immune cells or healthy tissues. Another limiting factor is the limited understanding of tumor-specific markers. While some tumor-specific proteins, RNAs, or other markers have been identified, there are still many unknowns regarding which markers on EVs are specific enough to reliably indicate the presence or progression of a particular type of cancer. Some markers may be expressed in multiple cancer types or in normal cells under certain conditions, complicating their use as definitive indicators.
This review aims to highlight the importance of the number of tEVs in liquid biopsy samples when single EV tools are employed. We use "single EV" to refer to the methodology used for the analysis of individual EVs, while "single EVs" refers to multiple vesicles analyzed individually, as opposed to EVs analysed in bulk. In the beginning of the article, we estimate the number of total EVs in liquid biopsy samples and perform simulations on the number of tEVs vs total EVs found in blood based on the number of cells, secretion rates, elimination rates, and uptake rates. We then discuss the use of different EV-containing biofluids. Next, we address EV isolation challenges, with a focus on the limited availability of tEVs and the need for enrichment methodologies specific for tEVs. In addition, we assess EV characterization methodologies for improved sensitivity and robustness. The discussion will then focus on the number of EVs required for accurate cancer detection, the limitations of single EV analysis techniques. Finally, we address the role of artificial intelligence (AI)-assisted data analysis and classification models in advancing the analysis of single EV analysis for cancer diagnosis by enabling automated and precise categorization of EVs based on their characteristics.
2. How Many EVs are Enough?
The total number of tEVs in analyzed samples is rarely evaluated and considered in single EV research. In a solid tumor, there may be hundreds of millions to billions of cancer cells [33]. Cancer cells tend to release EVs at significantly higher rates than normal cells [34, 35]. By using reported data of the number of EVs in 1 ml of blood of human prostate cancer patients, 104 are tumor-specific [36], and the average total number of EVs in ml of human blood is 1010 [33], we calculated that only 0.0001% of total EVs could be from cancer cells in a mixed sample (Figure 1). These estimations are supported by the findings of Auber et.al., indicating that only <1% of EVs are derived from solid organs, and the other 99% of EVs from plasma are derived from immune cells and non-immune cells, including platelets and erythrocytes [37].
To make the matter more complicated, 1 ml of blood contains up to 106 lipoproteins that may be misidentified as EVs, and up to 109 platelets that continue secreting EVs [37-39]. This makes detection of single tEVs in the blood even more challenging. In addition, a low abundance of tumor-specific biomarkers in tEVs can further hinder results interpretation. For example, a study of glioblastoma shows that less than one copy of most RNA species is present in tEVs. The levels of mRNAs that are most commonly mutated in glioblastoma were not higher than one copy per 100,000 EVs approximately, which indicates the necessity to appropriately interpret the data of EV RNA cancer biomarker research that is based on a single EV analysis [40].
In addition, estimating the rate of EV production is challenging because of the dynamic process associated with the de novo production and uptake of external EVs by any given cell type [41]. The estimation of tEV secretion rates is complicated by the heterogeneity of tumors that have been shown to vary in cellular composition, genetic mutations, and phenotypic characteristics. The number of cancer cells can vary widely depending on the type and stage of cancer, which is shown to be reflected in EV secretion rates [42, 43]. For example, the level of EVs in colorectal cancer and prostate cancer patients was statistically higher than that in healthy controls or benign tumor groups, and the numbers of EVs in plasma were associated with the degree of tumor differentiation and overall survival [44]. Breast cancer cells shed lower numbers of EVs (~60 to 65 per cell per hour) compared with tissue-matched, nontumorigenic cell line-derived EVs [45]. Several strategies and labeling methods are used to study EV secretion rates and their uptake, including fluorescent labeling, reporter gene systems, biotinylation, and stable isotope labeling. [41, 46-49]. While mechanisms of EV uptake and cargo delivery are incompletely characterized, these studies show that EV uptake is highly regulated by the surface composition of EVs, may occur at different rates, and depends on the type of recipient cells.
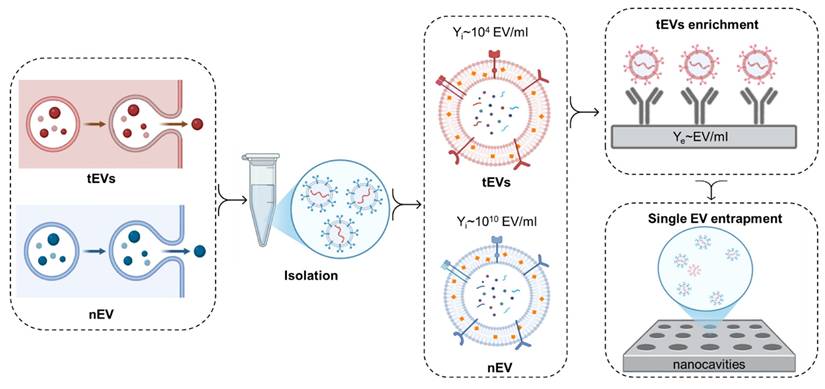
An overview of the critical factors affecting the efficacy of single EV analysis, such as EV release rates, isolation yields (Yi), and the importance of prior tEV enrichment. Cancer cell-derived EVs typically exhibit higher release rates, which can vary according to the tumor's type, grade, and microenvironmental factors. The release rates are quantified in terms of the number of EVs secreted per cell per day and are shown to correlate with tumor size and metabolic activity. Although total EV concentrations in 1 ml of body fluid can be in the range of 109 to 1010 particles per milliliter, the proportion of tEVs is often low (<0.1 %). Therefore, it is crucial to implement enrichment strategies and single EV entrapment methodologies for increasing the proportion of tEVs, which, in turn, improves the detection of cancer-specific biomarkers.
To compare the number of tEV with that of nEV we can use the following formula:
(Eq 1)
where Ntc and Nnc are number of tumor cells and normal cells, respectively, Srtc and Srnc are the EV secretion rates from tumor and normal cells, respectively, Urtc and Untc are the uptake rates for tumor and normal cells, respectively, Ertc and Entc are elimination rates for tumor and normal cells, respectively, and Yit and Yin are the isolation yields of tEVs and nEVs, respectively.
This formula gives a more nuanced understanding of the abundance of tEVs versus nEVs by considering both the biological factors, including the number of tumor and normal cells, their secretion rates, and technical aspects such as the isolation efficiency. Notably, the secretion rates and isolation yields may significantly vary between tumor and normal cells. The number of tEVs for single EV analysis can be potentially enhanced by over 90% by selectively enriching tEVs [29, 50, 51]. This has been demonstrated by S. Stott and collaborators, who achieved 94% tEV specificity in the detection of glioblastoma multiforme patients via immunocapture of EGFRvIII in plasma spiked with tEVs [50]. Moreover, Ferguson et al. achieved 100% specificity in the detection of stage 1 pancreatic cancer, below the minimal tumor size for imaging-based detection, via single EV analysis through the detection of mutated KRAS and P53 proteins in endogenous EVs [29]. Furthermore, Min et al. achieved, via single EV analysis of endogenous EVs, 100% specificity in detecting esophageal cancer via the CD36 marker, 95% specificity for stomach cancer via the TENM2 marker, 100% specificity for colorectal cancer via the CDH13 marker, 93% specificity for liver cancer via the TIMP2 marker, and 97% specificity for detecting lung cancer via the MUC1 marker [51]. Selective enrichment can therefore lead to a more accurate reflection of the tEV population in the analysis, improving both the sensitivity and reliability of the results (Figure 3).
The rates of EV uptake and clearance are influenced by various factors, including the size, composition, and surface markers of the EVs, as well as the target cell type and the presence of specific receptors [52]. EV uptake typically follows a dose-dependent pattern, meaning that larger quantities of EVs are more likely to be internalized, but the efficiency of uptake can vary based on the recipient cell's ability to recognize and internalize the vesicles. Immune cells, such as macrophages and dendritic cells, are particularly efficient at EV uptake due to their specialized role in maintaining immune homeostasis [53]. The rate of clearance from the bloodstream, on the other hand, is largely determined by the reticuloendothelial system (RES), primarily the liver and spleen, which filter out circulating EVs. Larger or more negatively charged EVs are generally cleared faster due to greater recognition by phagocytic cells in these organs [54]. In contrast, smaller EVs or those with camouflaged surface markers, such as those expressing certain proteins or lipid modifications, may evade rapid clearance, circulating for longer periods. Additionally, the half-life of EVs in circulation can vary depending on whether they are exposed to enzymatic degradation, endocytosis, or renal filtration. A study by Matsumoto et al. reported that the half-life of plasma-derived small EVs in circulation is about 7 min and was directly affected by the concentration of macrophages [53].
Simulations of EV numbers in blood. Simulations were performed using Eq 1 to demonstrate the impact of varying different biological factors on the number and progression of blood plasma tEVs originating from breast cancer tumors. Contributions from various non-cancerous blood cell types were included in the calculations, including red blood cells, platelets, monocytes, and memory cells, for which secretion rates (in EVs/cell/min) and cellular quantities, i.e. Sntc and Nnc, respectively, were acquired from Auber & Svenningsen [37]. While organ-derived EVs were not considered during the simulations, their contribution to the total plasma EV makeup has been demonstrated to account for only ~1% of all plasma EVs. Additionally, Bonsergent et al. demonstrated the normal uptake of EVs to be a low-yield process, with a spontaneous rate of approximately 1%/hour, 0.01 hour-1, or ~0.0001667 min-1 [41]. Pharmacokinetic analyses of blood plasma sEV (small EV) concentrations by Matsumoto et al. [53] also suggested an elimination half-life of 7 minutes, which can be converted into an elimination rate through pharmacokinetic considerations:
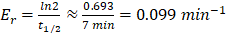 (Eq 2)
(Eq 2)
For the purposes of the simulation, identical isolation yields of 1 were assumed between cancerous and non-cancerous EVs, thereby permitting Yit and Yin to be disregarded from the analysis. Consequently, for any given cell type, with the consideration of the secretion rate in EVs/cell/min and the uptake/elimination rates in the units of EVs/min, the rate of plasma EV inflow per minute (Rinflow) can be found as follows:
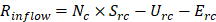 (Eq 3)
(Eq 3)
However, with the aforementioned secretion and uptake rates being found in the units of min-1, a time-dependent analysis is evidently required. Consequently, Rinflow at any given minute k can be found by computing the number of EVs secreted per minute, Nc ✕ Src, and subtracting from that the product between the total number of EVs at the previous time point and the sum between the elimination and uptake rates:
(Eq 4)
Note that the first and second terms of Eq. 4 both give values in the units of EVs/min, thereby giving Rinflow in EVs/min as well. As a result, the net number of plasma EVs at any given minute k can be found iteratively:
(Eq 5)
or
(Eq 6)
Considering the aforementioned cellular quantities and secretion rates from Auber & Svenningsen [37], as well as the normal elimination and uptake rates of ER = 0.099 min-1 and UR = 0.0001667 min-1, the progressions in the number of plasma EVs originating from non-cancerous blood cell types were simulated using Eq. 6. Furthermore, the progression in plasma EVs originating from breast cancer cells was also simulated, considering the previously mentioned secretion rate of ~65 EVs/cell/hour [45]. It is worth noting that this secretion rate approximation was acquired from breast cancer cells cultured in vitro. Consequently, this is likely an overestimation of the contribution to the total number of EVs by breast cancer cells in vivo, as in vivo secretion rates are likely to vary from those observed in vitro, and only a fraction of EVs secreted by tumors are transferred to plasma. Ultimately, our estimate constitutes an approximation for the purposes of demonstrating the effect of changing tumor size, tumor-EV uptake, and tumor-EV elimination. Notably, by estimating a tumor as a sphere with a density of 109 cells/cm3, the diameter of the breast tumor could be directly related to the number of tumor cells, Ntc. Furthermore, EV clearance from the blood has been demonstrated as being heavily dependent on phagocytic activity by immune cells [53]. Considering that tEVs have been demonstrated as being capable of evading immune cells and impairing their function, it is likely that tEVs also exhibit slower blood plasma elimination rates [55]. Consequently, elimination rates 0.1x and 0.01x the normal elimination rate were also tested to determine their effect on tEV progression. Finally, properties of the tumor microenvironment have been shown to promote tEV uptake, thereby suggesting that tEVs may exhibit higher than normal uptake rates, prompting an investigation into the effect of uptake rates 10x and 100x higher than normal on tEV progression [56].
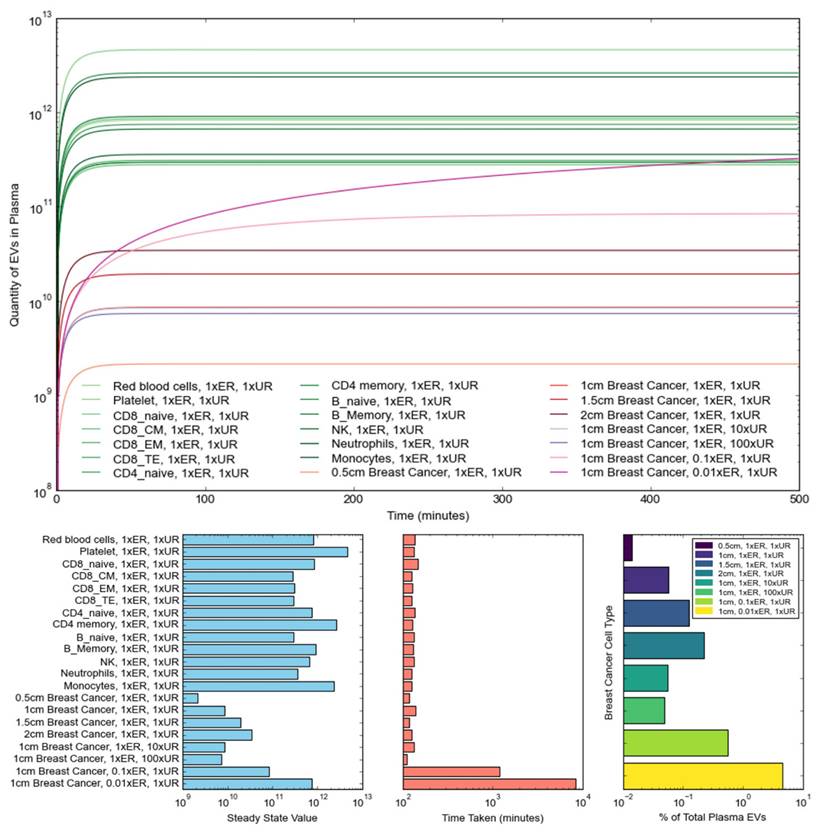
The results obtained from the simulated investigations are shown in Figure 4, where the top graph shows the progression, in the blood plasma, of EVs originating from normal cells and breast cancer cells with varying tumor sizes, uptake rates, and elimination rates. The three bar graphs respectively showcase the steady-state EV quantities reached for each simulated cell type, the number of minutes taken to reach steady states, and the steady-state percentage contributions of breast cancer tEVs to the total makeup of plasma EVs under various simulation conditions. Notably, with regards to non-tumor cells, platelets, CD4 memory cells, and monocytes make up the primary contributors of net EV quantities in the plasma, consistent with the observations made by Auber & Svenningsen [37]. Additionally, steady-state quantities were reached at relatively the same time for all normal cell types.
With regards to investigations performed in relation to breast-cancer-derived EVs, increasing tumor size had no significant impact on the speed at which steady state was reached, but instead shifted the curve upwards to higher steady state concentrations, likely as a result of the higher net secretion rates arising from the increased number of cells. Notably, increasing the tumor size from 0.5 cm in diameter to 2 cm in diameter resulted in an over ten-fold increase in the steady state number of tumor-derived EVs in the plasma. Opposingly, increasing uptake rate had no significant impact on both the steady-state number of tEVs and the speed at which the steady state was reached. Notably, there was a negligible decrease in the number of tEVs originating from a 1cm breast tumor when the uptake rate was increased from 1xUR to 100xUR, likely as a result of the basal uptake rate UR being relatively low to start with. Contrastingly, decreasing the elimination rate from 1xER to 0.01xER resulted in a significant increase in both the steady-state tEV quantity and the time taken to reach the steady state. As is showcased in Figure 4, each tenfold decrease in the elimination rate was accompanied by a tenfold increase in both the steady state tEV number and the time-to-steady-state, thereby suggesting that immune cell evasion and impairment is the primary means by which tumor EVs proliferate and promote tumor progression. Ultimately, the tumor size increase accompanied by tumor progression exacerbates tEV secretion further, with the increased tEV uptake rate having an almost strategically negligible impact on slowing tEV secretion in the blood plasma.
Simulated progression of EVs in blood plasma originating from different cell types. (Top) Variation in the quantity of EVs over 500 minutes, demonstrating the effect on EV progression of changing tumor size, uptake rate, and elimination rate for breast cancer cells; (Bottom-Left) Steady-state EV quantity reached for the different cell types; (Bottom-Middle) Time taken to reach steady-state EV quantities for different cell types; (Bottom-Right) Steady-state percentage contributions of breast cancer tEVs under different simulation conditions. Note that increasing tumor size shifts steady state concentrations of tEVs to higher values, without having discernible effects on the time taken to reach steady state. However, a ten-fold decrease of the elimination/clearance rate leads to a ten-fold increase in both the steady state concentrations of tEVs and the time taken to reach steady state. Uptake rates, on the other hand, have little effect on both the steady state tEV concentration and the time-to-steady-state.
Furthermore, to evaluate the contribution of tEVs to the total plasma EV makeup, percentage makeups of tEVs under the tested simulation conditions were calculated as follows:
(Eq 7)
In our case, NnEV, steady-state was estimated by adding up all the steady-state EV quantities of the simulated non-cancerous blood cell types. As previously mentioned, while these do not make up an exhaustive list of all sources of plasma EVs in the body, 99% of all plasma EVs are estimated to arise from these blood cells [37]. That being said, as shown in Figure 4, the percentage contribution of tEVs does not typically rise above 0.1%, usually falling around the mark of 0.06%. It is noteworthy, however, that actual percentage contributions of tEVs are likely to be much lower with the addition of organ-derived EVs and with the consideration that only a certain percentage of tEVs is likely to leave the tumor microenvironment to enter the bloodstream. Notably, the uptake of tEVs in the blood and by surrounding cells, tumor cells, and immune cells is influenced by several factors, including the enhanced permeability and retention (EPR) effect [57]. The EPR effect is a phenomenon that describes the irregular and leaky structure of tumor vasculature with large gaps between endothelial cells, leading to increased permeability [58]. It allows macromolecules and nanoparticles to accumulate more easily within tumor tissue compared to normal tissues [59]. Due to the leaky nature of tumor vasculature, EVs are more likely to accumulate in the tumor microenvironment, where they can be internalized by recipient cells through various mechanisms depending on the specific characteristics of the EVs and the type of target cell. This interaction can alter cellular behavior, promoting tumor cell proliferation, migration, and immune evasion. The EPR effect not only enhances the passive accumulation of EVs in the tumor but also improves their interaction with target cells in the tumor microenvironment, leading to a more effective cellular uptake [60].
Overall, the simulations presented in Figure 4 show that larger tumors result in a larger contribution of tEVs to the plasma, as generally expected. Additionally, it is once more notable that the strong impact of decreasing elimination rate on the proliferation of tEVs, with a 100x reduction in ER, causing the percentage contribution to rise to almost 5%. While such low tEV elimination rates are highly unlikely, the impact of immune cell evasion and impairment is once more demonstrated as being an indispensable contributor to a tumor's progression and growth.
3. Choosing the Optimal Biofluid for Single EV Analysis in Liquid Biopsy
The choice of the optimal biofluid for single EV analysis in liquid biopsy is critical to ensuring the sensitivity and accuracy of biomarker detection. Different biofluids, such as blood, urine, and saliva, contain varying concentrations and types of EVs, which can influence the detection of tEVs. The choice of biofluid must be carefully considered based on the stage of disease and the EV profile that best correlates with diagnostic and prognostic outcomes (Table 1).
While blood plasma in many cases is the biofluid of choice for tEV isolation due to its relatively higher EV yield and the ability to reflect systemic changes associated with cancer, urine and saliva are used to isolate EVs for prostate cancer detection and oral disease diagnosis, respectively. The anatomical proximity to the prostate makes urine a rational biofluid for prostate cancer diagnosis [66]. This excretory biofluid is highly dynamic in terms of composition and depends on diet and medications, and requires pre-analytical steps to ensure consistent experimental results. Microbial contamination of urine may influence EV quantitation. In addition, the reported total number of EVs isolated from urine (109 EVs/ml) is approximately the same as in blood [67].
Given its direct interaction with the oral environment, saliva is a biofluid of choice for oral cancer EV liquid biopsy. Reported concentrations of salivary EVs are slightly lower compared to blood, 108 EVs/ml [68]. As urine, salivary composition is highly dependent on medications and circadian rhythms [69].
Comparison of optimal body fluids for single EV analysis in liquid biopsy
| Biofluid | Advantages | Disadvantages | Applications | References |
|---|---|---|---|---|
| Plasma | high EV concentration rich in tEVs widely used clinically | high background of lipoproteins/proteins co-isolation of contaminants hemolysis may affect results | cancer diagnostics, biomarker discovery, monitoring treatment | [61] |
| Urine | non-invasive low protein background | low EV concentration diluted samples variable EV content depending on hydration | urological cancers | [62] |
| Saliva | non-invasive easy sampling reflects oral and systemic conditions | low EV concentration high bacterial content contamination with mucins, high viscosity challenges EV isolation | oral cancers | [63] |
| CSF | high relevance for CNS diseases | invasive collection low volume available | brain tumors | [64] |
| Pleural/Ascitic Fluids | abundance of tEVs relevant for metastatic cancers | invasive patient-specific variability | ovarian, lung, and gastrointestinal cancers | [65] |
4 Is the Enrichment of tEVs the Key to Unlocking the Potential of Single EV Analysis in Liquid Biopsy?
As our estimation shows, one significant challenge is the often-limited number of single tEVs that can be analyzed from a mixed sample of EVs in biofluids. Despite the high release rates of tEVs from cancer cells, the actual number of tEVs isolated and successfully analyzed may be low, particularly in early-stage diseases where the tumor burden is smaller. This limitation can result in insufficient data to make robust diagnostic conclusions or to capture the full spectrum of biomarkers present in EVs. Achieving high-purity tEV populations from complex biological fluids is critical for accurate analysis. Current isolation techniques, such as ultracentrifugation, polymer precipitation, and immunoaffinity capture, may lead to contamination with proteins and cellular debris, affecting the reliability of results. Inadequate purification can mask the true tEV population and hinder the detection of relevant biomarkers. Recently, new methods of isolation, including size exclusion chromatography, ultrafiltration, and immunocapture, showed higher purity in isolated EVs [70]. In the single EV analysis of biofluids, prior enrichment of tEVs by employing immunocapture techniques targeting cancer-related or tissue-specific molecules can enhance specificity. For example, Melan A and MICA for melanoma [71], PSA for prostate cancer [66], L1CAM for brain tissue [72], epithelial cell adhesion molecule EpCAM, and HER2 are enrichment markers used for breast cancer [30]. This approach of targeted enrichment of tEVs may yield over 90% tEV specificity across various cancer types, provided the correct biomarker is chosen [50, 73, 74]. These promising results highlight the potential of selective enrichment techniques in improving the accuracy and sensitivity of single EV analysis of liquid biopsy. Additionally, selective enrichment through specific tEV biomarkers can assist in prognosis, on top of diagnosis, by targeting specific surface markers during immunocapture techniques. Examples of such markers include TMPRSS2 for bladder cancer, CPNE3 for colorectal cancer, and GPC1 for pancreatic cancer [75]. However, the effectiveness of this approach can be challenged by the fact that the expression of tissue-specific and cancer-specific proteins in EVs is not fully understood. One of the major challenges in enriching tEVs for single EV analysis is the lack of a universal surface marker that can reliably distinguish these vesicles from others in the biofluid. While various surface markers mentioned above have been suggested for tEVs enrichment, none of these markers are universally expressed on all tEVs [76, 77]. The challenges of this heterogeneity on selective enrichment are further exacerbated by the presence of active proteases on the surface of some EVs. Notably, these proteases can cleave surface proteins of EVs in a sample, potentially leading to the shedding of important surface biomarkers before they can be immunocaptured [78]. Another potential type of tumor marker for tEV enrichment is neoantigens. Neoantigens are mutated proteins that arise from genetic alterations in tumor cells. These mutations, whether point mutations, insertions, deletions, or gene rearrangements, generate new peptide sequences that are absent in normal tissues. These altered peptides are displayed on major histocompatibility complex (MHC) molecules on the surface of cancer cells and can be recognized by the immune system as foreign. Neoantigens are particularly attractive for cancer immunotherapy because they are specific to the tumor and not present on normal cells. This makes them ideal targets for immune responses without causing widespread autoimmune effects. As tumors evolve, the landscape of neoantigens can change, which means that tumor-specific EVs could potentially carry different neoantigens depending on the tumor's stage, heterogeneity, and genetic evolution. Currently, EVs are mainly explored as immunotherapeutic agents for neoantigen loading and delivery [79]. Yet, neoantigens in EVs can be utilized as potential biomarkers, increasing the specificity of EV-based diagnostics. In addition to neoantigens, there are several new promising marker types that are being actively studied for applications in EV analysis, such as glycosylation patterns, membrane lipid components, and non-coding RNAs. Notably, these new marker types display significant heterogeneity from nEVs to tEVs across different cancer cell types, offering diagnostic potential that has been extensively reviewed elsewhere [80-83].
Tumors exhibit heterogeneity in their EV populations, with different cancer types and even stages within the same type showing variable marker expression [84]. Continued work on establishing universal and specific markers for enriching tEVs is essential to overcome a major hurdle that impacts the reproducibility and reliability of single EV analysis, particularly in liquid biopsy applications for early cancer detection. The lack of standardized markers for tEV enrichment is further complicated by the proteolytic cleavage of surface markers, which can lead to the loss or alteration of these markers on EVs, diminishing the effectiveness of enrichment strategies. This challenge underscores the need for more robust methods and markers that can consistently identify and isolate tumor-derived EVs, ensuring more accurate and reliable results in clinical applications [85].
Moreover, there is a pressing need for standardized protocols in EV isolation, characterization, and analysis. Variability in EV isolation methodologies can hinder reproducibility and the comparability of findings across different studies. Establishing consensus guidelines is essential to facilitate the adaptation of single EV analysis in clinical practice. The International Extracellular Vesicle Society addressed the initial lack of methodological consensus and reporting in the EV field by introducing several initiatives, such as MISEV guidelines and EV-TRACK platform.
The heterogeneity of EV populations complicates data interpretation. Single EV analysis can reveal diverse biomarker profiles, which may vary not only between different tumor types but also within the same tumor. A thorough understanding of the biological context of the analyzed EVs is essential for accurate diagnostic conclusions. Moreover, distinguishing between clinically relevant signals and background noise can be challenging, particularly with low-abundance biomarkers.
Beyond low-abundance and standardization issues, high-throughput single EV analysis faces other challenges to clinical translation. Notably, cost is variable depending on the methodology employed, where digital ELISA assays come at moderate costs compared to flow cytometry- and Raman spectroscopy-based high-throughput technologies, which incur high costs due to the need to purchase large, expensive equipment [86-88]. However, such single EV analysis technologies are nevertheless expected to incur less cost than liquid biopsies focusing on ctDNA of CTCs [89]. Liquid biopsies are typically slightly more expensive than traditional tissue biopsies, but often incur less cost compared to imaging techniques such as CT and MRI scans [90]. Additionally, with regards to time required to perform diagnostic tests, liquid biopsy-based single EV analysis techniques are typically time-consuming, particularly due to the time required for isolating EVs, on top of the time needed for measurement and data analysis/interpretation [91]. Ultimately, for single EV analysis to undergo widespread adoption as a non-invasive cancer diagnosis technique, further standardization is inevitably needed [91]. Moreover, rather than competing with alternative liquid biopsy techniques involving ctDNA and/or CTCs, combined analysis of EVs, ctDNA, and CTCs offers significant synergistic potential, but has minimal clinical evidence and standardization, as well as further challenges arising from handling of the multimodal data [92].
5. Balancing Robustness and Sensitivity in Single EV Characterization Methods
To detect one copy of certain cancer biomarkers, including miRNA, a large number of single EVs must be analyzed [26, 93]. This number varies depending on the biomarker of interest and the cancer stage. As shown previously, the levels of most commonly mutated mRNAs in glioblastoma (TP53 and PTEN) were approximately 1 molecule per 100,000 EVs, while most abundant mRNA species were present at 1 copy per 1,000 EVs [40]. This threshold helps ensure that the heterogeneity of EV populations is adequately represented and that rare biomarkers are detected with sufficient confidence. However, for certain applications, such as monitoring specific mutations or protein expressions associated with particular cancers, significantly higher numbers may be necessary to improve statistical power and diagnostic accuracy.
Existing single-particle analysis techniques, including electron microscopy, surface plasmon resonance imaging (SPRi), super-resolution microscopy, fluorescent microscopy, and label-free plasmonic sensors, while being sensitive, are low-throughput, require expensive instrumentation, and are time-consuming (Table 2) [25]. Therefore, the optimization of these methods is needed for the analysis of a large number of single EVs and to detect rare tEVs.
In addition, there are methods such as nanoparticle tracking analysis and flow cytometry that can analyze a large number of single EVs with high throughput. Flow cytometry is widely available in clinical laboratories, which makes it the most promising method for cancer EV liquid biopsy, as indicated in the recent review by Mizenko et al [111]. Yet, the sensitivity of the technology remains to be improved to eliminate swarm detection. While traditional flow cytometers lack sensitivity to small nanoparticles and EVs, recent advancements of nano flow cytometers address this shortcoming and have been applied to analyse small nanoparticles, single EVs, and viruses. Liu H. and colleagues employed this emerging technology to perform in-depth characterization of DNA associated with single EVs. The study showed that localization of the DNA depends on the size of the EVs, revealing attachment of the DNA to the surface of the small EVs (<100 nm) and luminal localization of DNA in larger EVs (80 nm - 200 nm) [96].
Single EV characterization methods
| Method | Advantages | Disadvantages | Detection Limit | References |
|---|---|---|---|---|
| flow cytometry | high-throughput multiparametric analysis surface marker-specific detection using fluorescent antibodies | limited sensitivity to small EVs (<100 nm), specificity depends on antibody quality and labeling signal overlap and background can affect accuracy | ~100-150 nm | [94-96] |
| NTA | size distribution of individual EVs high-throughput fluorescent mode allows limited marker-specific detection | low resolution for heterogeneous or small EVs requires large sample volumes | ~30-100 nm | [97, 98] |
| AFM | high spatial resolution label-free morphological analysis high specificity for physical properties (stiffness, size, surface structure) | low throughput technical expertise required | <10 nm | [99] |
| Raman spectroscopy/SERS | label-free chemical composition high biochemical specificity | low throughput sensitive to noise and sample preparation artifacts | [27, 100, 101] | |
| TEM | high morphological resolution high specificity for EV structure and morphology selective molecular identification via immunogold labeling enables | labor-intensive low throughput | <10 nm | [102] |
| Super-Resolution Microscopy | allows investigation of EVs functions in vivo/in situ with molecular resolution | requires labels | [103, 104] | |
| SPRi | real-time monitoring of EVs-ligand binding kinetics, small sample size, label free | labor intensive, limited by use of capturing molecules | [105, 106] | |
| Digital ELISA | detection of EV surface or cargo proteins | requires optimized antibody pairs | <100 EVs/mL | [95, 107] |
| Nano-FTIR | label-free quantitative and qualitative characterization of EV biochemical content, minimal preprocessing, small sample size | technically demanding low throughput | ~50-100 nm | [108] |
| qSMLM | Quantitative characterization of the size, shape and protein content | requires optimized antibody pairs, low throughput, complex sample preparation, and limited multiplexing capabilities | ~10 nm | [109, 110] |
In recent years, notable advancements were made in the efforts to tackle this fundamental challenge by developing innovative high-throughput single EV analysis platforms with the ability to analyze 20 million single vesicles per minute and limit of detection of 11 EVs/μl in blood [95]. This technology is based on digital ELISA that utilizes paramagnetic, fluorescent beads coated with CD81 antibodies to capture single EVs. Another interesting approach for analysing a single EV is quantitative single-molecule localization spectroscopy (qSMLM) [112]. This method had been applied to asses shape, size, tetraspanins content of single EV and explore their heterogeneity [109, 110]. C. Han and colleagues developed single EV imaging methods based on total internal reflection microscopy to explore EV subpopulations [113].
Emerging high-throughput technologies for single EV analysis require robust data analysis capabilities to fully unlock their potential. The sheer volume of data generated by these platforms, combined with the inherent complexity of extracellular vesicle populations, necessitates the application of advanced computational tools to ensure accurate and meaningful interpretation. By implementing advanced machine learning algorithms, AI can analyze complex datasets generated from single EV characterization, such as size, shape, and molecular composition. This capability enables the identification of specific biomarkers associated with cancer, facilitating early detection and more accurate diagnoses. Additionally, AI-driven image analysis and pattern recognition techniques can significantly improve the sensitivity of detecting rare cancer-related EVs in biological samples, thus enabling personalized treatment strategies. AI classification models are pivotal in advancing the analysis of single EV analysis for cancer diagnosis by enabling automated and precise categorization of EVs based on their characteristics. These models often developed using machine learning techniques such as support vector machines (SVM), random forests, convolutional neural networks (CNN), transfer learning, and deep learning algorithms, can learn from large datasets to identify patterns that distinguish cancer-related EVs from normal ones. For example, SVM has been used to classify EVs based on their protein profiles obtained through mass spectrometry. By training on labeled datasets, SVM can effectively differentiate between EVs from cancer patients and healthy controls, highlighting specific biomarkers associated with tumor presence [114]. CNNs have been applied to analyze imaging data from flow cytometry or nanoparticle tracking analysis. These models can automatically identify and classify EVs based on morphological features, enabling the detection of subtle differences that may indicate malignancy. CNNs have been used to classify EVs isolated from the 6 types of early-stage cancers based on Raman spectroscopy profile [115]. Random forests have been utilized to analyze multi-omics data from EVs isolated serum of colon cancer, integrating information from RNA sequencing and proteomics [116, 117]. Random Forests can classify EVs based on a combination of molecular signatures, providing a robust approach to identifying cancer-associated EVs. Next, advanced deep learning architectures, such as recurrent neural networks (RNNs) and autoencoders, have been explored for analyzing time-series data and high-dimensional feature sets from EVs [118]. These models can detect dynamic changes in EV composition that correlate with tumor evolution, aiding in real-time monitoring of cancer.
As single EV data becomes more complex with the advancement of high-throughput technologies and the integration of multi-omics approaches, incorporating unsupervised clustering and dimensionality reduction pre-processing steps in model pipelines has become essential. It is widely accepted that biological data often suffers from the “curse of dimensionality”, wherein the number of data points required for a model to effectively form generalizations increases exponentially with the number of features [119]. For biological datasets, such as transcriptomics and proteomics, the high dimensionality can significantly degrade model performance if proper pre-processing techniques like clustering and dimensionality reduction are not applied [119]. This is also relevant to single EV analysis data, which often demonstrates high dimensionality that benefits from treatment with unsupervised learning algorithms [120-122]. In essence, clustering methodologies identify instances with inherent similarities within the feature space, thereby partitioning the data to capture inherent subpopulations. Common clustering methodologies include K-Means Clustering, Hierarchical Clustering, and Expectation-Maximization Clustering. Notably, Yin et al. utilized K-Means clustering for their AI-assisted analysis of high-throughput SERS-based digital counting of single EVs for cancer diagnosis [120]. Moreover, Wen et al. applied hierarchical clustering to their algorithm for the detection of plasma EV protein biomarkers for Ewing Sarcoma diagnosis via microfluidic Topographically-Intensified Partition-less dELISA [121]. On the other hand, dimensionality reduction serves the purpose of transforming high-dimensional data into a low-dimensional representation that retains the most informative features, while discarding noise and redundancy. Some examples of dimensionality reduction techniques include linear methods such as Principal Component Analysis (PCA) and Independent Component Analysis (ICA), as well as non-linear techniques such as t-distributed Stochastic Neighbour Embedding (t-SNE) and Uniform Manifold Approximation and Projection (UMAP) [123]. In addition to K-Means clustering, Yin et al. also utilized PCA for distinguishing between patients with different cancer types via SERS-based digital counting of single EVs [120]. Moreover, Von Lersner et al. have applied UMAP in their machine-learning pipeline for analyzing and converting data from multiparametric single-vesicle flow cytometry into distinguishable EV fingerprints [122].
AI-assisted analysis techniques, when exploited in conjunction with high-throughput single EV detection methodologies, have also demonstrated significant potential in resolving heterogeneities within EVs. Min et al. trained a logistic regression model on single EV proteomic data of surface biomarkers from 100 cancer patients and 100 healthy patients, demonstrating the ability to distinguish between (i) healthy and cancerous patients, (ii) cancer type in cancerous patients (esophageal, stomach, colorectal, liver, or lung cancer), and (iii) intra-tumoral heterogeneity of EVs in colorectal cancer patients [51]. Moreover, von Lersner et al. combined high-throughput multiparametric single vesicle flow cytometry with AI-assisted dimensionality reduction and clustering techniques to form EV fingerprints that help discern the partitioning of molecular cargo across different EV subpopulations [122]. Furthermore, Yin et al. combined clustering and dimensionality reduction to create an algorithm that can potentially be used for distinguishing between patients with different cancer types via SERS-based digital counting of single EVs [120]. However, the main obstacle in developing high-performing and versatile algorithms for resolving heterogeneity in single EVs remains a lack of annotated data, especially when considering the high-dimensionality of newly developed high-throughput techniques.
In addition, single EVs contain a wealth of information, including proteins, lipids, mRNA, and DNA. AI models can integrate these multiple types of data, generating more comprehensive insights. Multi-omics approaches such as proteomics, transcriptomics, and genomics allow for deeper understanding and cross-validation of cancer markers at different molecular levels, helping to pinpoint the most informative biomarkers for early detection. AI can also be used to merge data from different experimental platforms (flow cytometry, mass spectrometry, RNA sequencing). This data fusion enables researchers to better understand the complexity of cancer-derived EVs and identify comprehensive molecular signatures that may indicate the presence of early-stage cancer. Moreover, AI can be used to enhance the real-time detection of tEVs using microfluidic platforms and biosensors. Such technologies could potentially isolate individual EVs and analyze them in real time, while AI models process the data to classify EVs as cancerous or normal immediately. This real-time analysis is especially valuable for monitoring treatment responses or detecting recurrences.
While there are exciting avenues for AI application in single EV research, a lack of large, annotated datasets for training machine learning models can lead to data imbalance and reduce model performance, especially for rare cancers or early-stage diseases. Also, it is essential to develop ML models with deeper integration with domain-specific biological knowledge.
Overall, the integration of AI in single EV analysis not only streamlines diagnostic workflows but also holds the potential to revolutionize cancer management through more targeted and effective therapeutic interventions.
6. Conclusions
Single EV analysis represents a groundbreaking advancement in liquid biopsy technology, offering unprecedented opportunities for disease diagnosis. By enhancing sensitivity and specificity, it has the potential to transform the diagnostic landscape for various diseases, including cancer. This non-invasive method may offer detection of tumor-specific biomarkers with high sensitivity and specificity. The ability to analyze individual EVs provides valuable insights into the diverse characteristics of tumors, facilitating the development of personalized treatment strategies based on specific molecular profiles. However, a critical challenge in implementing single EV analysis is the availability of a sufficient number of tEVs for analysis. Despite the promise shown by EVs, our analysis indicates that current studies based on the analysis of hundreds of single EVs may lack robust statistical power and accurate representation of EV heterogeneity. For certain applications, particularly those monitoring specific mutations or protein expressions, even higher numbers of single EVs may be necessary to enhance diagnostic accuracy. Given the vast number of EVs released from a variety of cell types, including the immune cells, platelets, and erythrocytes, the concentration of cancer-specific EVs in a typical plasma sample is extremely low.
To address this challenge, in this article, we introduced a simulation model that helps provide an estimate of the abundance of tEVs among the total number of EVs. This model accounts for variabilities in the EV secretion, elimination, and uptake rates, as well as for the cell type and isolation yields. Using this simulation model, we estimate that no more than ~0.1% of the total EVs in a blood sample might originate from cancer cells, making it exceptionally difficult to capture and identify a single tumor-derived EV in a mixed sample. This low abundance is further complicated by the heterogeneity of tumors and the dynamic nature of EV production and uptake by different cell types. One important question that still needs to be addressed is whether studies analyzing hundreds or thousands of single EVs are inherently biased towards the detection of soluble tumor-associated markers within the analyte, or do they more accurately reflect alterations in the immune landscape of cancer patients rather than detecting cancer biomarkers associated with tEVs.
Considering these factors, it is critical in single EV studies to analyze a large number of single EVs in order to reliably detect tumor-specific EVs and associated cancer biomarkers. Therefore, for the identification of the extremely small fraction of cancer EVs, large-scale EV analyses are necessary, with the number of single EVs to be analyzed depending on the expected concentration of tumor-derived EVs. In practice, the number of EVs to be analyzed (with single EV techniques) should be at least 1 million per test to ensure that rare, low-abundance cancer-specific EVs are sufficiently represented. This is especially important for ensuring accurate biomarker analysis, given the low levels of tumor-specific RNA or proteins often found within these EVs. The number we provide here can be further refined by performing additional, detailed, and specific simulations in order to capture the prevalence of tEVs in different diagnostic circumstances, such as other tumor types, cancer stage, or biofluids. While improvements to the simulation model will be necessary to perform more accurate estimates, the main challenge is the availability of accurate data obtained from in vivo measurements. In this context, EV subtype-specific rates that are measured in relevant environments will be needed.
Furthermore, similar recommendations apply for other biofluids such as urine and saliva, although factors like microbial contamination and the highly variable composition of these fluids may further influence EV quantification. Despite these challenges, increasing the number of single EVs analyzed remains the most effective strategy for overcoming the low abundance and variability inherent in tumor-derived EV detection, paving the way for more accurate liquid biopsy diagnostics for cancer.
To propel the field forward, it is imperative to improve EV isolation and enrichment techniques to capture adequate amounts of tEVs from complex biological fluids, as well as develop high-throughput characterization techniques. However, post-processing of high-throughput single EV data necessitates the incorporation of machine learning pipelines for efficient and thorough consideration of inter- and intra-tumor heterogeneity within EV subpopulations. As research continues to refine these methodologies, single EV analysis could transform cancer diagnosis and monitoring, offering hope for early detection and improved patient outcomes. Efforts should also be placed to overcome accessibility barriers to clinical translation, including diagnostic time frames, cost, and standardization. Ultimately, while single EV analysis is not yet fully realized as a standalone or complementary diagnostic tool, it holds significant promise as part of a comprehensive approach to cancer care.
Abbreviations
EV: extracellular vesicle; tEVs: tumor-derived EVs; ctDNA: circulating tumor DNA; CTCs: circulating tumor cells; cfRNA: cell-free RNA; EGFR: epidermal growth factor receptor; ESCRT: endosomal sorting complex required for transport; HER2: human epidermal growth factor 2; TME: tumor microenvironment; ECM: extracellular matrix; MHCI: major histocompatibility complex class I; HSP: heat shock protein; ANXA6: annexin A6+; VEGF: vascular endothelial growth factor; PD-L: programmed death-ligand 1; TGF-β: transforming growth factor-β; IL-10: interleukin-10; EpCAM: epithelial cell adhesion molecule; RES: reticuloendothelial system; Melan A: melanoma antigen; MICA: MHC class I chain related-proteins A; PSA: prostate specific antigen; TMPRSS2: transmembrane protease serine 2; CPNE3: copine 3; GPC: glypican-1; SPRi: surface plasmon resonance imaging; qSMLM: quantitative single-molecule localization spectroscopy; SERS: surface-enhanced Raman spectroscopy; SVM: support vector machines; CNN: convolutional neural networks; RNNs: recurrent neural networks; ELISA: enzyme-linked immunosorbent assay; PCA: principal component analysis; ICA: independent component analysis; t-SNE: t-distributed stochastic neighbour embedding; UMAP: uniform manifold approximation and projection.
Acknowledgements
This work was financially supported by the Natural Sciences and Engineering Research Council of Canada (NSERC), Discovery Grant RGPIN-2018-05675 (to S.W.-H.); M.S. thanks MEUSMA scholarship from the Faculty of Engineering at McGill University. The authors would like to thank Ms. Xinyue Hu for providing some comments in the very early stages of the manuscript.
Author contributions
SWH: conceptualization; data curation; writing - review and editing; supervision; financial support. MI: writing - original draft, review, and editing; visualization; formal analysis. MS: performed the simulations, prepared Fig. 4, wrote the simulation description and discussion, and contributed significantly to AI section and revisions of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Rorke LB. Pathologic diagnosis as the gold standard. Cancer. 1997;79:665-7
2. Nikanjam M, Kato S, Kurzrock R. Liquid biopsy: current technology and clinical applications. J Hematol Oncol. 2022;15:131
3. Roskams-Hieter B, Kim HJ, Anur P, Wagner JT, Callahan R, Spiliotopoulos E. et al. Plasma cell-free RNA profiling distinguishes cancers from pre-malignant conditions in solid and hematologic malignancies. NPJ Precis Oncol. 2022;6:28
4. Rostami P, Kashaninejad N, Moshksayan K, Saidi MS, Firoozabadi B, Nguyen N-T. Novel approaches in cancer management with circulating tumor cell clusters. J Sci Adv Mat Devices. 2019;4:1-18
5. Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R. et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545:446-51
6. Larson MH, Pan W, Kim HJ, Mauntz RE, Stuart SM, Pimentel M. et al. A comprehensive characterization of the cell-free transcriptome reveals tissue- and subtype-specific biomarkers for cancer detection. Nat Commun. 2021;12:2357
7. Deng Z, Wu S, Wang Y, Shi D. Circulating tumor cell isolation for cancer diagnosis and prognosis. eBioMedicine. 2022;83:104237
8. van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213-28
9. Irmer B, Chandrabalan S, Maas L, Bleckmann A, Menck K. Extracellular vesicles in liquid biopsies as biomarkers for solid tumors. Cancers (Basel). 2023;15:1307
10. Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A. et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619-24
11. Buzas EI. The roles of extracellular vesicles in the immune system. Nat Rev Immunol. 2023;23:236-50
12. van Niel G, Carter DRF, Clayton A, Lambert DW, Raposo G, Vader P. Challenges and directions in studying cell-cell communication by extracellular vesicles. Nat Rev Mol Cell Biol. 2022;23:369-82
13. Nagelkerke A, Ojansivu M, van der Koog L, Whittaker TE, Cunnane EM, Silva AM. et al. Extracellular vesicles for tissue repair and regeneration: evidence, challenges and opportunities. Adv Drug Deliv Rev. 2021;175:113775
14. Chen J, Fei X, Wang J, Cai Z. Tumor-derived extracellular vesicles: Regulators of tumor microenvironment and the enlightenment in tumor therapy. Pharmacol Res. 2020;159:105041
15. Kilinc S, Paisner R, Camarda R, Gupta S, Momcilovic O, Kohnz RA. et al. Oncogene-regulated release of extracellular vesicles. Dev Cell. 2021;56:1989-2006.e6
16. Seres M, Spacayova K, Sulova Z, Spaldova J, Breier A, Pavlikova L. Dynamic multilevel regulation of EGFR, KRAS, and MYC Oncogenes: driving cancer cell proliferation through (epi)genetic and post-transcriptional/translational pathways. Cancers. 2025;17(2):248
17. Kumar MA, Baba SK, Sadida HQ, Marzooqi SA, Jerobin J, Altemani FH. et al. Extracellular vesicles as tools and targets in therapy for diseases. Signal Transduct Target Ther. 2024;9:27
18. Li Q, Cai S, Li M, Salma KI, Zhou X, Han F. et al. Tumor-derived extracellular vesicles: their role in immune cells and immunotherapy. Int J Nanomedicine. 2021;16:5395-409
19. Njock M-S, O'Grady T, Nivelles O, Lion M, Jacques S, Cambier M. et al. Endothelial extracellular vesicles promote tumour growth by tumour-associated macrophage reprogramming. J Extracell Vesicles. 2022;11:e12228
20. Akbar N, Digby JE, Cahill TJ, Tavare AN, Corbin AL, Saluja S. et al. Endothelium-derived extracellular vesicles promote splenic monocyte mobilization in myocardial infarction. JCI Insight. 2017;2(17):e93344
21. Driedonks T, Jiang L, Carlson B, Han Z, Liu G, Queen SE. et al. Pharmacokinetics and biodistribution of extracellular vesicles administered intravenously and intranasally to Macaca nemestrina. J Extracell Biol. 2022;1:e59
22. Imanbekova M, Saridag AM, Kahraman M, Liu J, Caglayan H, Wachsmann-Hogiu S. Complementary Metal-Oxide-Semiconductor-Based sensing platform for trapping, imaging, and chemical characterization of biological samples. ACS Appl Opt Mater. 2022;1(1):329-339
23. Suarasan S, Liu J, Imanbekova M, Rojalin T, Hilt S, Voss JC. et al. Superhydrophobic bowl-like SERS substrates patterned from CMOS sensors for extracellular vesicle characterization. J Mater Chem B. 2020;8(38):8845-8852
24. Jalali M, del Real Mata C, Montermini L, Jeanne O, I.Hosseini I, Gu Z. et al. MoS2-plasmonic nanocavities for Raman spectra of single extracellular vesicles reveal molecular progression in glioblastoma. ACS Nano. 2023;17:12052-71
25. Imanbekova M, Suarasan S, Lu Y, Jurchuk S, Wachsmann-Hogiu S. Recent advances in optical label-free characterization of extracellular vesicles. Nanophotonics. 2022;11(12):2827-2863
26. Ferguson S, Yang KS, Weissleder R. Single extracellular vesicle analysis for early cancer detection. Trends Mol Med. 2022;28:681-92
27. Smith ZJ, Lee C, Rojalin T, Carney RP, Hazari S, Knudson A. et al. Single exosome study reveals subpopulations distributed among cell lines with variability related to membrane content. J Extracell Vesicles. 2015;4:28533
28. Kalluri R, McAndrews KM. The role of extracellular vesicles in cancer. Cell. 2023;186:1610-26
29. Ferguson S, Yang KS, Zelga P, Liss AS, Carlson JCT, Del Castillo CF. et al. Single-EV analysis (sEVA) of mutated proteins allows detection of stage 1 pancreatic cancer. Sci Adv. 2022;8:eabm3453
30. Tian F, Zhang S, Liu C, Han Z, Liu Y, Deng J. et al. Protein analysis of extracellular vesicles to monitor and predict therapeutic response in metastatic breast cancer. Nat Commun. 2021;12:2536
31. Witwer KW, Buzás EI, Bemis LT, Bora A, Lässer C, Lötvall J. et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013 2
32. De Sousa KP, Rossi I, Abdullahi M, Ramirez MI, Stratton D, Inal JM. Isolation and characterization of extracellular vesicles and future directions in diagnosis and therapy. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2023;15:e1835
33. Johnsen KB, Gudbergsson JM, Andresen TL, Simonsen JB. What is the blood concentration of extracellular vesicles? Implications for the use of extracellular vesicles as blood-borne biomarkers of cancer. Biochim Biophys Acta Rev Cancer. 2019;1871:109-16
34. Logozzi M, De Milito A, Lugini L, Borghi M, Calabrò L, Spada M. et al. High levels of exosomes expressing CD63 and Caveolin-1 in plasma of melanoma patients. PLOS ONE. 2009;4:e5219
35. Tavoosidana G, Ronquist G, Darmanis S, Yan J, Carlsson L, Wu D. et al. Multiple recognition assay reveals prostasomes as promising plasma biomarkers for prostate cancer. Proc Natl Acad Sci U S A. 2011;108:8809-14
36. Rikkert LG, Beekman P, Caro J, Coumans FAW, Enciso-Martinez A, Jenster G. et al. Cancer-ID: Toward identification of cancer by tumor-derived extracellular vesicles in blood. Front Oncol. 2020;10:608
37. Auber M, Svenningsen P. An estimate of extracellular vesicle secretion rates of human blood cells. J Extracell Biol. 2022;1:e46
38. Kuchinskiene Z, Carlson L. Composition, concentration, and size of low density lipoproteins and of subfractions of very low density lipoproteins from serum of normal men and women. J Lipid Res. 1982;23:762-9
39. Simonsen JB. What are we looking at? Extracellular vesicles, lipoproteins, or both? Circ Res. 2017;121:920-2
40. Wei Z, Batagov AO, Schinelli S, Wang J, Wang Y, El Fatimy R. et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat Commun. 2017;8:1145
41. Bonsergent E, Grisard E, Buchrieser J, Schwartz O, Théry C, Lavieu G. Quantitative characterization of extracellular vesicle uptake and content delivery within mammalian cells. Nat Commun. 2021;12:1864
42. Bebelman MP, Smit MJ, Pegtel DM, Baglio SR. Biogenesis and function of extracellular vesicles in cancer. Pharmacol Ther. 2018;188:1-11
43. Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208-15
44. Silva J, Garcia V, Rodriguez M, Compte M, Cisneros E, Veguillas P. et al. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer. 2012;51:409-18
45. Chiu YJ, Cai W, Shih YR, Lian I, Lo YH. A Single-Cell assay for time lapse studies of exosome secretion and cell behaviors. Small. 2016;12:3658-66
46. Koo D, Cheng X, Udani S, Baghdasarian S, Zhu D, Li J. et al. Optimizing cell therapy by sorting cells with high extracellular vesicle secretion. Nat Commun. 2024;15:4870
47. Mulcahy LA, Pink RC, Carter DRF. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3:24641
48. Li Y, Kanao E, Yamano T, Ishihama Y, Imami K. TurboID-EV: Proteomic mapping of recipient cellular proteins proximal to small extracellular vesicles. Anal Chem. 2023;95:14159-64
49. Royo F, Cossío U, de Angulo AR, Llop J, Falcon-Perez JM. Modification of the glycosylation of extracellular vesicles alters their biodistribution in mice. Nanoscale. 2019;11:1531-7
50. Reátegui E, van der Vos KE, Lai CP, Zeinali M, Atai NA, Aldikacti B. et al. Engineered nanointerfaces for microfluidic isolation and molecular profiling of tumor-specific extracellular vesicles. Nat Commun. 2018;9:175
51. Min Y, Deng W, Yuan H, Zhu D, Zhao R, Zhang P. et al. Single extracellular vesicle surface protein-based blood assay identifies potential biomarkers for detection and screening of five cancers. Mol Oncol. 2024;18:743-61
52. Liu Y-J, Wang C. A review of the regulatory mechanisms of extracellular vesicles-mediated intercellular communication. Cell Commun. Sig. 2023;21:77
53. Matsumoto A, Takahashi Y, Chang HY, Wu YW, Yamamoto A, Ishihama Y. et al. Blood concentrations of small extracellular vesicles are determined by a balance between abundant secretion and rapid clearance. J Extracell Vesicles. 2020;9:1696517
54. Watson DC, Bayik D, Srivatsan A, Bergamaschi C, Valentin A, Niu G. et al. Efficient production and enhanced tumor delivery of engineered extracellular vesicles. Biomaterials. 2016;105:195-205
55. Kuang L, Wu L, Li Y. Extracellular vesicles in tumor immunity: mechanisms and novel insights. Mol. Cancer. 2025;24:45
56. Fu X, Song J, Yan W, Downs BM, Wang W, Li J. The biological function of tumor-derived extracellular vesicles on metabolism. Cell Commun. Sig. 2023;21:150
57. Lopez K, Lai SWT, Lopez Gonzalez EDJ, Dávila RG, Shuck SC. Extracellular vesicles: A dive into their role in the tumor microenvironment and cancer progression. Front Cell Dev Biol. 2023;11:1154576
58. Shi Y, van der Meel R, Chen X, Lammers T. The EPR effect and beyond: Strategies to improve tumor targeting and cancer nanomedicine treatment efficacy. Theranostics. 2020;10:7921-4
59. Kim J, Cho H, Lim D-K, Joo MK, Kim K. Perspectives for improving the tumor targeting of nanomedicine via the EPR effect in clinical tumors. Int J Mol Sci. 2023;24:10082
60. Jiang Y, Lyu Z, Ralahy B, Liu J, Roussel T, Ding L. et al. Dendrimer nanosystems for adaptive tumor-assisted drug delivery via extracellular vesicle hijacking. Proc Natl Acad Sci U S A. 2023;120:e2215308120
61. Wei P, Wu F, Kang B, Sun X, Heskia F, Pachot A. et al. Plasma extracellular vesicles detected by Single Molecule array technology as a liquid biopsy for colorectal cancer. J Extracell Vesicles. 2020;9:1809765
62. Oshi M, Murthy V, Takahashi H, Huyser M, Okano M, Tokumaru Y. et al. Urine as a source of liquid biopsy for cancer. Cancers (Basel). 2021;13(2):2652
63. Kumar P, Gupta S, Das BC. Saliva as a potential non-invasive liquid biopsy for early and easy diagnosis/prognosis of head and neck cancer. Transl Oncol. 2024;40:101827
64. Norman M, Shami-shah A, D'Amaddio SC, Travis BG, Ter-Ovanesyan D, Dougan TJ. et al. Toward identification of markers for brain-derived extracellular vesicles in cerebrospinal fluid: a large-scale, unbiased analysis using proximity extension assays. J Extracell Vesicles. 2025;14:e70052
65. Kim IA, Hur JY, Kim HJ, Lee SE, Kim WS, Lee KY. Liquid biopsy using extracellular vesicle-derived DNA in lung adenocarcinoma. J Pathol Transl Med. 2020;54:453-61
66. Ramirez-Garrastacho M, Bajo-Santos C, Line A, Martens-Uzunova ES, de la Fuente JM, Moros M. et al. Extracellular vesicles as a source of prostate cancer biomarkers in liquid biopsies: a decade of research. Br J Cancer. 2022;126:331-50
67. Hinzman CP, Jayatilake M, Bansal S, Fish BL, Li Y, Zhang Y. et al. An optimized method for the isolation of urinary extracellular vesicles for molecular phenotyping: detection of biomarkers for radiation exposure. J Transl Med. 2022;20:199
68. Faur CI, Dinu C, Toma V, Jurj A, Mărginean R, Onaciu A. et al. A new detection method of oral and oropharyngeal squamous cell carcinoma based on multivariate analysis of surface-enhanced Raman spectra of salivary exosomes. J Pers Med. 2023;13(5):762
69. Pedersen A, Sørensen C, Proctor G, Carpenter G, Ekström J. Salivary secretion in health and disease. J Oral Rehabil. 2018;45:730-46
70. Poupardin R, Wolf M, Maeding N, Paniushkina L, Geissler S, Bergese P. et al. Advances in extracellular vesicle research over the past decade: source and isolation method are connected with cargo and function. Adv Healthc Mater. 2024;13:2303941
71. Andre F, Schartz NEC, Movassagh M, Flament C, Pautier P, Morice P. et al. Malignant effusions and immunogenic tumour-derived exosomes. Lancet. 2002;360:295-305
72. Nogueras-Ortiz CJ, Eren E, Yao P, Calzada E, Dunn C, Volpert O. et al. Single-extracellular vesicle (EV) analyses validate the use of L1 Cell Adhesion Molecule (L1CAM) as a reliable biomarker of neuron-derived EVs. J Extracell Vesicles. 2024;13:e12459
73. Yoh KE, Lowe CJ, Mahajan S, Suttmann R, Nguy T, Reichelt M. et al. Enrichment of circulating tumor-derived extracellular vesicles from human plasma. J Immunol Methods. 2021;490:112936
74. Jiang N, Saftics A, Romano E. et al. Multiparametric profiling of HER2-enriched extracellular vesicles in breast cancer using Single Extracellular VEsicle Nanoscopy. J Nanobiotechnol. 2024;22:589
75. Xue Y, Feng X, Fan X, Zhu G, McLaughlan J, Zhang W. et al. Extracellular vesicles for the diagnosis of cancers. Small Struct. 2022;3:2100096
76. Mizenko RR, Brostoff T, Rojalin T, Koster HJ, Swindell HS, Leiserowitz GS. et al. Tetraspanins are unevenly distributed across single extracellular vesicles and bias sensitivity to multiplexed cancer biomarkers. J Nanobiotechnology. 2021;19:250
77. Carney RP, Hazari S, Colquhoun M, Tran D, Hwang B, Mulligan MS. et al. Multispectral optical tweezers for biochemical fingerprinting of CD9-positive exosome subpopulations. Anal Chem. 2017;89:5357-63
78. Nawaz M, Shah N, Zanetti BR, Maugeri M, Silvestre RN, Fatima F. et al. Extracellular vesicles and matrix remodeling enzymes: the emerging roles in extracellular matrix remodeling, progression of diseases and tissue repair. Cells. 2018;7(10):167
79. Li J, Li J, Peng Y, Du Y, Yang Z, Qi X. Dendritic cell derived exosomes loaded neoantigens for personalized cancer immunotherapies. J Control Release. 2023;353:423-33
80. Abramowicz A, Story MD. The Long and Short of It: The emerging roles of non-coding RNA in small extracellular vesicles. Cancers. 2020;12(6):1445
81. Wu L, Gao C. Comprehensive overview the role of glycosylation of extracellular vesicles in cancers. ACS Omega. 2023;8:47380-92
82. Williams C, Pazos R, Royo F, González E, Roura-Ferrer M, Martinez A. et al. Assessing the role of surface glycans of extracellular vesicles on cellular uptake. Sci Rep. 2019;9:11920
83. Ghadami S, Dellinger K. The lipid composition of extracellular vesicles: applications in diagnostics and therapeutic delivery. Front Mol Biosci. 2023;10:1198044
84. Willms E, Cabañas C, Mäger I, Wood MJA, Vader P. Extracellular vesicle heterogeneity: subpopulations, isolation techniques, and diverse functions in cancer progression. Front Immunol. 2018;9:738
85. Rupp A-K, Rupp C, Keller S, Brase JC, Ehehalt R, Fogel M. et al. Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol Oncol. 2011;122:437-46
86. Hilton SH, White IM. Advances in the analysis of single extracellular vesicles: A critical review. Sens Actuators Rep. 2021;3:100052
87. Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New technologies for analysis of extracellular vesicles. Chem Rev. 2018;118:1917-50
88. Zhang J, Wu J, Wang G, He L, Zheng Z, Wu M. et al. Extracellular vesicles: techniques and biomedical applications related to single vesicle analysis. ACS Nano. 2023;17:17668-98
89. Wang X, Wang L, Lin H, Zhu Y, Huang D, Lai M. et al. Research progress of CTC, ctDNA, and EVs in cancer liquid biopsy. Front Oncol. 2024;14:1303335
90. Sheriff S, Saba M, Patel R, Fisher G, Schroeder T, Arnolda G. et al. A scoping review of factors influencing the implementation of liquid biopsy for cancer care. J Exp Clin Cancer Res. 2025;44:50
91. Salmond N, Williams KC. Isolation and characterization of extracellular vesicles for clinical applications in cancer - time for standardization? Nanoscale Adv. 2021;3:1830-52
92. Keup C, Kimmig R, Kasimir-Bauer S. Combinatorial power of cfDNA, CTCs and EVs in oncology. Diagnostics. 2022;12(4):870
93. Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM. et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A. 2014;111:14888-93
94. Silva AM, Lázaro-Ibáñez E, Gunnarsson A, Dhande A, Daaboul G, Peacock B. et al. Quantification of protein cargo loading into engineered extracellular vesicles at single-vesicle and single-molecule resolution. J Extracell Vesicles. 2021;10:e12130
95. Yang Z, Atiyas Y, Shen H, Siedlik MJ, Wu J, Beard K. et al. Ultrasensitive single extracellular vesicle detection using high throughput droplet digital Enzyme-Linked Immunosorbent Assay. Nano Letters. 2022;22:4315-24
96. Liu H, Tian Y, Xue C, Niu Q, Chen C, Yan X. Analysis of extracellular vesicle DNA at the single-vesicle level by nano-flow cytometry. J Extracell Vesicles. 2022;11:e12206
97. Filipe V, Hawe A, Jiskoot W. Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein Aggregates. Pharm Res. 2010;27:796-810
98. Arab T, Mallick ER, Huang Y, Dong L, Liao Z, Zhao Z. et al. Characterization of extracellular vesicles and synthetic nanoparticles with four orthogonal single-particle analysis platforms. J Extracell Vesicles. 2021;10:e12079
99. Kim SY, Khanal D, Kalionis B, Chrzanowski W. High-fidelity probing of the structure and heterogeneity of extracellular vesicles by resonance-enhanced atomic force microscopy infrared spectroscopy. Nat Protoc. 2019;14:576-93
100. Enciso-Martinez A, Van Der Pol E, Hau CM, Nieuwland R, Van Leeuwen TG, Terstappen LWMM. et al. Label-free identification and chemical characterisation of single extracellular vesicles and lipoproteins by synchronous Rayleigh and Raman scattering. J Extracell Vesicles. 2020;9:1730134
101. Li J, Sina AAI, Antaw F, Fielding D, Möller A, Lobb R. et al. Digital decoding of single extracellular vesicle phenotype differentiates early malignant and benign lung lesions. Adv Sci. 2023;10:2204207
102. Rikkert LG, Nieuwland R, Terstappen L, Coumans FAW. Quality of extracellular vesicle images by transmission electron microscopy is operator and protocol dependent. J Extracell Vesicles. 2019;8:1555419
103. Huang G, Liu Y, Wang D, Zhu Y, Wen S, Ruan J. et al. Upconversion nanoparticles for super-resolution quantification of single small extracellular vesicles. eLight. 2022;2:20
104. Dechantsreiter S, Ambrose AR, Worboys JD, Lim JME, Liu S, Shah R. et al. Heterogeneity in extracellular vesicle secretion by single human macrophages revealed by super-resolution microscopy. J Extracell Vesicles. 2022;11:e12215
105. Raghu D, Christodoulides JA, Christophersen M, Liu JL, Anderson GP, Robitaille M. et al. Nanoplasmonic pillars engineered for single exosome detection. PLOS ONE. 2018;13:e0202773
106. Yang Y, Shen G, Wang H. et al. Interferometric plasmonic imaging and detection of single exosomes. Proc Natl Acad Sci USA. 2018;115(41):10275-10280
107. Reynolds DE, Pan M, Yang J, Galanis G, Roh YH, Morales R-TT. et al. Double digital assay for single extracellular vesicle and single molecule detection. Adv Sci. 2023;10:2303619
108. Polito R, Musto M, Temperini ME, Ballerini L, Ortolani M, Baldassarre L. et al. Infrared nanospectroscopy of individual extracellular microvesicles. Molecules. 2021;26(4):887
109. Saftics A, Abuelreich S, Romano E, Ghaeli I, Jiang N, Spanos M. et al. Single extracellular vesicle nanoscopy. J Extracell Vesicles. 2023;12:12346
110. Lennon KM, Wakefield DL, Maddox AL, Brehove MS, Willner AN, Garcia-Mansfield K. et al. Single molecule characterization of individual extracellular vesicles from pancreatic cancer. J Extracell Vesicles. 2019;8:1685634
111. Mizenko RR, Feaver M, Bozkurt BT, Lowe N, Nguyen B, Huang K-W. et al. A critical systematic review of extracellular vesicle clinical trials. J Extracell Vesicles. 2024;13:e12510
112. Lennon KM, Wakefield DL, Maddox AL, Brehove MS, Willner AN, Garcia-Mansfield K. et al. Single molecule characterization of individual extracellular vesicles from pancreatic cancer. J Extracell Vesicles. 2019;8(1):1685634
113. Han C, Kang H, Yi J, Kang M, Lee H, Kwon Y. et al. Single-vesicle imaging and co-localization analysis for tetraspanin profiling of individual extracellular vesicles. J Extracell Vesicles. 2021;10:e12047
114. Huang S, Cai N, Pacheco PP, Narrandes S, Wang Y, Xu W. Applications of Support Vector Machine (SVM) Learning in Cancer Genomics. Cancer Genomics Proteomics. 2018;15:41-51
115. Shin H, Choi BH, Shim O, Kim J, Park Y, Cho SK. et al. Single test-based diagnosis of multiple cancer types using Exosome-SERS-AI for early stage cancers. Nat Commun. 2023;14:1644
116. Yin H, Xie J, Xing S, Lu X, Yu Y, Ren Y. et al. Machine learning-based analysis identifies and validates serum exosomal proteomic signatures for the diagnosis of colorectal cancer. Cell Rep Med. 2024;5(8):101689
117. Jakobsen KR, Paulsen BS, Bæk R, Varming K, Sorensen BS, Jørgensen MM. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J Extracell Vesicles. 2015;4:26659
118. Hinestrosa JP, Kurzrock R, Lewis JM, Schork NJ, Schroeder G, Kamat AM. et al. Early-stage multi-cancer detection using an extracellular vesicle protein-based blood test. Commun Med (Lond). 2022;2:29
119. Simpson C, Tabatsky E, Rahil Z, Eddins DJ, Tkachev S, Georgescauld F. et al. Lifting the curse from high-dimensional data: automated projection pursuit clustering for a variety of biological data modalities. Gigascience. 2025;14:giaf052
120. Yin L, Han X, Guo F, Zou Y, Xie Q, Wang J. et al. An all-in-one nanohole array for size-exclusive trapping and high-throughput digital counting of single extracellular vesicles for non-invasive cancer screening. Angew Chem Int Ed Engl. 2025;64:e202506744
121. Wen Y, Li Y, Cheng S, Crow J, Samuel G, Vishwakarma V. et al. Partition-less digital immunoassay using configurable topographic nanoarrays for extracellular vesicle diagnosis of ewing sarcoma. ACS Nano. 2025;19:11973-86
122. von Lersner AK, Fernandes F, Ozawa PMM, Jackson M, Masureel M, Ho H. et al. Multiparametric single-vesicle flow cytometry resolves extracellular vesicle heterogeneity and reveals selective regulation of biogenesis and cargo distribution. ACS Nano. 2024;18:10464-84
123. Xiang R, Wang W, Yang L, Wang S, Xu C, Chen X. A Comparison for dimensionality reduction methods of single-cell RNA-seq data. Front Genet. 2021;12:646936
Author contact
![]() Corresponding author: sebastian.wachsmannhogiuca.
Corresponding author: sebastian.wachsmannhogiuca.
 Global reach, higher impact
Global reach, higher impact